The epigenetic landscape of exercise in cardiac health and disease
Guiling Wu,Xing Zhang*,Feng Gao*
School of Aerospace Medicine,Fourth Military Medical University,Xi’an 710032,China
Abstract With the rising incidence of cardiovascular diseases,the concomitant mortality and morbidity impose huge burdens on quality of life and societal costs.It is generally accepted that physical inactivity is one of the major risk factors for cardiac disease and that exercise benefits the heart in both physiological and pathologic conditions. However, the molecular mechanisms governing the cardioprotective effects exerted by exercise remain incompletely understood.Most recently,an increasing number of studies indicate the involvement of epigenetic modifications in the promotion of cardiac health and prevention of cardiac disease.Exercise and other lifestyle factors extensively induce epigenetic modifications,including DNA/RNA methylation,histone post-translational modifications,and non-coding RNAs in multiple tissues,which may contribute to their positive effects in human health and diseases.In addition, several studies have shown that maternal or paternal exercise prevents age-associated or high-fat dietinduced metabolic dysfunction in the offspring,reinforcing the importance of epigenetics in mediating the beneficial effects of exercise.It has been shown that exercise can directly modify cardiac epigenetics to promote cardiac health and protect the heart against various pathological processes,or it can modify epigenetics in other tissues,which reduces the risk of cardiac disease and affords cardioprotection through exerkines.An in-depth understanding of the epigenetic landscape of cardioprotective response to exercise will provide new therapeutic targets for cardiac diseases. This review,therefore,aimed to acquaint the cardiac community with the rapidly advancing and evolving field of exercise and epigenetics.
Keywords: Cardioprotection;DNA methylation;Epigenetics;Exercise;Histone post-translational modifications;Non-coding RNAs
1. Introduction
With an aging population continuously growing worldwide,the increasing incidence of cardiac disease increases mortality and morbidity and imposes major burdens on quality of life and societal costs. Physical inactivity is suggested as one of the major risk factors for numerous non-communicable diseases, including cardiac disease.1Additionally, exercise is implicated in benefiting heart health. Numerous studies have documented an inverse relationship between physical activity(self-reported or objectively measured)and cardiovascular disease (CVD) or all-cause mortality.2-4Even a lower dose of exercise than that recommended by the World Health Organization yields cardiac benefits;it improves cardiometabolic biomarkers in adolescents5and decreases all-cause mortality in adults.6,7For instance, in an Asian adult population8just 15 min of daily exercise effectively reduced risk of all-cause mortality by 14% compared with physical inactivity, and running for 5-10 min daily at slow speeds (<6 miles per hour)led to a considerably reduced risk of death from CVD in U.S.adults.9Intervention trials have also shown that exercise plays a protective role against various cardiac diseases.10
Although the biological mechanisms underlying these exercise-induced benefits have been thoroughly investigated, the molecular mechanisms governing the cardioprotective effects of exercise are not yet completely understood. Several recent studies have indicated the involvement of epigenetic modifications in exercise-exerted cardioprotection.11-14Epigenetics refers to almost all biochemical changes that influence gene expression independent of genetic code changes.Lifestyle and environmental factors have been shown to influence health via epigenetic modifications,and exercise is one of the critical factors responsible for playing a positive role in human health and preventing disease.15Furthermore, recent studies have shown that maternal or paternal exercise prevents age-associated and high-fat, diet-induced metabolic dysfunction in offspring.Given that metabolic dysfunction is one of the risk factors for CVD, this suggests that exercise-induced benefits can be inherited.16These findings reinforce the importance of epigenetics in mediating the beneficial effects of exercise. In this review, therefore, we specifically discuss the epigenetic landscape of exercise in cardiac health and diseases.
2. Epigenetic modification
Epigenetic mechanisms include 3 interconnected categories: DNA/RNA methylation, non-coding RNAs (ncRNAs),and histone post-translational modifications. DNA methylation, the only recognized epigenetic mechanism affecting DNA directly, includes the addition of a methyl group at the fifth carbon of cytosine nucleotides(5mC),the sixth carbon of adenine to form N6-methyladenine (6mA), and the seventh carbon of guanine to form 7-methylguanine (7mG). Among these types of DNA methylation,5mC is the most widely studied. Whether the expression of the gene with methylation is enhanced or reduced is largely dependent on the distribution of methylation.17,18It has been suggested that methylation at a gene’s promoter or“enhancer location”will interfere with the transcription of the gene; however, methylation in intergenic regions is generally believed to enhance gene expression.18The enzymes responsible for importing and/or sustaining DNA methylation are DNA methyltransferases(DNMTs).The DNMT family consists of DNMT1, DNMT2, DNMT3A,DNMT3B,and DNMT3L.DNMT1,DNMT3A,and DNMT3B are canonical DNMTs that catalyze the addition of methylation marks to genomic DNA. By contrast, DNMT2 and DNMT3L are non-canonical family members, since they do not possess catalytic DNMT activity. DNMT3A and DMMT3B are responsible for de-novo methylation and modification of unmethylated DNA,19whereas DNMT1 is required to maintain DNA methylation.20DNA methylation is relatively static in adults,but there are 2 distinct phases of demethylation during gametogenesis and embryogenesis.21,22The exact mechanisms concerning DNA methylation reprogramming remain to be elucidated.
RNA methylation has been found in most eukaryotic mRNA and ncRNA, which epigenetically impacts numerous biological processes.23N6-methyladenosine(m6A)is the most common and abundant methylation modification in RNA molecules present in eukaryotes and accounts for more than 80%of all RNA methylation.24Methyltransferase-like 3, methyltransferase-like 14, and Wilms’ tumor 1-associating protein constitute m6A methyltransferases. The fat mass and obesity-associated protein (FTO) and alkB homolog 5 are m6A demethylase.25,26The m6A regulates target gene expression and RNA translation, degradation, splicing,export, and folding, thus influencing the corresponding cell processes and function.27
A histone modification is a covalent post-translational modification to histone proteins,including methylation,phosphorylation, acetylation, ubiquitylation, sumoylation, biotinylation,and ADP-ribosylation.28,29The modification can alter gene expression through the regulation of chromatin structure or recruitment of histone modifiers. Though the specific mechanisms remain unclear, both the distribution and number of modifications are critical for regulating genetic expression by histone modification.30Histone is acetylated by histone acetyltransferases and deacetylated by histone deacetylases(HDACs),31which are classified into 4 classes: Class I(HDACs 1-3 and HDAC 8),Class II(HDACs 4-7,HDAC 9,and HDAC 10),Class III(sirtuin(SIRT)family),and Class IV(HDAC 11).32Histone methylation is catalyzed by histone methyltransferases and converted by histone demethylases,which include members of the Jumonji family as well as lysine-specific demethylase-1.
The ncRNAs are also involved in epigenetic regulation.Among ncRNAs, microRNAs (miRNAs), and long ncRNAs(lncRNAs) have been extensively studied. It is well-established that miRNAs modulate gene expression mainly by base-pairing with the 3′-untranslated region or the 5′-untranslated region of target mRNAs,33thus inhibiting translation or promoting degradation of target mRNAs. The regulation of gene expression by lncRNAs is relatively more complex because it involves multiple steps, including chromatin structure remodeling, transcription machinery recruitment, mRNA processing transportation,and interaction with miRNAs.34
3. Role of epigenetics in cardiac health and diseases
3.1. Epigenetics in cardiac development
The heart is the first functional organ to develop in the fetus. Cardiac development during the fetal period is influenced by both genetic and epigenetic processes. Numerous studies have shown that DNA methylation, histone modification, and ncRNAs play a significant role in heart development.35-37The role of exercise-induced epigenetic modulation in heart development or congenital heart disease has not been well-documented.Several studies have suggested that exercise-modulated epigenetics may influence cardiac development. For instance, the human fetus’s heart may be responsive to maternal exercise, given that the intensity of maternal physical activity is negatively associated with heart rate and the duration of physical activity is positively associated with fetal heart rate variability.38Saiyin et al.39have shown that maternal voluntary exercise mitigated the incidence of congenital heart defects in pre-gestational diabetes.Saiyin et al.39found that cardiac gene expression of Notch1,Snail1, Gata4, and Cyclin D1 was significantly higher in the embryos of exercised diabetic mice compared to non-exercised mice. Although the specific epigenetic regulation of these genes was not elucidated,these results suggest that epigenome modification of germ cells by exercise may be involved in cardiac development and congenital heart disease.
3.2. Epigenetics in cardiac diseases
Epigenetic modifications from environmental factors including diet, exercise, smoking, atmospheric pollutants, and other factors.These factors may positively or negatively influence cardiac diseases through cardiac risk factors including aging, dyslipidemia, obesity, and diabetes, or by directly perturbing cardiac homeostasis.For example,alterations of epigenetic patterns in aging include decreased global DNA methylation,40decreased acetylation of histone 3 at lysine 9 and trimethylation of histone 3 at lysine 9 and lysine 27,41as well as disrupted expression of miR-29, miR-34a, miR-217,and miR-146.42,43These age-induced epigenetic alterations contribute to the cardiac dysfunction through disrupting selfrenewing hematopoietic stem cell dysfunction and lessening angiogenic potential.41,44Epigenetic alterations also directly influence cardiac function. Haas et al.45examined genome-wide cardiac DNA methylation in patients with idiopathic dilated cardiomyopathy(DCM)and controls.These researchers identified methylation differences in pathways correlated with cardiac disease, as well as in genes with yet unknown function in DCM and heart failure.45The detailed mechanisms of DNA methylation in modulating cardiac pathologic processes have not yet been clarified. RNA methylation m6A has also been reported to be involved in cardiac diseases. For example, it has been reported that m6A altered in cardiac hypertrophy and heart failure leads to changes in protein abundance,which contributes to the development of heart failure.46The FTO is decreased in the failing hearts of humans, pigs, and mice, as well as in mouse cardiomyocytes under hypoxia.47Upregulation of FTO attenuates ischemia-induced cardiac remodeling and improves cardiac contractility.47With regard to the significance of histone modification in cardiac pathologic processes, Nguyen et al.48found that the heart-specific loss of the disruptor of telomere silencing 1-like, a histone methyltransferase catalyzing H3K79 methylation in mammals, caused a DCM-like phenotype in mice. It has been demonstrated that specific deletion of Dicer, a miRNA biogenesis enzyme,leads to progress of heart failure.49Corsi et al.50recently identified different methylations of mitochondrial DNA(mtDNA)including cytochrome-C-oxidase I,cytochrome-C-oxidase III, and transfer RNA leucine 1 in platelets prior to CVD development in a population of adults with overweight and obesity. Collectively, the epigenetic regulation of gene expression affects cardiac function and serves as a link between lifestyle and environmental factors and cardiac health and disease.
4. Exercise and epigenetics
4.1. Exercise induces epigenetic modulation
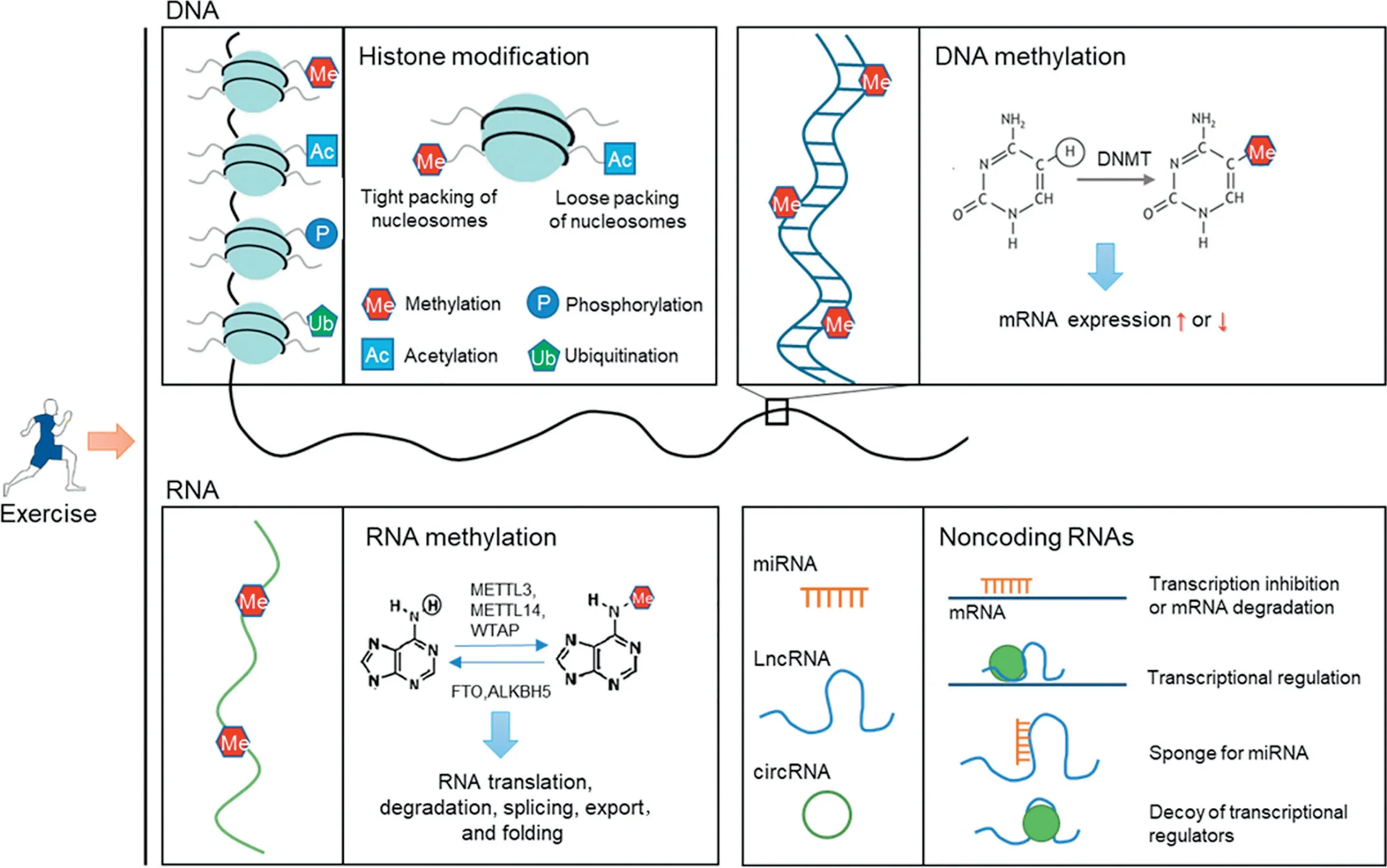
Fig.1. Exercise induces profound alterations in epigenetics in multiple tissues.Exercise induces alterations in histone modification,DNA/RNA methylation,and non-coding RNA expression in multiple tissues, which underlies its beneficial effects on health promotion and disease prevention. ALKBH5=alkB homolog 5;circRNA=circular RNA; DNMT=DNA methyltransferases; FTO=the fat mass and obesity-associated protein; LncRNA=long non-coding RNA;METTL3=methyltransferase-like 3;METTL14=methyltransferase-like 14;miRNA=micro RNA;WTAP=Wilms’tumor 1-associating protein.
Exercise induces profound alterations in epigenetics,which may underlie its beneficial effects (Fig. 1). Numerous studies have reported the influence of exercise on the genome-wide DNA methylation in different tissues including skeletal muscle,adipose tissue and blood.In skeletal muscle,it is noteworthy that epigenetic-modified genes mainly comprise key metabolic genes.51Exercise is responsible for dose-dependent upregulation of peroxisome proliferator-activated receptor γ coactivator 1α (PPARGC1A, encoding PGC-1α), pyruvate dehydrogenase kinase 4(PDK4),and PPARδ(PPARD,encoding PPARδ),which is consistent with marked hypomethylation on promoters of these genes.51Most of the methylationmodified genes in adipose tissues show upregulated methylation and downregulated expression, which is opposite to the expression pattern in skeletal muscle,where most of the identified genes show downregulated methylation and upregulated expression.52Ras like proto-oncogene A binding protein 1(RALBP1), a gene involved in the progress of metabolic syndrome and interference of glucose transporter 4 (GLUT4)transfer,53is hypermethylated by exercise in adipose tissue.52Global DNA methylation in the blood following exercise was shown to be upregulated.54Moreover, exercise modified genome-wide leukocyte DNA methylation after 4 weeks of sprint interval training.55Among these modified genes, genes enriched for pathways responsible for cardiovascular fitness include mitogen-activated protein kinase(MAPK)and phosphatidylinositol 3 phosphate kinase/protein kinase B (PI3K/Akt)signaling pathways. Whether exercise regulates RNA methylation has not been reported.
Exercise also regulates histone modifications including histone acetylation and methylation. As for histone acetylation,early studies have addressed the implication of HDACs after exercise in modifying expression of genes involved in metabolic pathways through transcription factor myocyte enhancer factor 2(MEF2).56For instance,a study observed that H3K9/14 acetylation increased after swimming. The specific hyperacetylation occurs at the MEF2 binding site on the GLUT4 gene, which promotes the MEF2 binding and GLUT4 expression.56Another study indicated that exercise triggered disassociation of HDAC5 from MEF2A, which may promote MEF2A expression.57Histone modifications are also responsible for mitigating neurological and psychiatric disorders following exercise.58In addition,exercise also altered activity of histone acetyltransferases.For instance,a study showed that acute exercise increased histone acetyltransferase activity, leading to a hyperacetylation in frontal cortices of rats.59As for histone methylation, a previous study found that H3K4 trimethylation is decreased in human skeletal muscle by high-intensity resistance training.60However,the specific polycomb repressive complex responsible for this process was not identified.
The alterations in ncRNA induced by exercise also constitute a large part of epigenetic modification after exercise. In hearts of healthy subjects, exercise modulates the expression of numerous miRNAs, including miR-27a,miR-27b, miR208, miR-21, miR-144, miR-124, miR-126,and miR-222, which mainly induce cardiomyocyte growth and left ventricular hypertrophy.12,61-64Significantly,exercise can also restore the miRNAs of hearts with various diseases. For instance, swimming restores the aberrant expression of miR-214, miR-1, miR-29a, and miR-29c in myocardial infarction (MI) hearts.65,66All these restorations modulate downstream targets that are implicated in alleviating cardiac dysfunction. Exercise can also induce changes of miRNAs in other tissues that may affect cardiac function. For example, circulating miRNAs derived from a variety of tissues, including blood cells, adipose tissue,skeletal muscle, the liver and blood vessels, are modulated by exercise, which constitute messengers to regulate gene expression in the heart.11
4.2. Exercise and epigenetic inheritance
Emerging human and animal studies have shown that paternal or maternal exercise exerts intergenerational beneficial effects. Stanford et al.67found that maternal exercise before and during pregnancy mitigated the offspring’s metabolic disorders induced by a maternal high-fat diet,decreasing the risk of obesity and insulin resistance,which is the common soil for numerous non-communicable diseases.The associated mechanism is that maternal exercise prevented the promoter of PPARGC1A from hypermethylation and increased its expression.68Stanford et al.69also recently reported that paternal exercise improved glucose metabolism in adult offspring.These results demonstrate the inheritance of exercise-induced benefits. It has been reported that if mothers begin voluntary exercise either early in life or in later life,maternal age-associated risk of congenital heart risk in newborns is mitigated.70Another study of humans showed that maternal exercise for obesity improved the cardiovascular health of adult male offspring and enhanced levels of calcium-handling proteins.71
Epigenome modification of germ cells induced by exercise may be passed to the next generation, which constitutes the underlying mechanism of exercise-induced intergenerational inheritance.It has been reported that global and genome-wide sperm DNA methylation in humans was affected by 3 months of exercise.72The reactive oxygen species generated during exercise promote ten-eleven translocation (TET) enzymemediated hydroxy methylation of cells,including gametes and primordial germ cells.73Additionally, exercise intervention in obese fathers has been shown to normalize their sperm miRNA profile and alleviate metabolic syndrome in female offspring.74Whether these epigenetic changes can be inherited and modulate the phenotype of offspring remains to be examined, but these results indicate the potential role of germ cell epigenetics in mediating intergenerational effects of exercise.
5. Epigenetic landscape of exercise-induced cardiac protection
5.1. Molecular mechanisms of exercise-induced cardioprotection
Although it is well-established that exercise promotes a cardioprotective phenotype,a detailed understanding of the cellular mechanisms responsible for its beneficial effects remains incomplete.The mechanisms that have been proposed include a decreased risk of cardiac diseases, intrinsic myocardial changes and exerkine-mediated cardioprotection.75-77In addition to decreasing traditional risk factors of cardiac diseases,including hypertension, dyslipidemia, diabetes, obesity, and inflammation, exercise exerts direct effects on hearts, such as increasing anti-oxidative and anti-inflammatory capacity,remodeling mitochondrial structure and function, activating survival signaling, remodeling cardiac structure, increasing cardiac reserve,and decreasing cardiac vulnerability.78Recent studies have also shown that exercise protects the heart through inducing secretion of exerkines from multiple organs and tissues. For example, exercise induces secretion of interlukin-6, irisin, apelin, and other myokines from skeletal muscle, which contributes to cardiac protection induced by exercise.79-81Evidence has shown that epigenetic modulations are involved in these mechanisms. For example, we found that exercise-induced exosomal miR-342-5p secreted mainly by vascular endothelial cells activates cardiac survival signaling,which decreases cardiac vulnerability in myocardial ischemia/reperfusion injury.11
5.2. DNA methylation
Cardiac DNA methylation regulated by exercise has not yet been documented, whereas it has been reported that exercise-modulated DNA methylation in other tissues prevents inflammation and remodels metabolism, consequently reducing the incidence of cardiac diseases.It has been reported that exercise increases the methylation of the adaptor protein, apoptosis-associated speck-like protein containing a caspase recruitment domain (ASC), which is necessary for inflammasome activation of interleukin-1β.13ASC with higher methylation and lower expression is positively related to the outcome of heart failure.82Even though the study did not clarify the causal link between ASC methylation and heart failure, the results suggest the potential of ASC methylation as a target regulated by exercise to mitigate cardiac diseases.82Another study tracking the difference in modified methylation in peripheral blood between controls and elite athletes found accelerated hypermethylation of Kruppel-like factor 14 in elite athletes, which is correlated with anti-inflammatory activity.83Long-term exercise also induces DNA methylation of src homology 2 domain-containing transforming protein C1 in mononuclear cells, which promotes healthy microvascular aging and decreases risk of cardiac disease.84Genome-wide DNA methylation in leukocytes in healthy young men following 4 weeks of sprint interval training has been reported.55The results from the study indicate that DNA methylation of genes in MAPK, calcium, and PI3K/Akt signaling networks,which are involved in cardiac health, was changed after exercise.55These findings suggest that exercise-induced DNA methylation may contribute to cardiac protection.
In 2 major metabolic tissues,skeletal muscle and adipose tissue, exercise-induced DNA methylation may be significant in mitigating metabolic syndrome. Barr`es et al.51tested the promoter methylation of human skeletal muscle after a single bout of exercise and found that exercise induced a dose-dependent expression of PGC-1α, with a marked hypomethylation on its promoter. Interestingly, cardiac-specific PGC-1α-deficient mice underwent accelerated heart failure, indicating the significance of PGC-1α in heart development and function.85However,studies concerning exercise-modulated PPARGC1A methylation in cardiac tissue have not been conducted.In addition to PGC-1α,both methylation and expression of PDK4 and PPARD are modulated by acute exercise.86Because PDK4 and PPARδ play critical role in regulation of glucose and fatty acid metabolism as well as insulin secretion and sensitivity,it is possible that acute exercise benefits whole-body metabolism through regulation of these genes’ epigenetics.86Additionally, Sailani et al.87investigated the role of lifelong exercise in DNA methylation and gene expression and found that among differentially methylated 714 promoters,promoters for genes encoding critical insulin-responsive enzymes in glycogen metabolism,glycolysis,and tricarboxylic acid cycle were more hypomethylated in active men compared with inactive men. Another 2 genes, nuclear respiratory factor 1 (NRF1) and nuclear receptor subfamily 4 group A member 1 (NR4A1), which are encoding transcription factors related to insulin sensitivity, were epigenetically regulated by exercise.88,89In a population of diabetic patients, 16 weeks of exercise hypomethylated NRF1, with a corresponding increase of mitochondrial density, indicating the potential involvement of NRF1 methylation in exercise-induced positive metabolic effects.88The inherited hypomethylation and upregulated expression of NR4A1 in skeletal muscle,as well as higher insulin levels induced by a maternal high-fat diet, were normalized by exercise in offspring.89For adipocyte, a study showed that upregulated methylation of RALBP1 promoter and decreased expression of RALBP1 occurred after exercise.53RALBP1 has also been shown to play a significant role in the development of metabolic syndrome.90Therefore, exercise-modulated methylation of RALBP1 may have the potential to alleviate metabolic syndrome and reduce the risk of cardiac diseases. Collectively, these studies suggest that exercise-induced modifications of DNA methylation are involved in cardiac protection (Fig. 2).
5.3. Histone modifications
5.3.1. Exercise-induced cardiac-specific histone modification
Recently, studies examining exercise-regulated cardiacspecific histone modification are emerging.Most of the studies concentrate on exercise-induced modifications of histone acetylation by HDACs in the heart. For instance, it has been reported that the HDAC4 N-terminal fragment that exercise yielded was involved in preventing the heart from failure.14The underlying mechanism includes regulation of the hexosamine biosynthetic pathway with downstream influence on the protein O-GlcNAcylation and calcium handling.91HDAC1 and HDAC2 have also been shown to play a remarkable role in beneficial cardiac effects when modulated by exercise.92One study has shown that exercise promotes the interaction between mammalian switch-independent 3A (mSin3A)/HDAC1/HDAC2 complex and O-linked β-N-acetylglucosamine transferase(OGT), reversing the pathologic process of the diabetic heart.92mSin3A/HDAC1/HDAC2 complex modulate hypertrophic genes through connection with the DNA-binding proteins repressor element-1 silencing transcription factor and OGT. Collectively, these studies suggest that histone modifications induced by exercise prevent cardiac diseases,including heart failure and diabetic and obesity cardiomyopathy (Fig. 3). However, the specific role that HDACs play in exercise-induced cardiac protective response remains to be further clarified, and the significance of other histone modifications (in addition to histone acetylation such as methylation, phosphorylation, and ubiquitination) in beneficial cardiac effects induced by exercise should be examined.
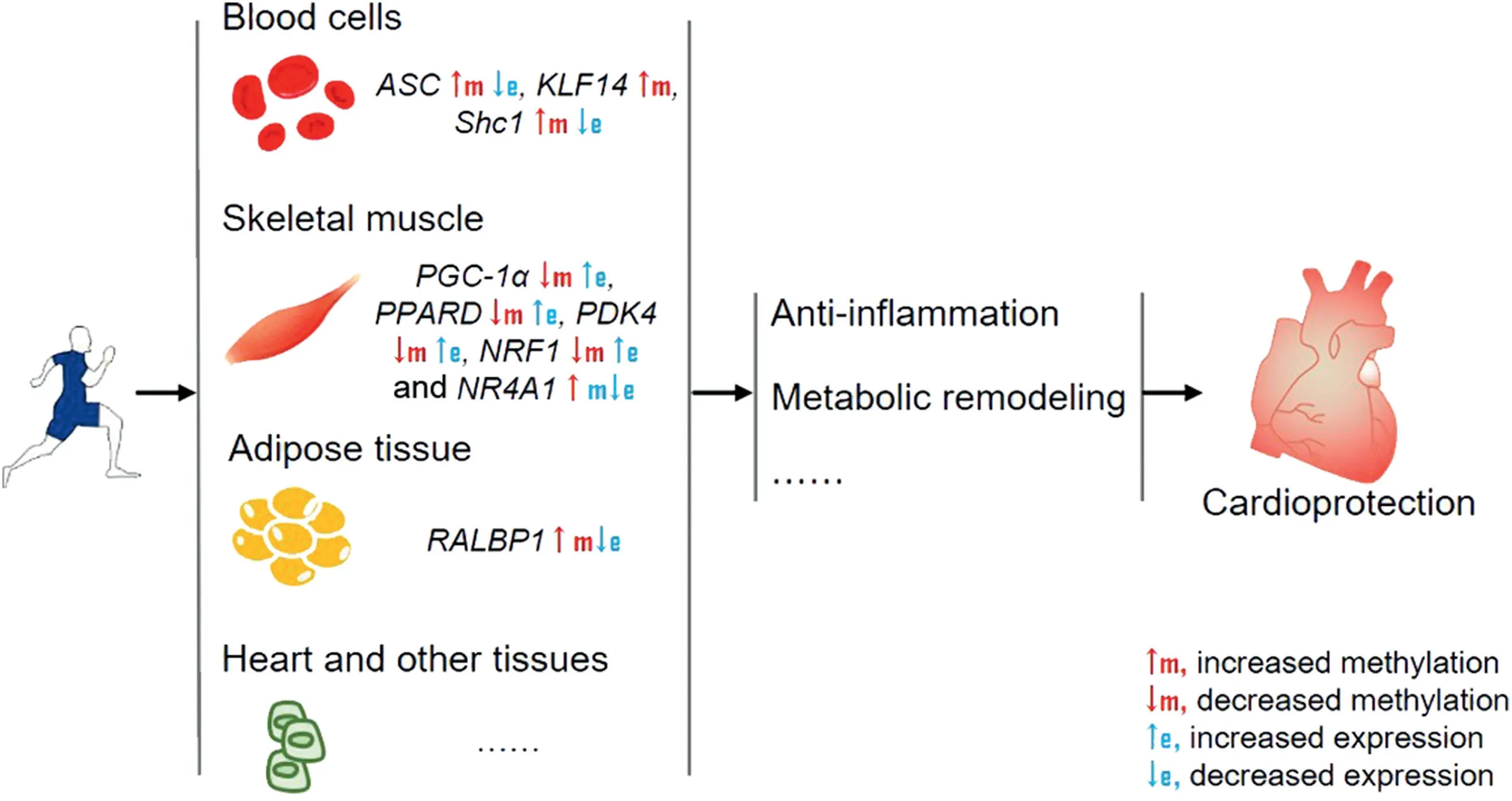
Fig.2. Exercise induces DNA methylation alterations in multiple tissues,which may contribute to cardioprotection through anti-inflammation,metabolic remodeling,and other mechanisms.ASC=apoptosis-associated speck-like protein containing a caspase recruitment domain;e=expression;KLF14=Kruppel like factor 14;m=methylation;NRF1=nuclear respiratory factor 1; NR4A1=nuclear receptor subfamily 4 group A member 1; PDK4=pyruvate dehydrogenase kinase 4;PGC-1α=peroxisome proliferator activated receptor γ coactivator 1 α; PPARD=peroxisome proliferator activated receptor δ; RALBP1=Ras like proto-oncogene A binding protein 1;Shc1=src homology 2 domain-containing transforming protein C1.
5.3.2. Exercise-induced histone modification in other tissues exerting indirect protective effects on the heart
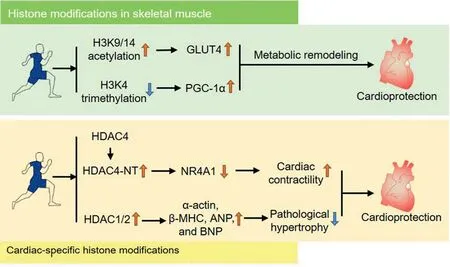
Fig. 3. Exercise induces histone modification in heart and skeletal muscle, which may contribute to cardioprotection exerted by exercise. ANP=atrial natriuretic peptides; BNP=brain natriuretic peptides; β-MHC=β-myosin heavy chain; GLUT4=glucose transporter 4; H3K4=histone H3 lysine 4; H3K9/14=histone H3 lysine 9/14; HDAC=histone deacetylases; HDAC4-NT=HDAC4 N-terminal fragment; NR4A1=nuclear receptor subfamily 4 group A member 1;PGC-1α=peroxisome proliferator activated receptor γ coactivator 1 α.
Some studies have also suggested that histone modifications induced by exercise in other tissues,including skeletal muscle,can alleviate cardiac disease risk through metabolism remodeling (Fig. 3). The studies examining exercise-induced cardiacspecific histone modification did not identify the specific gene regions at which histone modifications occurred. However,some researchers have recently examined exercise-induced histone modulations at specific gene loci in skeletal muscle,and these genes were identified as participants in positive metabolic response, which is an indicator of reduced cardiac risk.For instance,researchers in 1 study using a rat swimming model observed an increase in H3K9/14 acetylation, which regulates GLUT4 on a transcriptional level through action on its promoter.56The specific hyperacetylation occurs at the MEF2 binding site on the GLUT4 gene, which promotes MEF2 binding and GLUT4 expression.MEF2A is a transcription factor and plays a central role in the transmission of extracellular signals to the genome. It causes modified expressions of genes that control multiple aspects of different tissues, and its binding activity with the binding region of GLUT4 is necessary for regulation of the GLUT4 gene promoter in muscle and adipose tissues.93Furthermore,exercise promotes disassociation of HDAC5 from MEF2A, which may promote MEF2A expression.57Additionally, it has been observed that compounds that disrupt Class IIa HDAC function promote a positive metabolic response and protect subjects from chronic diseases driven by physical inactivity,which further elucidates the pivotal role of Class IIa HDACs in metabolic health.94SIRTs have been frequently associated with an equivocal role in the pathologic process of aging-associated pathophysiological conditions,including cancer and cardiovascular,muscular,neurodegenerative, bone, and respiratory diseases.95-98Specifically, numerous studies have demonstrated that SIRTdependent pathways are activated by exercise, which modulates chromatin structure, gene expression and mitochondrial function that lead to beneficial outcomes.99In addition to DNA methylation,histone trimethylation in histone-3 lysine-4 after exercise modulates PGC-1α, which upregulates PGC-1α and remodels metabolism of skeletal muscle.100These findings suggest that exercise-induced histone modifications may contribute to cardiac protection.
5.4. ncRNAs
The ncRNAs have been recognized as critical regulators governing gene expression in numerous physiological processes and appear to be pivotal modulators in a myriad of cardiovascular pathological conditions. Recently, studies concerning the role of ncRNA expression in response to exercise are emerging.
5.4.1. Cardiac-specific ncRNAs in cardioprotection induced by exercise
Numerous miRNAs in the heart are altered by exercise and have been shown to be involved in manipulating many processes,including angiogenesis, cardiac physiological hypertrophy, and cardiomyocyte survival. Several studies have profiled differentially expressed cardiac miRNAs after exercise.12,62,101However,because these researchers used different exercise models and the exercise period varied, the numbers and differentials of expressed candidates also varied.In 1 study comparing sedentary controls with a group receiving 10 weeks of exercise, 87 differentially expressed miRNAs were identified among 349 cardiac miRNAs, including 48 upregulated and 39 downregulated miRNAs.62It has also been reported that 35 miRNAs were differentially expressed after short-term exercise.101Another study using 2 exercise models,swimming and wheel-run,found that 124 miRNAs were differentially expressed in the heart among wheel-run mice, and 55 miRNAs were differentially expressed in the heart among swimming mice in comparison to sedentary controls.12Among these differentially expressed cardiac miRNAs,several miRNAs have been shown to be critical modulators in cardiac physiological and pathological processes after exercise. Physiological cardiac hypertrophy is the most common phenotype provoked by exercise, and the implication of altered cardiac miRNAs in this process has been wellestablished. For instance, aerobic exercise induced cardiac hypertrophy in rats through regulation of cardiac miRNAs,including miR-27a, miR-27b, and miR-143, which modify renin-angiotensin system genes.61Ma et al.63found that cardiac miRNAs(miR-21,miR-124,miR-144,and miR-145)promote cardiac hypertrophy through activation of the PI3K/Akt mechanistic target of rapamycin kinase pathway in rats undergoing chronic swimming.MiR-133 and miR-1 are involved in promoting cardiac pathologic hypertrophy.Exercise downregulates the expression of miR-133 and miR-1 and improves cardiac function in rats.102MiR-150,upregulated by exercise,is involved in exercise-provoked physiological left ventricular hypertrophy.101In addition, miR-208a and miR-208b are involved in regulating several targets that repress the β-myosin heavy chain gene and inhibit physiological cardiac hypertrophy.103Exercise downregulates miR-208a and miR-208b, which promote cardiac hypertrophy.104Additionally,cardiac-specific miR-26b-5p,miR-204-5p,and miR-497-3p are downregulated by exercise and play a role in cardiac hypertrophy by augmenting autophagy.105
In addition,miRNAs are also implicated in cardiac growth,survival and prevention of pathological cardiac remodeling.Significantly, Liu et al.12profiled cardiac miRNA expression in 2 distinct models including swimming and wheel-run.They found that miR-222 was upregulated in both models. Downstream miR-222 targets were identified, including homeodomain-interacting protein kinase 1 and Homeobox-1, which promote cardiomyocyte growth and proliferation and protect the heart from ischemic injury. Another cardiac-derived miRNA, miR-17-3p, also contributes to exercise-induced vulnerability resistance against myocardial ischemia/reperfusion(I/R) injury through targeting tissue inhibitor of metalloproteases metallopeptidase inhibitor 3.106Additionally, exerciseregulated cardiac miRNAs have been implicated in other cardiac protective processes, such as reduced cardiac fibrosis,increased angiogenesis and enhanced contractility. Swimming-increased miR-29 downregulates the collagen gene and mitigates cardiac fibrosis, promoting left ventricular compliance.62Swimming also upregulates cardiac miR-126, which targets Sprouty-related protein 1 and phosphoinositol-3 kinase regulatory subunit 2, which are inhibitors of the vascular endothelial growth factor pathway and can thus induce cardiac angiogenesis.64In addition to aerobic exercise, resistance exercise decreases cardiac miR-214 and increases cardiomyocyte contractility through upregulation of sarcoplasmic reticulum Ca2+-ATPase.107Exercise also mitigates the diabetic heart through upregulation of miR-29b and miR-455, which bind to the 3′region of matrix metallopeptidase 9 (MMP9) and downregulate its expression, thus mitigating the deleterious downstream effects of matrix metallopeptidase 9 (MMP9).108
Moreover,lncRNAs and circular RNAs(circRNAs)participate in the development of cardiac diseases. For instance,lncRNA myosin heavy-chain-associated RNA transcript and Braveheart,as well as circ-angiomotin Like 1 and heart-related circRNA, have been shown to be associated with cardiac hypertrophy.109-112circRNA_010567 has been observed to inhibit cardiac fibrosis,113and lncRNA zinc finger nuclear transcription factor, X-Box binding 1-type containing 1 antisense RNA 1 has been shown to be markedly increased in MI mice,thereby inducing contractility dysfunction.114However,the effects of exercise on these lncRNAs and circRNAs have not been documented.Collectively,exercise-provoked cardiac-specific miRNAs have been widely researched in recent years,inspiring novel clinical therapies in treating cardiac disease and reinforcing the importance of exercise in promoting cardiac health and preventing cardiac diseases.
5.4.2. Circulating ncRNAs in cardioprotection induced by exercise
The ncRNAs are shuttled by exosomes,microvesicles and apoptotic bodies secreted from cells into the extracellular environment, which are then transferred from 1 tissue to another,conferring crosstalk among distal tissues.115Exercise-modulated,circulating ncRNAs are secreted from multiple tissues, which may act as exerkines assembled mainly in exosomes. These ncRNAs play a significant role in directly promoting cardiac function and indirectly reducing CVD risks.116Studies have shown that exercise-modulated exosomes can be specifically taken up by the hearts11(Fig. 4). Our group recently found that exercise-derived plasma exosomes exerted protective effects against myocardial ischemia/reperfusion injury.11In identifying these exercise-derived exosomes,miR-342-5p,which derives from multiple tissues was stood out as the most potent cardioprotective molecule through pro-survival and anti-apoptosis.11Other studies have shown that exercise-modulated ncRNAs are derived from multiple tissues. Remarkably, many skeletal-muscle-derived miRNAs (including miR-1, miR-133a, miR-133b, miR-206, and miR-761) have been shown to participate in regulating metabolism-related pathways such as MAPK and PGC-1α.117,118It has also been reported that the expression of several miRNAs in neutrophils,including miR-16,miR-17,miR-18a,miR-20a,miR-20b, miR-106a, and miR-223, was altered in response to exercise.119Bioinformatic analysis has revealed that these miRNAs are associated with 3 significant pathways that are key mechanisms for inflammation: the ubiquitin-mediated proteolysis pathway, the Janus kinase-signal transducer and activator of transcription signaling pathway and the Hedgehog signaling pathway.119
There is evidence implicating exercise-induced circRNAs and lncRNAs in mitigating insulin resistance and aging-associated disorders.For example,a study found that lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1)expression, which is elevated in diabetes, was downregulated by exercise, and that MALAT1 contributes to insulin resistance in diabetic mice through targeting the miR-382-3p/resistin axis.120Additionally,a recent study found that circular Bardet-Biedl syndrome 9(circBBS9)(decreased in aging)was elevated after exercise.121Bioinformatic analysis has also found that circular Bardet-Biedl syndrome 9 (circBBS9)may play a positive role in muscle aging. Collectively, these results implicate both local and circulating ncRNAs in exercise-induced cardioprotection (Fig. 5) and provide potential novel therapeutic targets in treating metabolic disorders and preventing cardiac diseases.
5.5. Epigenetic landscape of extreme exercise in induction of cardiac risks
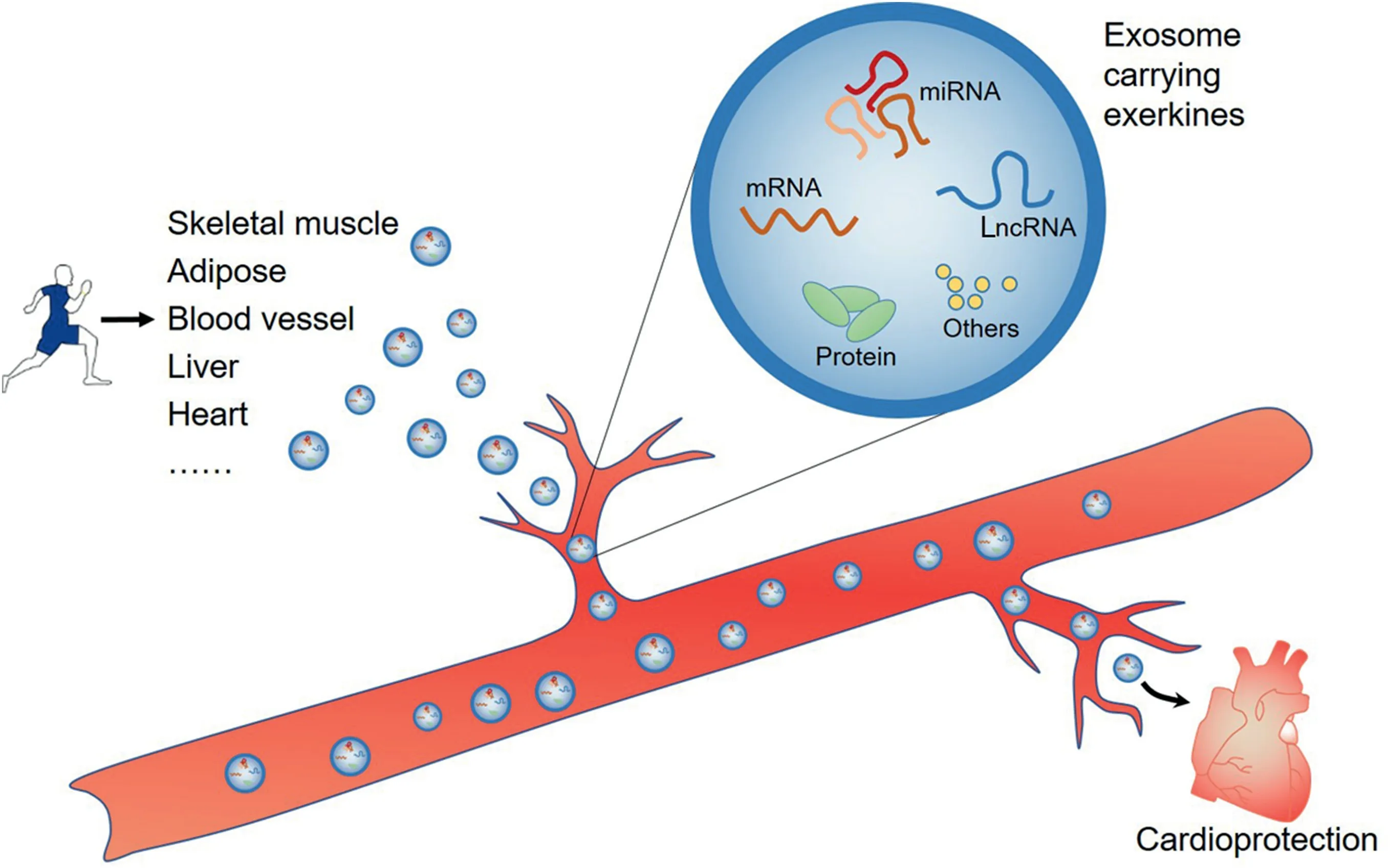
Fig. 4. Exercise-modulated, circulating non-codingRNAs are secreted from multiple tissues, which may act as exerkines shuttled by exosomes that are taken up specifically by the heart, directly promoting cardiac function and indirectly reducing the risk of cardiac diseases. LncRNA=long non-coding RNA; miRNA=microRNA.
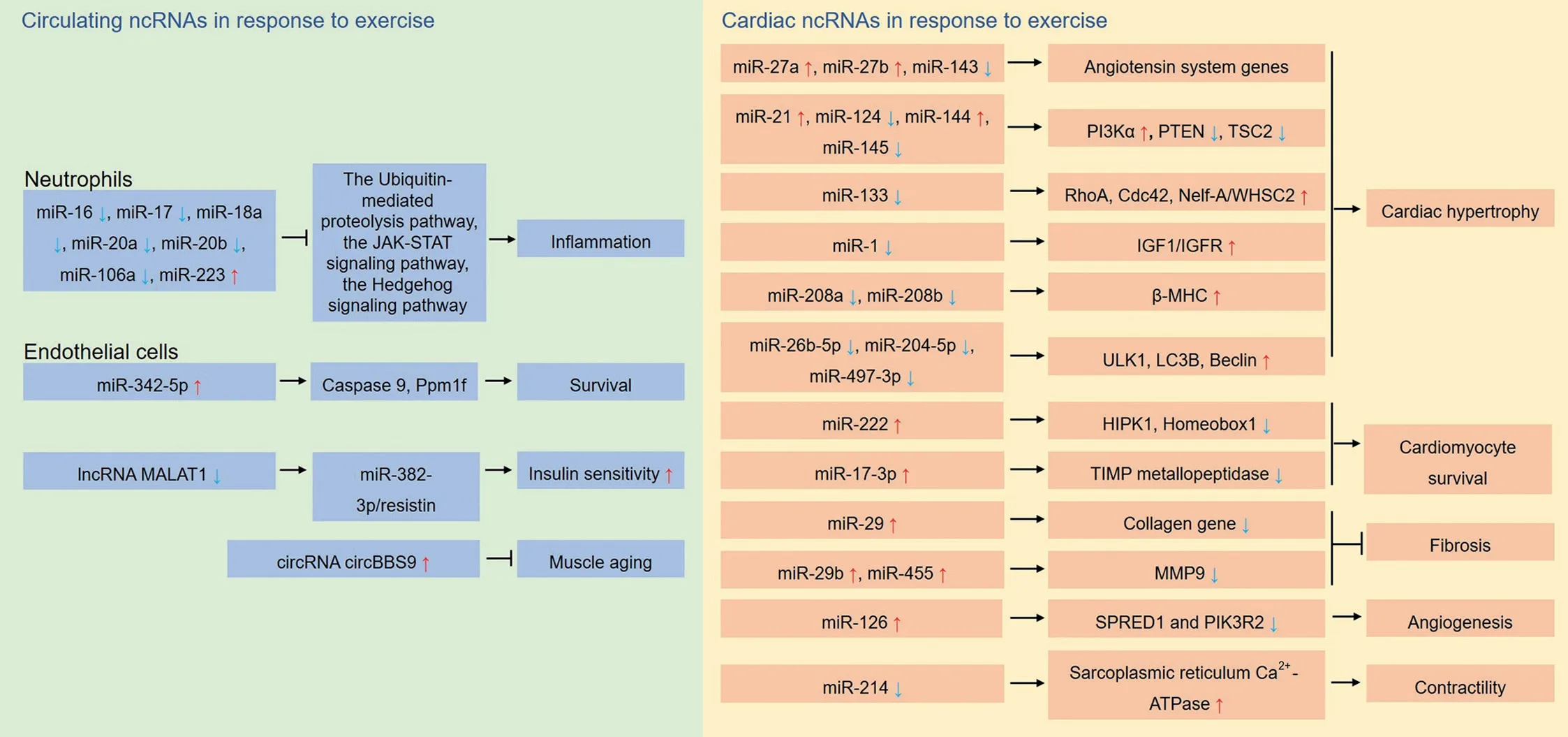
Fig.5. Exercise exerts cardioprotection through regulation of ncRNAs.The ncRNAs are identified as potential exerkines in promoting cardiac health and preventing cardiac diseases. These ncRNAs are either from the heart or are circulated after secretion by multiple tissues. β-MHC=β-myosin heavy chain;Cdc42=cell division cycle 42; circBBS9=circular Bardet-Biedl syndrome 9; circRNA=circular RNA; EVH1=Ena- vasodilator-stimulated phosphoprotein homology 1;HIPK1=homeodomain interacting protein kinase 1; IGF1=insulin-like growth factor 1; IGFR=insulin-like growth factor receptor; JAK=Janus kinase;LC3B=microtubule associated protein 1 light-chain 3β; lncRNA=long non-coding RNA; MALAT1=metastasis-associated lung adenocarcinoma transcript;miR=microRNA; MMP9=matrix metallopeptidase 9; ncRNA=non-coding RNA; Nelf-A/WHSC2=negative elongation factor A; PI3K=phosphatidylinositol 3-kinase; PIK3R2=phosphoinositide-3-kinase regulatory subunit 2; Ppm1f=protein phosphatase, Mg2+/Mn2+ dependent 1F; PTEN=phosphatase and tensin homolog;SPRED1=Sprouty-related EVH1 domain-containing 1;STAT=signal transducer and activator of transcription;TIMP=tissue inhibitor of metalloproteases;TSC=tuberous sclerosis complex;ULK1=unc-51-like autophagy activating kinase 1;WHSC2=Wolf-Hirschhorn syndrome candidate 2.
Intense physical training is commonly used to enhance physical performance during an athletic career. The “athlete’s heart”exhibits a hypertrophied left ventricle, which is a myocardial adaptation in response to chronic exercise for the enhancement of cardiac function. However, intense physical training may contribute to acute heart failure or promote a preexisting ventricular dysfunction in athletes.122The underlying mechanisms include pro-inflammatory cytokines released from exercising skeletal muscle and an inflammatory process in a distant organ or system, such as a respiratory tract infection or untreated post-exercise trauma.123An excessive release of catecholamines can be induced by mental stress and hyperactivity of the sympathetic nervous system during sport competition.124However, epidemiologic evidence for cardiac dysfunction induced by intense physical training is limited.For example,it has been reported that ultra-endurance exercise leads to acute myocardial damage and decreased right ventricular function, but almost all abnormalities were transient and not evident a week later.125In addition, it has been reported that extreme and uninterrupted endurance training over long periods of time (up to 17 years) among Olympic athletes was not associated with deterioration in left ventricular function,significant changes in left ventricular morphology or the occurrence of cardiovascular symptoms or events.126Although direct evidence of epigenetics in cardiac adaptations induced by intense physical training is still lacking,exercise may nonetheless regulate adaptive molecular expressions through epigenetics modulation.
6. Future directions
Despite the well-established evidence of benefits of exercise in cardioprotection, less is known about the epigenetic mechanisms involved in mediating its beneficial effects. Most studies have identified the epigenetic alterations that may affect some aspects of cardiac health and diseases,but only a few studies have investigated other aspects of exercise-provoked epigenetic responses, such as cardiac DNA methylation and histone modification. Further investigation of exercise-induced epigenetic modulations regarding different exercise modules and different epigenetic modifications in different tissues will broaden our understanding of the beneficial effects of exercise. At this point in time, most findings have only shown correlations between modified DNA methylation,histone modifications,and ncRNAs and exercise’s positive phenotype. Evidence for a causal relationship between epigenetics and the beneficial effects of exercise is lacking.Elucidating the underlying mechanisms, from an epigenetic perspective, of the beneficial effects of exercise will help provide novel targets for treating cardiac diseases and promoting cardiac health.Furthermore,intergenerational inheritance of exercise-induced cardioprotection and its underlying mechanisms should be further studied.Such research should specifically include whether epigenetic modifications of germ cells afforded by exercise plays an important role in the cardiac function of subsequent generations.
In summary, there is growing evidence that exercise profoundly induces epigenetic modifications that may contribute to its beneficial effects on the promotion of cardiac health and the prevention of cardiac diseases.The epigenetic landscape induced by exercise in cardioprotection is in its infancy. Elucidating the epigenetic mechanisms of the link between exercise and cardiac health will advance our understanding of exercise cardioprotection and help us to identify novel biomarkers that will promote translational research and personalized exercise prescriptions.
Acknowledgment
This study was supported by grants from the National Key Research and Development Project (2019YFF0301600) and National Natural Science Foundation of China (31930055,81870273,and 31871146).
Authors’contributions
FG conceptualized and revised the manuscript;GW and XZ drew the figures. All authors performed the literature search,drafted the manuscript, and contributed to the writing of the manuscript. All the authors have read and approved the final version of the manuscript,and agree with the order of presentation of the authors.
Competing interests
The authors declare that they have no competing interests.
 Journal of Sport and Health Science2021年6期
Journal of Sport and Health Science2021年6期
- Journal of Sport and Health Science的其它文章
- The association of grip strength with cardiovascular diseases and all-cause mortality in people with hypertension:Findings from the Prospective Urban Rural Epidemiology China Study
- Cardiorespiratory fitness measured with cardiopulmonary exercise testing and mortality in patients with cardiovascular disease:A systematic review and meta-analysis
- The relationships between step count and all-cause mortality and cardiovascular events:A dose-response meta-analysis
- IGF1-PI3K-induced physiological cardiac hypertrophy:Implications for new heart failure therapies,biomarkers,and predicting cardiotoxicity
- Exploring the impact of COVID-19 on the movement behaviors of children and youth:A scoping review of evidence after the first year
- No independent associations between physical activity and clinical outcomes among hospitalized patients with moderate to severe COVID-19
