Construction and validation of an immune-related lncRNA prognostic model for rectal adenocarcinomas*
Danni Jian,Yi Cheng,Jing Zhang,Kai Qin (✉)
1 Department of Otolaryngology,Union Hospital,Tongji Medical College,Huazhong University of Science and Technology,Wuhan 430022,China
2 Department of Oncology,Tongji Hospital,Tongji Medical College,Huazhong University of Science and Technology,Wuhan 430030,China
Abstract Objective This study aimed to construct a prognostic model for rectal adenocarcinomas based on immune-related long noncoding RNAs (lncRNAs) and verify its prediction efficiency.Methods Transcript data and clinical data of rectal adenocarcinomas were downloaded from The Cancer Genome Atlas (TCGA) database.Perl software (strawberry version) and R language (version 3.6.1) were used to analyze the immune-related genes and immune-related lncRNAs of rectal adenocarcinomas,and the differentially expressed immune-related lncRNAs were screened according to the criteria |log2FC|> 1 and P < 0.05.The key immune-related lncRNAs were screened using single-factor Cox regression analysis and lasso regression analysis.Multivariate Cox regression analysis was performed to construct an immune-related lncRNA prognostic model using the risk scores.Next,we evaluated the effectiveness of the model through Kaplan-Meier (K-M) survival analysis,ROC curve analysis,and independent prognostic analysis of clinical features.In addition,prognostic biomarkers of immune-related lncRNAs in the model were analyzed by K-M survival analysis.Results In this study,we obtained gene expression profile matrices of 89 rectal adenocarcinomas and 2 paracancerous specimens from TCGA database and applied immunologic signatures to these transcripts.Through R and Perl software analysis,we obtained 847 immune-related lncRNAs and 331 protein-encoded immune-related genes in rectal adenocarcinomas.Eight important immune-related lncRNAs related to the prognosis of rectal adenocarcinomas were identified using univariate Cox regression and lasso regression analysis.Furthermore,four immune-related lncRNAs were identified as prognostic markers of rectal adenocarcinomas via multivariate Cox regression analysis.The prognostic risk model was as follows:risk score=(-4.084) * expression LINC01871+(3.112) * expression AL158152.2+(7.616) * expression PXNAS1+(-0.867) * expression HCP5.The independent prognostic effect of the rectal adenocarcinoma risk score model was revealed through K-M analysis,ROC curve analysis,and univariate,and multivariate Cox regression analysis (P=0.035).LINC01871 (P=0.006),PXN-AS1 (P=0.008),and AL158152.2 (P=0.0386) were closely correlated with the prognosis of rectal adenocarcinomas through the K-M survival analysis.Conclusion We constructed a prognostic model of rectal adenocarcinomas based on four immunerelated lncRNAs by analyzing the data based on TCGA database,with high prediction accuracy.We also identified two biomarkers with poor prognosis (PXN-AS1 and AL158152.2) and one biomarker with good prognosis (LINC01871).
Key words: rectal adenocarcinoma;immune-related lncRNA;prognostic model;The Cancer Genome Atlas (TCGA) database
Rectal cancer is the eighth most common cancer in the world and the tenth leading cause of cancer-related death.In 2018,there were 704,376 new cases and 310,394 deaths[1].In recent years,the incidence of rectal cancer has increased in China[2].Although the widespread comprehensive treatment involving total mesorectal excision (TME) surgery and chemoradiotherapy has made progress in patient survival,the long-term survival rate is still unsatisfactory,especially for patients with locally advanced and distant metastases,where the overall 5-year survival rate patients with rectal cancer is about 53%[3].Therefore,there is an urgent need to identify new biomarkers to predict the prognosis of patients and guide precise treatment.
Immune-related long noncoding RNAs (lncRNAs),which are located near or overlapping the coding gene clusters of immune-related proteins,play an important role in guiding the development,differentiation,and activation of a variety of immune cells[4].However,to date,only a few immune-related lncRNAs have been implicated in cancer[5].Therefore,it is of great significance to study the role of immune-related lncRNAs in immune regulation.Although some reports have shown that an lncRNA is recognized as a biomarker to predict the prognosis of rectal adenocarcinomas[6-8],there are few studies on immune-related lncRNAs in rectal adenocarcinomas.In this study,immune-related lncRNAs in rectal adenocarcinomas were obtained by analyzing the transcripts and immune-related gene sets in The Cancer Genome Atlas (TCGA) database.We used univariate/multivariate Cox regression analysis to screen immune-related lncRNAs associated with the prognosis of rectal adenocarcinomas.We constructed a prognostic model composed of four immune-related lncRNAs and identified prognostic biomarkers for rectal adenocarcinoma.
Materials and methods
Patients and datasets of rectal adenocarcinomas
The transcripts and clinical data of rectal adenocarcinomas were downloaded from TCGA database(https://portal.gdc.cancer.gov/) on March 13,2020.The screening conditions were as follows:(a) primary tumor site:rectal carcinoma;(b) project:TCGA-READ;(c) disease type:adenocarcinoma or adenoma;(d) data classification:transcriptome profiling;(e) data type:quantitative data of gene expression;and (f) workflow type:HTSeq-FPKM.The transcription data of rectal adenocarcinomas were sorted and transformed into a matrix according to the Strawberry Perl software (version 5.30.1.1).The corresponding clinical data of rectal adenocarcinomas were obtained from TCGA program (including patient number,sex,clinical stage,survival time,survival status,and TNM stage).
Immune-related genes and lncRNAs of rectal adenocarcinomas
The mRNA matrix and long noncoding RNA matrix of the coding protein were obtained by sorting the previous matrix (gene and sample names) using Perl software.We searched the immune-related gene set in the MSigDB database (http://software.broadinstitute.org/gsea/msigdb): IMMUNE_RESPONSE (M19817)and IMMUNE_SYSTEM_PROCESS (M13664),which was used to extract the immune-related genes encoding the protein.The R (version 3.6.1) and Bioconductor(https://www.bioconductor.org/) packages were used for data processing and analysis to obtain immune-related lncRNAs.
Establishment and evaluation of the prognostic model and independent prognostic analysis of clinical characteristics
The differential expression of immune-related lncRNAs in rectal adenocarcinomas was analyzed using Software Package EdgeR (http://bioconductor.org/packages/release/bioc/html/edgeR.html),filtered by the criteria |log2 FC (fold change)| > 1 and false discovery rate(FDR) < 0.05.Clinical data from TCGA were analyzed using univariate Cox proportional hazard regression(PHR),and survival-related lncRNAs were screened according toP< 0.001.Furthermore,through lasso-Cox analysis,the lncRNAs most related to overall survival were determined and cross-validation was performed to prevent overfitting.Then,multivariate Cox-PHR analysis was used to construct prognostic indicators and calculate risk scores.According to the median risk score,patients with rectal adenocarcinomas were divided into high-and low-risk groups.Kaplan-Meier (K-M) analysis was used to compare the differences in survival rate between the two groups.The risk score of each patient was calculated according to the expression levels of lncRNAs.The risk score model was calculated using the following formula:
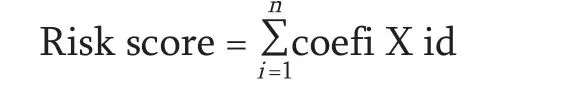
To determine if the risk score could be driven by other clinical cofactors,we used a multivariate model(Cox proportional hazards) to account for age,sex,grade,clinical stage,and T stage in TCGA cohort.Receiver operating characteristic (ROC) and area under the curve(AUC) of 5-year overall survival rate and other clinical characteristics (gender,stage,TNM,and risk score) were calculated by R-package“survival ROC.”Furthermore,K-M survival analysis was performed to identify lncRNAs associated with prognosis and to explore predictive lncRNAs.
Results
Differentially expressed lncRNAs in rectal adenocarcinomas
The clinical data of 90 rectal adenocarcinomas were downloaded from TCGA database (Table 1).We obtained the gene expression matrix of 89 cases of rectaladenocarcinomas;there were 2 cases of paracancerous specimens and 56754 genes were expressed.A total of 847 immune-related lncRNAs and 331 protein-encoded immune-related genes were processed and analyzed using the R language and the corresponding data packet.Using the edgeR package,47 differentially expressed immunerelated lncRNAs were screened with a threshold of|log2FC| > 1 and FDR < 0.05,including 11 upregulated and 36 downregulated lncRNAs.Eight key lncRNAs related to prognosis were identified using lasso regression analysis and univariate Cox regression (Table 2).
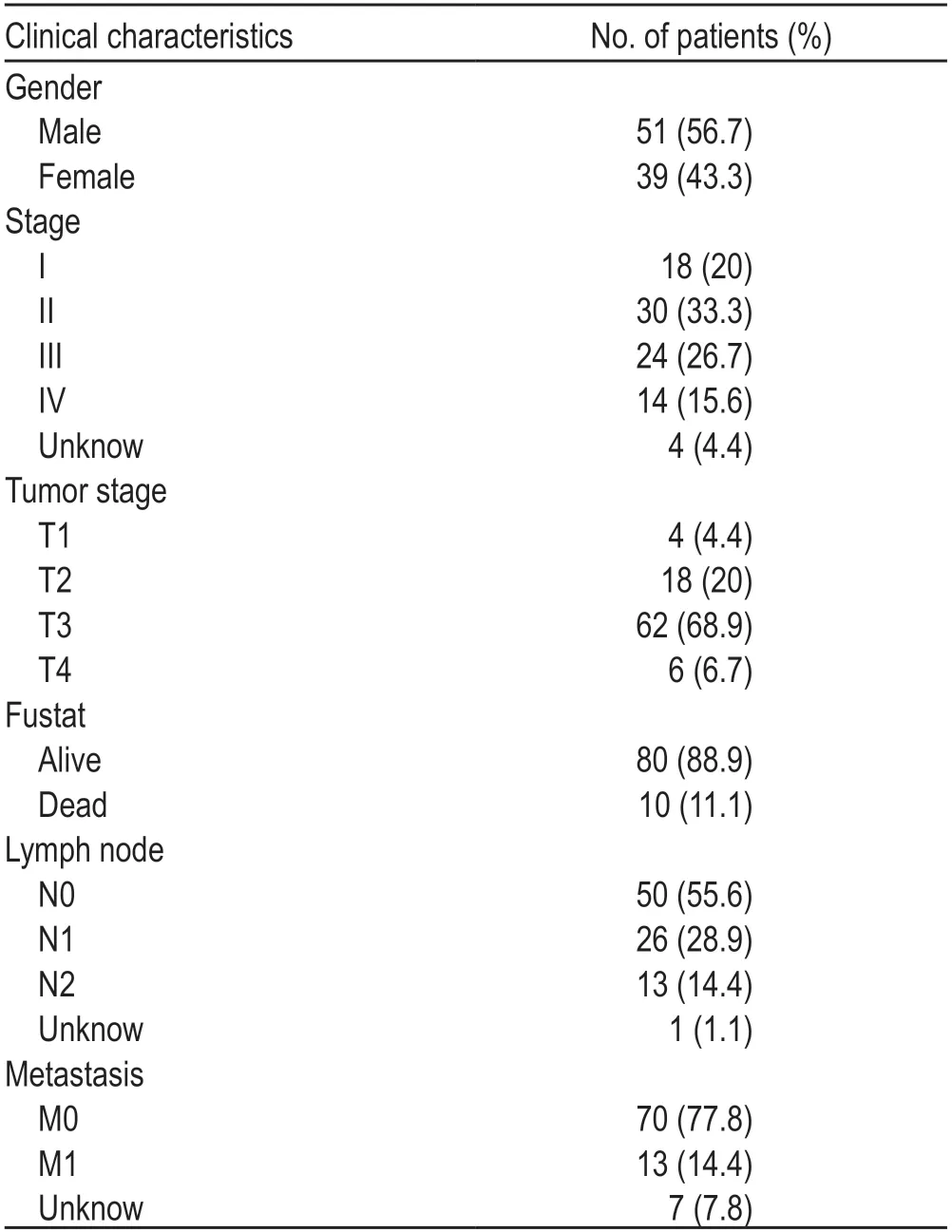
Table 1 Clinical characteristics of rectal adenocarcinomas

Table 2 Immune-related lncRNAs in rectal adenocarcinomas identified by univariate Cox regression analysis
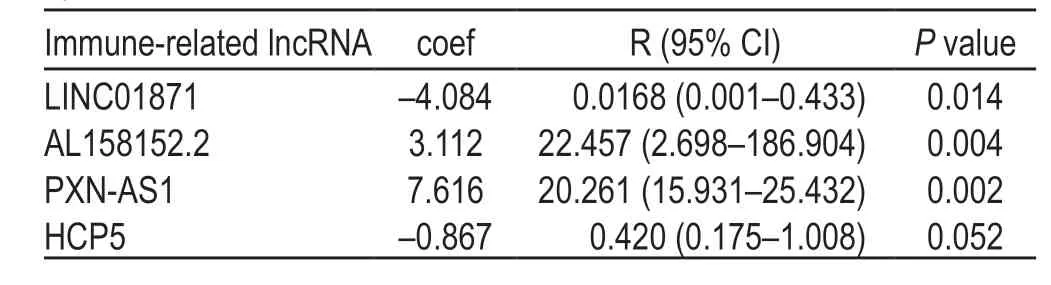
Table 3 Immune-related lncRNAs in rectal adenocarcinomas identified by multivariate Cox
Establishment and verification of the prognostic model of rectal adenocarcinomas
Eight key immune-related lncRNAs,obtained by univariate Cox regression analysis,were used in the multivariate Cox-PHR regression analysis to calculate the prognosis risk score of each patient,and we constructed a risk score model consisting of four lncRNAs (Table 3):risk score=(-4.084) * expression LINC01871+(3.112) *expression AL158152.2+(7.616) * expression PXN-AS1+(-0.867) * expression HCP5.K-M analysis comparing the survival difference between the high-and low-risk groups showed that the total survival time of patients in the low-risk group was significantly longer than that in the high-risk group (P=1.93e-03;Fig.1a).The area under the ROC curve was 0.957 (Fig.2).Univariate/multivariate independent prognostic analysis of clinical traits showed that the prognostic risk score had an independent prognostic risk effect on rectal adenocarcinomas (P=0.035;Table 4).The heat map,risk score,and scatter plots of survival time with respect to immune-related lncRNAs in rectal adenocarcinomas showed that the higher the risk score,the shorter the survival time and the more the death (Fig.3).

Fig.1 Three prognostic immune-related lncRNAs identified by the multivariate Cox regression.(a) LINC01871;(b) AL158152.2;(c) PXN-AS1 and K-M survival curves of the rectal adenocarcinoma prognostic model (d)
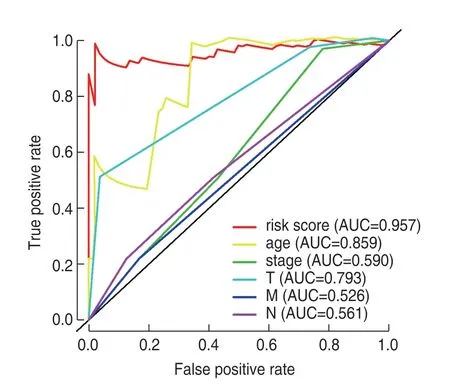
Fig.2 Operating characteristic (ROC) curve of clinical parameters in rectal adenocarcinomas
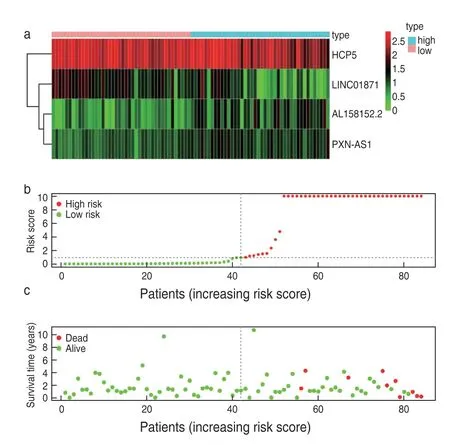
Fig.3 Heat map (a),risk score (b) and scatter plots of survival time (c)of immune-related lncRNAs in rectal adenocarcinoma

Table 4 Univariate and multivariate Cox regression analyses of clinical characters in rectal adenocarcinomas
Correlation between clinical features and key immune-related lncRNAs in the prognosis of rectal adenocarcinomas
Correlation with lymph node staging
We assessed the expression of the four immune-related lncRNAs in different lymph node stages of rectal cancer(AJCC,8th edition) and found that the expression of AL158152.2 was positively correlated with the lymph node stage and the difference was statistically significant(P< 0.05).The expression of HCP5,LINC01871,and PXN-AS1 was not significantly correlated with lymph node staging (P> 0.05;Fig.4a).
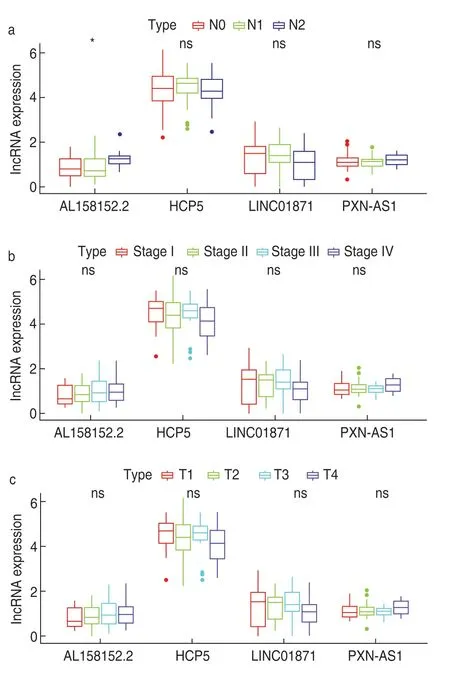
Fig.4 The lymph node stage was positively correlated with the expression of AL158152.2,not the expressions of HCP5,LINC01871 and PXN-AS1 (a),there were no correlations between T stage,clinical stage of rectal adenocarcinomas and 4 key immune-related lncRNAs (b and c)
Correlation with the T stage and clinical stage of rectal adenocarcinomas
The results showed that there was no significant correlation among immune-related lncRNA,clinical stage,and T stage (P> 0.05;Figs.4b and 4c).
Prognostic biomarkers of rectal adenocarcinomas
LINC01871 (P=0.001),PXN-AS1 (P=0.003),and AL158152.2 (P=0.018) were associated with the prognosis in the prognostic model,as per the K-M survival analysis.LINC01871,PXN-AS1,and AL158152.2 may be independent prognostic factors,while LINC01871 may be a protective prognostic factor for rectal adenocarcinomas(Figs.1b-1d).
Discussion
Immune-related lncRNAs are important regulators of gene expression in the immune system and play an important role in the occurrence and development of tumors[4-5,9].Liet alfound that immune-related lncRNAs,which have high tissue specificity,are highly expressed in B cells and T cells[5].Yuet alfound that lncRNAs can be used as biomarkers to mark different stages of cancer immunity to adjust tumor immunity[10].In recent years,increasing evidence has shown that immunerelated lncRNAs can be used as prognostic biomarkers for malignant tumors.At present,a prognostic model based on immune-related lncRNAs has been successfully constructed in a few malignant tumors,including breast cancer,head and neck squamous cell carcinoma,glioma,pancreatic cancer,and renal clear cell carcinoma[11-16].There are many prognostic factors in rectal cancer,among which are common clinical related factors,such as surgical methods and R0 resection[3];however,there are few reports on the prognostic role of immunerelated lncRNAs.In this study,we aimed to construct a prognostic risk model of rectal adenocarcinoma based on immune-related lncRNAs and further analyze and obtain prognostic markers.
In the present study,a prognostic model was constructed by analyzing rectal adenocarcinoma samples from TCGA database.Risk score=(-4.084) * expression LINC01871+(3.112) * expression AL158152.2+(7.616)* expression PXN-AS1+(-0.867) * expression HCP5.It had a high accuracy.LINC01871 is a protective immunerelated lncRNA,while PXN-AS1 and AL158152.2 are harmful prognostic markers.At present,there are few reports on the prognostic immune-related lncRNAs in rectal adenocarcinomas.Taoet alfound that NKILA,an immune-related lncRNA encoded by a gene on chromosome 20q13,was expressed at low levels in various human tumors,such as breast,lung,and rectal cancers.NKILA inhibits proliferation,migration,and invasion of rectal cancer cells by inhibiting NF-κB signaling,which is related to clinical progress and prognosis[17].Zhaoet alobtained a prognostic risk model of rectal cancer consisting of five lncRNAs (AC079789.1,AC106900.2,AL121987.1,AP004609.1,and LINC02163),in which AC106900.2 and LINC02163 are immune-related lncRNAs,but their functions are still unknown[7].Heet alfound that HCP5,EPB41L4A-AS1,SNHG12,and LINC00649 are significantly related to the occurrence and prognosis of colorectal cancer through the competitive endogenous RNA network mediated by lncRNA,among which HCP5 and SNHG12 are immune-related lncRNAs.Studies have shown that SNHG12 can increase the expression of cell cycle-related proteins and inhibit the expression of caspase-3 in colorectal cancer and human osteosarcoma.In addition,silencing the expression of SNHG12 inhibited the proliferation of triple-negative breast cancer cells.It was found that SNHG12,as miR-199a/b-5p,regulates the expression of MLK3 in hepatocellular carcinoma and affects the activation of the NF-κB pathway[18].Therefore,SNHG12 may be a potential biomarker.The lncRNA HLA complex P5 (HCP5) is located at 6P21.33,which is homologous to the retrovirus gene sequence[19].HCP5,which is considered a susceptibility gene site for HCVrelated liver cancer,is downregulated in ovarian cancer,.HCP5 targets miR-139-5p and inhibits the expression of miR-139-5p.The miR-139-5p/ZEB1/Wnt signaling pathway is involved in the occurrence and development of EMT in CRC[20].It was reported that HCP5 is highly expressed in glioma tissues and can promote the proliferation,migration,and invasion of glioma cells,inhibit apoptosis,and promote malignant biological behavior of glioma cells[18].The lncRNA PXN-AS1-L is upregulated in hepatocellular carcinoma,nasopharyngeal carcinoma,lung cancer,and glioma,and promotes tumor occurrence by upregulating PXN[16,18,20-22].In addition,we know nothing about the function and mechanism of LINC01871 and AL158152.2.However,the conclusion of this study was based on TCGA database,and we lacked domestic data to verify the prediction model and markers of immune-related lncRNAs in rectal adenocarcinomas.
In summary,we constructed a prognostic model of rectal adenocarcinomas based on the expression levels of four immune-related lncRNAs (LINC01871,PXN-AS1,HCP5,and AL158152.2) by analyzing TCGA database and immune-related gene sets,which have high prediction accuracy.Two negative prognostic biomarkers (PXNAS1 and AL158152.2) and positive prognostic biomarker(LINC01871) were identified.However,the mechanism of action in rectal adenocarcinomas needs to be further explored.
Conflicts of interest
The authors indicated no potential conflicts of interest.
 Oncology and Translational Medicine2021年3期
Oncology and Translational Medicine2021年3期
- Oncology and Translational Medicine的其它文章
- Antitumor and vascular effects of apatinib combined with chemotherapy in mice with non-small-cell lung cancer
- A study of the potential adverse effects of electrosurgical smoke on medical staff during malignant tumor surgery*
- Diagnostic value of lncRNAs as potential biomarkers for oral squamous cell carcinoma diagnosis:a meta-analysis*
- Recombinant human vascular endostatin injection to synchronize craniospinal radiotherapy for the treatment of recurrent medulloblastoma in children:A retrospective clinical study*
- Seroprevalence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in patients with cancer and the impact of anti-tumor treatment on antibodies*
- Future of targeted therapy for gastrointestinal cancer:Claudin 18.2
