Integrated transcriptome and metabolite profiling highlights the role of benzoxazinoids in wheat resistance against Fusarium crown rot
Shuonn Dun, Jingjing Jin, Yutin Go, Chnglin Jin, Junyi Mu, Wenho Zhen, Qixin Sun,Chojie Xie, Jun M,*
a College of Agronomy and Biotechnology, China Agricultural University, Beijing 100193, China
b Institute of Genetics and Physiology, Hebei Academy of Agriculture and Forestry Sciences, Shijiazhuang 050051, Hebei, China
c College of Agronomy, Hebei Agricultural University, Baoding 071001, Hebei, China
d Hebei Plant Protection and Quarantine General Station, Shijiazhuang 050051, Hebei, China
Keywords:Fusarium crown rot Wheat Metabolites Transcript Benzoxazinoid
ABSTRACT Fusarium crown rot (FCR), caused by Fusarium spp., is a chronic and severe plant disease worldwide. In the last years,the incidence and severity of FCR in China has increased to the point that it is now considered a threat to local wheat crops.In this study,for the first time,the metabolites and transcripts responsive to FCR infection in the partial resistant wheat cultivar 04 Zhong 36(04z36)and susceptible cultivar Xinmai 26(XM)were investigated and compared at 20 and 25 days post inoculation(dpi).A total of 443 metabolites were detected, of which 102 were significantly changed because of pathogen colonization.Most of these 102 metabolites belonged to the flavonoid,phenolic acid,amino acid and derivative classes.Some metabolites, such as proline betaine, lauric acid, ribitol, and arabitol, were stably induced by Fusarium pseudograminearum(Fp)infection at two time points and may have important roles in FCR resistance. In line with the reduced seedling height of 04z36 and XM plants, RNA-seq analysis revealed that FCR infection significantly affected the photosynthesis activities in two cultivars. Furthermore, 15 jasmonate ZIM-domain genes (JAZ) in the significantly enriched ‘regulation of jasmonic acid mediated signaling pathway’ in 04z36 were down-regulated. The down-regulation of these JAZ genes in 04z36 may cause a strong activation of the jasmonate signaling pathway.Based on combined data from gene expression and metabolite profiles, two metabolites, benzoxazolin-2-one (BOA) and 6-methoxy-benzoxazolin-2-one(MBOA),involved in the benzoxazinoid-biosynthesis pathway,were tested for their effects on FCR resistance. Both BOA and MBOA significantly reduced fungal growth in vitro and in vivo, and, thus, a higher content of BOA and MBOA in 04z36 may contribute to FCR resistance.Above all,the current analysis extends our understanding of the molecular mechanisms of FCR resistance/susceptibility in wheat and will benefit further efforts for the genetic improvement of disease resistance.
1. Introduction
Fusarium crown rot(FCR)is a severe and chronic soil-borne disease in many semiarid regions worldwide [1]. The disease can be caused by a range of Fusarium species,of which Fp and F.culmorum are the most prevalent [2–4]. FCR infected wheat plants show browning discoloration of leaf sheath or stem base and formation of white heads, which can be completely devoid of grains or present shriveled grains [5]. Significant yield losses due to FCR have been documented worldwide [1]. For example, in Australia, the annual yield loss caused by FCR is estimated to be approximately$79 million Australian dollars, whereas yield loss of up to 61%was reported in some sites in the Pacific Northwest of the US[3,6].In China,the incidence and severity of FCR have progressively increased in major wheat-producing provinces in the last few years,and FCR became a threat to the local wheat industry.Several studies have been conducted to identify resistant cultivars of Chinese wheat.However,similar to other countries,a limited number of elite cultivars in China have shown acceptable resistance levels to FCR [7].
Globally various management practices have been applied to reduce FCR incidence, including stubble management, tillage, soil fertility management, crop rotation, biocontrol control, and fungicides[1].However,these approaches are not always compatible with practical and environmental considerations. Growing resistant wheat cultivars has long been recognized as the most effective way to minimize disease damage.To date,genotypes with complete resistance to FCR have not been observed in wheat, and only a few genotypes with partial resistance are available[5].Some of the partial resistant genotypes, such as Sunco, Kukri and 2–49 have been used in breeding programs for FCR improvement in Pacific Northwest of USA [8]. A large number of quantitative trait loci(QTL) associated with FCR resistance were identified in those partial resistance genotypes through bi-parental mapping or genomewide association studies(GWAS)analysis[5,7,9].Significant efforts were made to fine map a FCR QTL on chromosome 3BL and to search for candidate genes [10].
In addition to QTL identification and gene mapping, functional omicsrepresentsapowerfulapproachtorevealthediseaseresistance mechanisms of plants during colonization and aid in the identification of potential resistant genes.In wheat,several transcript studies have been performed to investigate the gene expression profile after FCRcolonizationviagenechiporRNA-seqtechnology[11–13].Genes involved in antimicrobial defense,oxidative stress,primary and secondarymetabolism,pathogensensingandsignalingwereinducedby FCR. For example, nine 12-oxophytodienoate synthase genes involved in the biosynthesis of jasmonates acid(JA),a well-known plant hormone related to necrotrophic pathogen resistance, were inducedbyFCRcolonization.AnincreaseofJAlevelintheFCRinfected plants was observed at three days after colonization[13].Previous studieshavealsoshownthatmanyFp-responsivegeneswerealtered by the mycotoxins deoxynivalenol(DON)and JA[1,14].
Conversely, although omics studies have been proven to have great potential to substantially improve our understanding of plant-pathogen interactions in various crops and diseases [15–17], information on metabolite changes in wheat after FCR colonization is limited. Some metabolites have been shown to play an important role in FCR resistance. For example, Powell et al.[13]reported that the levels of tryptamine and serotonin were significantly increased in a moderate susceptible wheat cultivar after FCR colonization. Among a vast array of secondary metabolites,benzoxazinoids represent protective and allelopathic metabolites that are found in many Poaceae species, including wheat [18].Among benzoxazinoids, 2,4-dihydroxy-1,4-benzoxazin-3-one(DIBOA) and 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one(DIMBOA) are potent defense compounds that confer innate immunity against pests and diseases [19]. DIBOA and DIMBOA are stored in vacuoles and can decompose rapidly to BOA and MBOA, which exhibit antifungal, bacteriostatic and antifeeding activity against insects upon infection or physical damage[20,21]. The antifungal function of BOA and MBOA against Fusarium spp. has been observed in previous studies, and the degradation of plant benzoxazinoids is important for the virulence of FCR pathogens towards wheat [22,23].
In this study, two Chinese wheat cultivars with different FCR resistance level were used to compare the metabolic changes between these genotypes in response to FCR colonization via UPLC–MS/MS.The transcript data obtained through RNA-seq were also analyzed. The results of this study will improve our understanding of host-pathogen interactions and aid future disease improvement strategies against this important disease.
2. Materials and methods
2.1. Inoculum preparation
The highly aggressive and prevalent Fp isolate WZ-8A from the Yellow and Huai wheat regions of China, kindly provided by Prof.Honglian Li from the College of Plant Protection of Henan Agricultural University[24],was used for inoculation in this study.WZ-8A was first incubated on half-strength potato dextrose agar (PDA)plates for seven days at room temperature. Then, 20 pieces of mycelial discs(5 mm)were taken out from the margin of the plates and transferred into a 500 mL conical flask containing 200 mL of liquid carboxymethylcellulose medium (CMC). The conical flask was placed in a shaking incubator at 200 r min-1and 25 °C for approximately 4 days.After this period,the spores were suspended in sterile water,and the concentration of the spore suspension was adjusted to 1×106spores mL-1.Tween 20 was added to the spore suspension to a final concentration of 0.1% v/v before inoculation.
2.2. Plant material and pathogen inoculation
A FCR-susceptible and a partially resistant wheat cultivar, XM and 04z36, respectively, were used in this study [9]. The seeds of 04z36 and XM were surface-sterilized with 75% ethanol, rinsed twice with sterile water, and germinated in Petri dishes at 25 °C for approximately three days. The germinated seedlings were immersed in the spore suspension (inoculated) or water (control)for 1 min before sowing in 4 cm2punnets containing autoclaved potting mix. The plants were grown in an environmentally controlled glasshouse at (25 ± 2) °C / (20 ± 2) °C (day/night) and 65%/80%±5%(day/night)relative humidity under natural sunlight.To estimate fungal biomass and perform metabolic and transcriptional analyses,the stem bases between 0 and 2 cm above the roots of the plants were cut at 5,10,15,20,25,30 and 35 days post inoculation (dpi), and the height of the plants from the bottom of the stem base to the top of the leaves was also recorded.Three biological replicates, each containing seven seedlings, were collected at each time point.
2.3. Total DNA extraction and estimation of fungal biomass by RTqPCR
The total DNA of 2 cm infected stem bases was extracted using a modified cetyltrimethylammonium bromide (CTAB) method [25].DNA was eluted into 100 ng μL-1of sterile water and stored at- 20°C. The biomass of Fp was calculated based on the proportion of fungal DNA to wheat DNA using real-time quantitative PCR(RT-qPCR). The Tri5 gene primer and wheat actin-binding protein(WABP) primers were used to estimate fungal and wheat biomass(Table S1)[26].Three biological replicates were used,each containing seven seedlings.Each biological replicate was isolated and estimated using three technical replicates.
2.4.Metabolic analysis of inoculated and uninoculated 04z36 and XM plants
XM and 04z36 plants at 20 and 25 dpi were used for metabolite extraction. Similar to the fungal biomass estimation, three biological replicates,each containing 2 cm shoot bases of 15 seedlings of inoculated or uninoculated XM and 04z36 plants were collected at each time point. UPLC–MS/MS analysis was performed using a widely targeted metabolome method based on the Metware database (Metware Biotechnology Co., Ltd., Wuhan, Hubei, China) [27].The annotation of metabolites in the database was performed by matching the fragmentation pattern, accurate m/z value, and retention time. Three biological replicates of each sample were freeze-dried and ground using a mixer mill (MM 400, Retsch).The powder (100 mg) of each sample was infused in 1.2 mL of 70% aqueous methanol at 4 °C overnight. After 10 min 10000×g centrifugation, the supernatant was collected and filtered for UPLC–MS/MS analysis. Metabolite quantification was performed using a specific set of MRM modes of the QQQ mass spectrometer for each period. The retention time and peak type of metabolite mass spectrometry data were corrected for qualitative and quantitative analyses. Unsupervised principal component analysis (PCA)was performed using the statistical function ‘prcomp’ within R(www.r-project.org). Significantly different metabolites between sample groups were determined by VIP ≥1 and absolute log2Fold change ≥1. Metabolites KEGG annotation were annotated based on KEGG compound database (http://www.kegg.jp/kegg/compound/), and P-values were calculated.
2.5. Transcriptional analysis of inoculated and uninoculated XM and 04z36 plants
The same plant materials used for metabolic analysis were used for the RNA-seq analysis. The plant tissues were stored in dry ice and delivered to Metware Biotechnology Co., Ltd., Wuhan, Hubei,China, for RNA-seq analysis. Total RNA was extracted from stem bases using the TaKaRa MiniBEST Plant RNA Extraction Kit(TaKaRa, Japan) according to the manufacturer’s instructions.RNA integrity was evaluated using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system(Agilent Technologies,Santa Clara,CA, USA). Sequencing libraries were generated from 3 μg RNA per sample using the NEBNext Ultra RNA Library Prep Kit for Illumina(NEB, Ipswich, MA, USA). Fragments of the library were purified with AMPure XP system (Beckman Coulter, Beverly, CA, USA) to preferentially select 150–200 bp cDNA fragments. The quality of the library was assessed using the Agilent Bioanalyzer 2100 system. The library was sequenced on an Illumina Hiseq platform and 125 bp/150 bp paired-end reads were generated.
Low-quality reads and reads with adapter or poly-N were removed. Clean paired-end reads were aligned to the wheat reference genome (RefSeq 1.0) using TopHat v2.0.12 [28]. The read numbers mapped to each gene were counted using HTSeq v.0.6.1. The fragments per kilobase of transcript per million (FPKM)mapped was calculated based on the length of the gene and read counts was mapped to the corresponding gene to reflect the expression level. The DESeq R package was used to performed differential expression analysis of two conditions (three biological replicates per condition) [29]. Different expressed gene (DEGs)were assigned as genes with P-value <0.05,using DESeq R package.GOseq R package and KOBAS software were used in the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes(KEGG) enrichment pathway analysis. The corrected Pvalue <0.05 was used as the threshold for significance [30,31].
2.6. RT-qPCR analysis
To confirm transcriptional changes in XM and 04z36, the same samples for the RNA-seq analysis were used for RT-qPCR analysis.Total RNA was isolated and cDNA synthesized of each biological replicate using RNAprep pure Plant kit (Tiangen) and FastKing RT kit (Tiangen), according to the manufacturer’s instructions. RTqPCR was performed on a CFX96 Real-Time PCR Detection System(Bio-Rad) using TB Green Premix Ex Taq II (TaKaRa). The cDNA(100 ng) from was used in a 10-uL reaction with three technical replicates. The thermocycler settings were as follows: 30 s at 95 °C, followed by 40 cycles of 5 s at 95 °C, 30 s at 60 °C, and 10 s at 72 °C. Melt curves were run for all primer pairs(Table S1), and the WABP (U58278) gene was used as a reference.Data were analyzed using the BioRadCFXManager software.
2.7. LC–MS quantification of BOA and MBOA
Same to the metabolomics analysis, three biological replicates,each containing 1 cm shoot bases of 15 seedlings of inoculated or uninoculated XM and 04z36 plants were collected and mixed at 5,10,15,20,25,30 and 35 dpi.Samples were freeze-dried and sent to Metware Biotechnology Co., Ltd, Wuhan, Hubei, China for the quantification of BOA and MBOA. The samples were homogenized and powdered in a mill.Metabolites were extracted by mixing the dried powder (50 mg) with 500 μL of methanol. The extract was vortexed and ultrasonicated for 5 min at room temperature. The supernatants were collected after centrifugation. The residue was re-extracted repeating the steps above. A volume of 100 μL of the combined supernatants was used for further LC–MS analysis.The sample extracts were analyzed using a UPLC–ESI–MS/MS system(UPLC,ExionLC AD;MS,Applied Biosystems 6500 Triple Quadrupole). The analytical conditions were as follows: LC: column,Waters ACQUITY UPLC HSS T3 C18 (100 mm × 2.1 mm i.d.,1.8 μm);solvent system,water with 0.04%acetic acid(A),acetonitrile with 0.04% acetic acid (B); gradient program, started at 5% B(0 min), increased to 95% B (0.1–10 min), 95% B (10–11 min),finally ramped back to 5% B (11.01–14 min); flow rate,0.35 mL min-1; temperature, 40 °C; injection volume, 2 μL. Standards for BOA and MBOA were obtained from Sigma–Aldrich.
2.8. Chemical sensitivity assays
The effects of BOA and MBOA on the growth of fungal mycelia were assessed on PDA solid medium supplemented with the metabolites dissolved in 1%DMSO.Medium amended with solvent(1% DMSO) alone was used as a control. BOA and MBOA were tested at 0.25, 0.5, and 1 mg mL-1in half-strength PDA medium in 90 mm Petri dishes.The center of the plates was inoculated with a 5 mm circle agar disc from the growing edge of a fungal colony plate.The plates were incubated in the dark at 28°C.The mean colony diameter was measured daily by subtracting the initial agar disc diameter. A circle was drawn along the growing edge of the fungal mycelium on the plate for five days. The rate of inhibition was calculated and compared with that of the DMSO control.
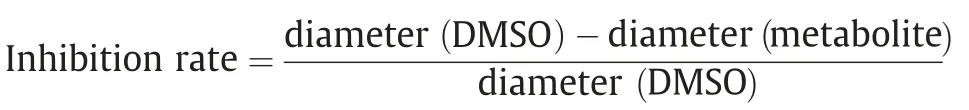
To assess the antifungal activity of BOA and MBOA, the inoculated XM and 04z36 seedlings were first sprayed with BOA and MBOA dissolved in 1% DMSO (1 mg mL-1) on the surface of the stem base at 10 dpi.The control group was sprayed with 1%DMSO.After the first spraying, the seedlings were sprayed every three days until 25 dpi. The wheat stem bases of each group were harvested at 25 dpi for fungal biomass estimation. Three replicates of this experiment were used, and each replicate contained 15 seedlings.
3. Results
3.1. FCR symptoms and effects on seedling height and fungal biomass
FCR-disease symptoms emerged on both XM and 04z36 seedlings at 10 dpi. The symptoms developed rapidly with time in the inoculated groups. 04z36 plants showed higher resistance levels in terms of degree of wilt, lesions, and brown discoloration at the investigated time points.Additionally,FCR infection resulted in significant height reduction in inoculated XM seedlings at 10,15,20, 30, and 35 dpi (P <0.05). The FCR infection caused a slight height reduction in XM plants at 25 dpi,which did not reach a significant level.A trend of height reduction was also observed in the 04z36 plants during the same period. However, the height difference between the inoculated and control 04z36 plants was only significant at 25 dpi. The differences at 5, 10, 15, 20, 30, and 35 dpi were not significant (Fig. 1A). In line with the phenotypic results described, the relative fungal biomass in inoculated seedlings of both XM and 04z36 also continuously increased 125- and 88-fold over 35 dpi, respectively (Fig. 1B). The relative fungal biomass of XM and 04z36 were similar before 20 dpi.Then,the fungal biomass in XM plants sharply increased 86-fold at 30 dpi,whereas in 04z36 plants, the fungal biomass increased approximately 45-fold.The fungal quantities in XM and 04z36 started to differentiate after 20 dpi(P <0.05).As a result,samples of XM and 04z36 taken at 20 and 25 dpi were used for further metabolic and transcriptional analysis,as this time interval may be a key phase for differences between the two cultivars in the defense process (Fig. S1).
3.2. Metabolites involved in FCR colonization
A total of four pairwise comparisons were made to characterize metabolic changes in XM and 04z36 plants after FCR infection using UPLC–MS/MS. These comparisons included XMCK20 vs.XMFP20, XMCK25 vs. XMFP25, 04z36CK20 vs. 04z36FP20, and 04z36CK25 vs. 04z36FP25. PCA was conducted to investigate the global metabolic changes in the four comparative groups that occurred in response to Fp colonization(Fig.2A).The first two principal components(PC1 and PC2)explained 45.85%of the variation.PC1 discriminated the inoculated samples from the uninoculated samples, and PC2 discriminated XM from 04z36. The PCA analysis indicated high consistency within each biological replicate group and separation between different sample groups.
A total of 443 metabolites were detected in the current analysis(Table S2).The number of significantly changed metabolites in the four comparative groups were listed in Table 1. Most of these metabolites were up-regulated after Fp colonization. The 102 non-redundant significantly changed metabolites from the four comparative groups after colonization are listed in Table S3. All these metabolites showed the same change tendency in the two cultivars at 20 and 25 dpi.Most of these metabolites are flavonoids(17), phenolic acids (16), amino acids and derivatives (14), alkaloids(10),organic acids(7),saccharides and alcohols(7),and phenolamines (6). Significant metabolic reprogramming was induced by Fp colonization in XM and 04z36.
We compared the overlapped metabolites in the four pairwise groups across two time points (Fig. 2B, C). We found that 17 metabolites, including arabitol, tryptamine, N-feruloyl serotonin,proline betaine, and DON, were consistently induced by FCR in both XM and 04z36 at 20 and 25 dpi. In contrast, chlorogenic acid and xylopyranosyljasminoside were consistently reduced by FCR infection in 04z36 and XM plants at 20 and 25 dpi. Some metabolites were specifically affected by FCR colonization in either XM or 04z36.For example,three metabolites,1,2-N-methylpipecolic acid,lauric acid, and N-feruloyl tryptamine, were induced, and two metabolites were reduced in 04z36 at two time points; whereas 10 metabolites were specifically induced and three metabolites were specifically reduced in XM (Fig. 2B, C). These significantly changed metabolites at 20 and 25 dpi indicate a stronger metabolic reprogram caused by Fp colonization in XM than in 04z36.

Fig. 2. PCA analysis and Venn diagram of two cultivars at 20 and 25 days post inoculation (dpi). (A) Principal component analysis (PCA) score plots for metabolites in the control and Fp-inoculated 04z36 and XM seedlings. The three roundness with different color in each ellipse represents three biological replicates. (B) Venn diagram of upregulated metabolites.(C)Venn diagram of down-regulated metabolites.The black numbers indicate the numbers of significant changed metabolites.XMCK20,uninoculated Xinmai 26 seedlings at 20 dpi; XMCK25, uninoculated Xinmai 26 seedlings at 25 dpi; XMFP20, inoculated Xinmai 26 seedlings at 20 dpi; XMFP25, inoculated Xinmai 26 seedlings at 25 dpi;04z36CK20,uninoculated 04 Zhong 36 seedlings at 20 dpi;04z36CK25,uninoculated 04 Zhong 36 seedlings at 25 dpi;04z36FP20,inoculated 04 Zhong 36 seedlings at 20 dpi; 04z36FP25, inoculated 04 Zhong 36 seedlings at 25 dpi.
To investigate the metabolites most sensitive to Fp colonization in the two wheat cultivars, the top 10-fold-change (increase and decrease)metabolites in XM and 04z36 are listed in Table S4.Interestingly, four butyric acids (2-hydroxy-2-methylbutyric acid,α-hydroxyisobutyric acid, (S)-2-hydroxybutanoicacid, and 2-hydroxybutanoic acid) were the most significantly increased metabolites in both 04z36 and XM plants at 20 and 25 dpi.Proline betaine and chlorogenic acid were among the top 10 changed metabolites. Two other metabolites (N-acetyl-5-hydroxytryptamine and N-feruloyl serotonin), which are serotonin-biosynthesis derivatives, were significantly increased after inoculation, in three pairwise comparisons (XMCK20 vs.XMFP20, XMCK25 vs. XMFP25, 04z36CK20 vs. 04z36FP20)(Table S4). KEGG enrichment of significantly changed metabolites showed that tryptophan metabolism was significantly enriched in all pairwise comparative groups,and indole alkaloid biosynthesis was specifically enriched in two 04z36 comparative groups.The enriched pathways are listed in Table S5.
3.3. Transcriptional analysis of wheat response to FCR colonization
The FCR-infected XM plants showed an obvious lesion on the stem bases at 25 dpi, which caused severe degradation of RNA.As a result, only the transcriptional result at 20 dpi was used for further analysis. Two pairwise comparisons were conducted,including XMCK20 vs. XMFP20 and 04z36CK20 vs. 04z36FP20.The DEGs (false discovery rate <0.05) identified at 20 dpi were 688 and 570 in 04z36 and XM, respectively (Table S6). Of the 688 DEGs in the 04z36 comparative group, 331 were upregulated and 357 were down-regulated, whereas the 570 DEGs in the XM comparative group included 467 up-regulated and 103 down-regulated genes.

Table 1 Significantly changed metabolites in XM and 04z36 at 20 and 25 days post inoculation (dpi).

Fig.1. The effects of FCR on seedling height and fungal biomass of two wheat cultivars.(A) The height (cm) of XM and 04z36 seedlings was measured at seven time points after inoculation.CK,uninoculated;FP,Fp inoculated.Data are presented as mean±SD of three experimental replicates;*,significant at P <0.05.(B)Relative Fp biomass in XM and 04z36 seedlings at seven time points after inoculation. The relative fungal biomass was estimated by comparing the fungal TRI5 gene with the WABP gene. Data are presented as mean ± SD of three experiments.
To provide insights into the pathway responses to FCR infection,KEGG and GO enrichment analyses were performed. A total of 10 and 6 KEGG pathways were enriched in XMCK20 vs. XMFP20 and 04z36CK20 vs. 04z36FP20, respectively (P <0.05) (Table S7). Of these pathways, four pathways including photosynthesis, metabolic pathways, starch and sucrose metabolism, amino sugar and nucleotide sugar metabolism were common for XM and 04z36.The plant hormone signal transduction pathway was specifically enriched in 04z36.
GO enrichment analysis showed that GO terms associated with photosystems were the most significantly enriched in XM and 04z36; photosystem I was enriched in both cultivars, whereas chlorophyll binding and photosynthesis light harvesting were specifically enriched in XM. The GO term jasmonic acid mediated signaling pathway was specifically enriched in 04z36, including 15 JAZ genes that act as JA repressors (P < 0.05) (Table S8)[32,33]. An adverse expression pattern of these genes was observed between XM and 04z36 after inoculation, as shown in Fig. 3A. All JAZs genes were significantly down-regulated in 04z36 after inoculation, whereas increased expression of these genes was observed in inoculated XM. In addition, DEGs of XM and 04z36 were also found in the benzoxazinoid biosynthesis pathway and phenylpropanoid biosynthesis pathway, the derivatives of which are potent secondary pathogen-related metabolites.
To validate the transcriptome expression data, three phenylpropanoid-pathway genes phenylalanine ammonia lyase(PAL): TraesCS4A02G401300, TraesCS6A02G222700,TraesCS1B02G048300, and three benzoxazinoid-biosynthesis pathway genes benzoxazinoids (BX): TraesCS2B02G076400,TraesCS4B02G207000, TraesCSU02G156500, and four JAZ genes(TraesCS4A02G007800, TraesCS4D02G296000, TraesCS7A02G201100,and TraesCS7A02G201200) were selected for RT-qPCR analysis(Fig. 3B). The expression patterns of RT-qPCR and transcriptional profile were similar in the two wheat cultivars at 20 dpi.PAL genes were strongly induced in XM and were slightly reduced in 04z36.BX genes were up-regulated in both XM and 04z36, whereas 04z36 had a relatively high expression level after inoculation.Four JAZ genes showed an adverse expression pattern between XM and 04z36; JAZs in XM and 04z36 were up- and down-regulated after inoculation, respectively.

Fig. 3. Relative expression levels of DEGs at 20 dpi in RNA-seq and RT-qPCR analysis. (A) Expression levels of 15 JAZs (jasmonate ZIM-domain genes) in XM and 04z36 seedlings at 20 dpi based on RNA-seq analysis.Bar plots represent the relative gene expression based on fragments per kilobase of transcript per million fragments mapped(FPKM).Values are the mean±SD(n=3).(B)Ten selected genes validated by RT-qPCR.Data were normalized to WABP expression level.PAL,phenylalanine ammonia lyase;BX, benzoxazinoids; JAZ, jasmonate ZIM-domain gene. Data are presented as mean ± SD (n = 3). Different letters indicate significant difference at P <0.05.
3.4. The antifungal activity of BOA and MBOA in vitro and vivo
RNA-seq analysis revealed that several genes in the benzoxazinoid biosynthesis pathway were differentially expressed in 04z36 and XM seedlings. As gene expression may be changed prior to metabolites, three significantly changed BXs in RNA-seq were also analyzed at 5, 10, and 15 dpi (Fig. S2). TraesCS4B02G207000 and TraesCSU02G156500 were both induced by Fp infection in XM and 04z36 (Fig. S2B, C). TraesCS2B02G076400 was only induced in 04z36 by Fp infection,and it was hardly detected in XM seedlings.BOA and MBOA are the predominant benzoxazinoids known to be involved in fungal resistance[21].As a result,we further measured the content of endogenous BOA and MBOA in 04z36 and XM stem bases at seven time points(Fig.4).As expected,compared with the control samples, the content of BOA and MBOA in both 04z36 and XM increased after inoculation at most of the seven sampling time points,especially during the early stage(5–25 dpi).The contents of BOA and MBOA were much higher in inoculated and uninoculated 04z36 samples than in XM at all time points. In addition, the content of MBOA (785–3380 ng mL-1) was much higher than that of BOA(0.6–11 ng mL-1)in the wheat stem bases of the two cultivars(Fig.4).Based on these results,BOA and MBOA were chosen to test their effects on FCR in vitro and in vivo.
A Fp disc was inoculated and incubated on different solid media for five days until the mycelium reached the edge of the PDA medium (Fig. 5A). The mycelium growth inhibition rate revealed that DMSO did not significantly affect mycelium growth rate by itself (P >0.05), but BOA or MBOA-amended PDA medium significantly inhibited the growth of Fp mycelium compared with the DMSO amended medium (P <0.05) (Table S9). In addition,there was a positive correlation between the concentration of BOA, MBOA, and the inhibition ratio of mycelium. MBOA had a higher inhibition ability to Fp growth than that of BOA, as the inhibition rate of BOA was 20.4% and 31.6% at 0.25 mg mL-1and 0.5 mg mL-1, whereas the inhibition rate of MBOA reached 34.5% and 97.4%, respectively (Table S9). At the highest concentration (1 mg mL-1) of BOA and MBOA, no mycelium of the fungus was observed after one month.
The antifungal activity of BOA and MBOA in vivo was investigated using the relative fungal biomass of the stem bases sprayed with DMSO, BOA, and MBOA at 25 dpi. Spraying BOA and MBOA(1 mg mL-1) could not completely inhibit Fp growth, as the DNA of Fp could be detected in the stem bases of all test groups(Fig.5B).The higher fungal biomass in XM sprayed with DMSO solvent compared with that in 04z36 was in line with its high susceptibility. Significantly lower fungal biomass was found in XM and 04z36 sprayed with BOA and MBOA than in those sprayed with DMSO (P <0.05) (Fig. 5B). These results revealed that BOA and MBOA exhibited antifungal activity during Fp infection in wheat.Interestingly, 04z36 seedlings sprayed with BOA had lower fungal biomass than sprayed with MBOA, whereas XM plants sprayed with MBOA had the lowest fungal biomass.

Fig. 4. The content of endogenous BOA and MBOA in XM and 04z36 seedlings quantified by LC–MS at seven time points after inoculation. (A) The content of BOA. (B) The content of MBOA. CK, control; FP, inoculated by Fp.

Fig. 5. The effects of BOA and MBOA on Fp growth in vitro and in vivo. (A) Inhibition of mycelium growth by BOA and MBOA in vitro at 5 days post inoculation (dpi). The numbers on the left represent different biological replicates.The rings marked on the plates indicate the daily hyphal-growth edge positions.All compounds were tested at three concentrations(0.25,0.5,and 1 mg mL-1).Half-strength PDA and half-strength PDA amended with 1%DMSO were used as controls.(B)Relative fungal biomass of XM and 04z36 seedlings sprayed with DMSO, BOA, and MBOA. Fungal biomass was calculated by RT-qPCR. Bar plots represent the relative Fp biomass. Data are presented as mean ± SD (n = 3). *, significant at P <0.05.
4. Discussion
4.1. FCR infection limited the growth and development of XM and 04z36 seedlings at early stages
During the seedling stage,environmental factors may have significant effects on plant growth and development [34–36]. In this study, compared with control plants, both XM and 04z36 inoculated seedlings showed various levels of height reduction between 10 and 35 dpi.The number of tillers was also reduced in inoculated XM and 04z36 seedlings at 35 dpi (data not shown). These results were consistent with a previous study that also reported a reduction in tiller height caused by FCR infection in adult wheat in the field [3]. A possible reason for this result is that the FCR pathogen damaged wheat stems and leaf sheaths at a very early stage, thus,affecting plant nutrient and water intake. Unlike XM plants, the height changes of partial resistant cultivar 04z36 were not significant at six of the seven investigated time points, except at 25 dpi.The DON content and fungal biomass reflected Fp infection development in the two cultivars.A relatively low DON content and fungal biomass were detected in 04z36, indicating that 04z36 was more resistant to FCR and,thus,suffered less damage from the disease(Fig.6A).Other factors,such as the completeness of resources between photosynthesis and defense reactions, may also contribute to the observed height reduction in the two cultivars.During photosynthesis, plants harvest luminous energy to generate ATP and reduce power in the form of NADPH to produce assimilates for various biological processes[37].Previous studies showed that the photosynthetic rates in the infected areas of plants were reduced after pathogen attack [16,38–40]. In our study, photosynthesis pathways were also significantly interrupted by FCR infection at 20 dpi in both XM and 04z36 plants based on the KEGG and GO analyses of the DEGs (Tables S7 and S8). All 46 DEGs in the photosynthesis pathway of XM plants were down-regulated after FCR infection. Among the nine DEGs associated with this pathway in 04z36 plants, eight were down-regulated.

Fig. 6. Quantitative analysis of metabolites extracted from metabolomics data. (A) Deoxynivalenol, (B) proline betaine, (C) ribitol, (D) arabitol, (E) lauric acid, and (F)chlorogenic acid relative contents in control and inoculated plants of 04z36 and XM at 20 and 25 days post inoculation. Data are presented as mean ± SD (n = 3).
4.2. Metabolites related to the FCR resistance
Compared with the assessment method based on the discoloration of the leaf sheath or stem base, the estimation of relative fungal biomass using RT-qPCR provided an alternative way to assess the resistance level at the early developmental stage when the symptoms were not obvious [41]. Positive correlations were observed between pathogen biomass and stem base browning in both glasshouse and field trials[42,43].In this study,we quantified fungal accumulation in the susceptible cultivar XM and partial resistant cultivar 04z36 using RT-qPCR at seven time points, and the results also showed a high correlation with disease symptoms(Figs. 1B, S1). As expected, the fungal biomass in 04z36 across all seven time points was much lower than that in XM, which was in line with the higher FCR resistance of 04z36 in a previous study[9].Based on RT-qPCR analysis,the fungal biomass of the two cultivars started to differentiate at 20 dpi, indicating that this time point may be a key phase during infection, causing the FCR resistance difference. We collected samples at 20 and 25 dpi for metabolic and transcriptional analysis to investigate the genes and metabolites involved in FCR resistance.
Of the 102 significantly changed metabolites in XM and 04z36 at 20 and 25 dpi, many metabolites are involved in resistance to various wheat fungal diseases,including FCR.For example,tryptophan metabolism was significantly enriched in both XM and 04z36 at 20 and 25 dpi. The levels of four metabolites in this pathway,namely tryptamine, N-feruloyl serotonin, N-feruloyl tryptamine,and N-acetyl-5-hydroxytryptamine,significantly increased in both XM and 04z36 after FCR infection(Table S3).Tryptamine can attenuate biotic stress through a hypersensitive response and scavenge ROS, and it is known to be responsive to FCR colonization[13,44,45]. The 17 flavonoid metabolites may also have vital roles in FCR resistance, considering the potential roles of flavonoids in plant defense, including UV protection, auxin transport inhibition,allelopathy,and flower coloring[46].Following flavonoid metabolites, 16 significantly changed phenolic acid metabolites. Phenolic acids have been proven to have defense functions through direct interference with the fungus or by reinforcing plant structural components that act as a mechanical barrier against the pathogen[47].
Proline betaine, significantly induced in two cultivars after Fp inoculation (Fig. 6B), could increase resistance to abiotic stress by protecting plasma membrane and inducing antioxidant defense[48,49]. Ribitol and arabitol are considered biomarkers of fungal infection because these two major fungus storage carbohydrates were present in rust-infected wheat[50].Increased ribitol and arabitol were also found in Vitis vinifera berries infected by the necrotrophic pathogen Botrytis cinerea and, thus, are considered biomarkers of B. cinerea infection [51]. In line with these results,ribitol and arabitol in 04z36 and XM significantly increased after FCR colonization,indicating that they could also be used as potential metabolic markers for FCR infection (Fig. 6C, D). The DEG TraesCS3D02G035900 associated with the interconversion of Larabitol and L-arabinose was up-regulated in 04z36 plants after inoculation.Lauric acid,which was specifically increased in inoculated 04z36 plants at 20 and 25 dpi, is considered a natural pesticide and fungicide because of its high insecticidal antimicrobial properties [52,53]. Lauric acid significantly inhibited mycelial growth of Rhizoctonia solani and Pythium ultimum in agar culture and reduced the colonization in barley leaves[54].The higher content of lauric acid in inoculated 04z36 may contribute to its higher resistance to FCR than that of XM (Fig. 6E).
Other metabolites that arose our interest included chlorogenic acid and D-sorbitol.It was reported that Fusarium pathogens could degrade chlorogenic acid and exogenous chlorogenic acid inhibited the production of mycotoxin by Fusarium species [55], and mycotoxin is a major component of virulence for the pathogens[13,56]. The consumption of chlorogenic acid was observed in maize plants infected with F. graminearum, suggesting that these metabolites may be involved in the resistance to the Fusarium pathogens [57]. In the current analysis, a higher level of chlorogenic acid was found in 04z36 control plants compared with XM plants.In the inoculated plants of 04z36 and XM,chlorogenic acid can only be detected in 04z36 plants at 20 dpi(Fig.6F).In line with this, two genes included TraesCS2B02G171200 and TraesCS2D02G150400 which catalyzed production of caffeoyl-CoA from chlorogenic acid were up-regulated in XM after inoculation.These results suggested the potential roles of chlorogenic acid in FCR resistance. D-sorbitol is a key metabolite in the fructose and mannose metabolism pathway.It was reported that sorbital could modulate disease resistance in apple by regulating the expression of an NLR resistance gene [58]. In this study, the level of Dsorbitol significantly increased in both of 04z36 and XM plants after inoculation. Based on RNA-seq analysis, the expression of TraesCS3D02G035900,which encoded aldehyde reductase that catalyze the reduction of D-glucose to D-sorbitol, was significantly increased in 04z36 plants.
4.3.JAZ genes are differentially expressed between XM and 04z36 after inoculation
GO enrichment analysis showed that regulation of JA mediated signaling pathway was significantly enriched in 04z36, which included 15 JAZ genes(Table S8).JAZ has been reported to mediate defense and growth status of plants by interacting with DELLA proteins and could inhibit the activity of transcriptional regulators of JA responses by physically binding to them [59]. The degradation of JAZs can release JAZ-bound transcription factors, leading to the activation of downstream JA responses upon insect herbivores or necrotrophic pathogens. JA signaling pathway plays a dominant role in regulating defense responses against necrotrophic pathogens [60,61]. A previous study revealed that JA was a primary responsive signal during Fp colonization in wheat,and the application of exogenous JA could significantly delay the development of necrotic symptoms in both partial resistant and susceptible wheat cultivars[14].Previous transcriptome results also showed that JArelated genes were up-regulated by FCR infection at 3 and 5 dpi[11]. In contrast, JA-biosynthesis pathway was not significantly enriched in this study, which is consistent with the fact that JA content was not significantly changed by Fp infection at 1, 3, and 7 dpi [13]. Three down-regulated JAZs were found in rice infected by F. fujikuroi in early stage [62]. In the current analysis, the 15 JAZ genes showed differential expression patterns between XM and 04z36 (Fig. 3A). The down-regulation of these genes in 04z36 might promote JA signaling pathway and result in increased resistance to FCR, whereas the up-regulation of JAZs genes in XM might contribute to its susceptibility. Similar results were also reported in sugarcane infected by Sporisorium scitamineum, in which JAZs were up-regulated in the susceptible genotype and down-regulated in the resistant genotype [63]. Additionally,up-regulated and down-regulated JAZs were found in a resistant Brassica oleracea line infected by F. oxysporum. From this perspective, the expression patterns of JAZs may depend on the pathogen species [64].
4.4.Treatment of BOA and MBOA significantly inhibited fungal growth
Tryptophan metabolism was significantly enriched in 04z36 at 20dpi.In this pathway,all of the significantly changed metabolites were increased after Fp infection,including N-feruloyl tryptamine,N-feruloyl serotonin, tryptamine, N-acetyl-5-hydroxytryptamine and anthranilic acid at 20 dpi. Benzoxazinones are secondary metabolites derived from tryptophan pathway and play important roles in the defense systems of members of Poaceae family [65].Studies have shown that the stem base of maize contains a higher benzoxazinone content and benzoxazinones are biocidal to some soil-borne bacteria and fungi and also act as recruitment signals[66–68]. Benzoxazolinones could enhance the resistance to maize Fusarium ear rots caused by Fg [23]. In the current analysis, DEGs in the benzoxazinoid biosynthesis pathway were also up-regulated in 04z36 plants after inoculation (Fig. 3B). Together, these results suggest a potential role of benzoxazolinones in FCR resistance in wheat. We tested the antifungal function of BOA and MBOA, the predominant benzoxazinoids in plants in vitro and in vivo (Fig. 5).Both BOA and MBOA showed antifungal activity in vitro at different concentrations in terms of the inhibition rate, and the antifungal activity of MBOA was higher than that of BOA(Table S9).Previous studies have also shown that MBOA contributes to resistance against Cephalosporium gramineum, Gaeumannomyces graminis var.graminis,and F.culmorum[69].As degradation of benzoxazolinones is an important strategy adopted by Fp to overcome host-derived chemical defenses, the better performance of MBOA compared with that of BOA may be because this chemical is more difficult to be degraded by pathogens[70].The failure growth of Fp on BOA and MBOA(1 mg mL-1)media for one month indicates that this concentration may beyond the degradation capability of Fp.The spraying of BOA and MBOA on the stem base of FCR-infected wheat seedlings also significantly reduced Fp biomass,which indicates the fungicidal function of BOA and MBOA to Fp in vivo.
The contents of endogenous BOA and MBOA were quantified to confirm whether BOA and MBOA participate in FCR defense(Fig.4).The results showed that BOA and MBOA were induced by Fp infection in the two cultivars, but higher levels of BOA and MBOA detected in 04z36 than in XM may contribute to the improved resistance of 04z36 plants. Additionally, TraesCS2B02G076400,which was only detected and induced in 04z36, might lead to a higher content of BOA and MBOA in 04z36 than in XM. The quantity of MOBA was much higher than that of BOA in both cultivars.One possible reason is that MBOA is the main hydroxamic acid present in wheat.The accumulation of BOA and MBOA starts to rise as soon as the seedling emerges, which is the same time the FCR pathogens start the infection [71]. Together with the observed antifungal activities of BOA and MBOA in in vitro and in vivo experiments,the modification of genes related to these two metabolites may improve FCR resistance.
5. Conclusions
In this study,for the first time,a comprehensive analysis of the metabolic and transcriptional profiles of the susceptible cultivar XM and partial resistant cultivar 04z36 were performed.In general,there was a high similarity in the transcriptional and metabolic reprogramming of the two cultivars against Fp infection. Metabolites in flavonoid, phenolic acids and amino acids and derivatives classes changed significantly after Fp infection, and tryptophan metabolism was the most significantly enriched metabolic pathway in the two cultivars. Some metabolites, such as proline betaine, lauric acid, and chlorogenic acid, were responsive to FCR infection and may have important roles in disease resistance.Increase levels of ribitol and arabitol might be considered biomarkers of necrotrophic pathogen infection. Transcriptional analysis identified 688 and 570 pathogen-responsive DEGs in XM and 04z36, respectively. Genes involved in the regulation of JAmediated signaling and benzoxazinoid biosynthesis pathways were responsive to FCR infection. Two predominant benzoxazinoids (BOA and MBOA) levels increased after FCR infection in wheat plants. BOA and MBOA also showed antifungal activities against Fp infection in vitro and in vivo. The higher benzoxazinoid content of 04z36 may contribute to a better FCR resistance, when compared with XM.
CRediT authorship contribution statement
Shuonan Duan:Conceptualization, Investigation, Validation,Formal analysis, Writing – original draft, Writing – review & editing, Funding acquisition.Jingjing Jin:Investigation, Validation.Yutian Gao:Investigation, Validation.Changlin Jin:Resources.Junyi Mu:Investigation, Validation.Wenchao Zhen:Writing –review & editing.Qixin Sun:Writing – review & editing.Chaojie Xie:Writing–review&editing.Jun Ma:Conceptualization,Formal analysis,Writing–original draft,Writing–review&editing,Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31872865), Central University Basic Scientific Research Program (2018QC158), Program for Modern Agriculture of Hebei Province (494-0402-JBN-S2XB), and the Basic Operating Foundation of Hebei Academy of Agriculture and Forestry Sciences(2018110102).
Appendix A. Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2021.06.004.
- The Crop Journal的其它文章
- Brief Guide for Authors
- Cob color, an indicator of grain dehydration and agronomic traits in maize hybrids
- Effects of sgRNA length and number on gene editing efficiency and predicted mutations generated in rice
- Imbalance between nitrogen and potassium fertilization influences potassium deficiency symptoms in winter oilseed rape(Brassica napus L.)leaves
- ZmWRKY104 positively regulates salt tolerance by modulating ZmSOD4 expression in maize
- A novel genomic prediction method combining randomized Haseman-Elston regression with a modified algorithm for Proven and Young for large genomic data

