Diagnostic value of lncRNAs as potential biomarkers for oral squamous cell carcinoma diagnosis:a meta-analysis*
Yuxue Wei,Hua Yang,Xiaoqiu Liu (✉)
1 Department of Stomatology,People's Hospital of Lanling County,Linyi 276000,China
2 Department of Stomatology,Jilin University,Changchun 130000,China
Abstract Objective Several studies have revealed the critical role of long non-coding RNAs (lncRNAs) as biomarkers for diagnosing oral squamous cell carcinoma (OSCC).However,the data remain inconsistent.This meta-analysis was performed to summarize the potential of lncRNAs as OSCC biomarkers.Methods We searched PubMed,Cochrane Library,Web of Science,and China National Knowledge Infrastructure databases for literature published until December 10,2020.Study quality was assessed using Quality Assessment for Studies of Diagnostic Accuracy-2,and sensitivity,specificity,and other measures regarding lncRNAs for OSCC diagnosis were pooled using bivariate meta-analysis models.Data analyses were performed using STATA 14.0.Results Overall,8 studies with 981 cases and 585 controls were included in the pooled analysis.The pooled sensitivity,specificity,positive likelihood ratio,negative likelihood ratio,diagnostic odds ratio,and area under the receiver operating characteristic curve values were as follows:0.76 [95% confidence interval (CI),0.65-0.84],0.90 (95% CI,0.82-0.95),7.5 (95% CI,4.20-13.40),0.27 (95% CI,0.18-0.39),28 (95% CI,13.00-58.00),0.90 (95% CI,0.87-0.93),respectively.Deeks' funnel plot asymmetry test (P =0.56) indicated no potential publication bias.Conclusion Our meta-analytical evidence suggests that lncRNAs could be employed as a potential noninvasive diagnostic tool for OSCC.
Key words: lncRNA;oral squamous cell carcinoma;diagnosis;meta-analysis
Oral squamous cell carcinoma (OSCC) is the eighth most common cancer worldwide[1-2].OSCC accounts for 95% of all oral malignancies,with a global five-year survival rate that ranges between 50% and 60%,especially in patients diagnosed at advanced stages[3-4].Most clinical examinations and imaging techniques currently used to detect OSCC fail to reliably distinguish early OSCC[5-6].Therefore,early diagnosis and monitoring of OSCC progression remain crucial.
Long non-coding RNAs (lncRNAs) are novel noncoding RNAs with a length of more than 200 nucleotides,which do not encode proteins[7-8].LncRNAs are characterized by low expression,moderate sequence conservation,and high tissue specificity and play an important role in several fundamental biological processes,including transcriptional regulation,cell differentiation,and chromatin modification[9].Notably,growing evidence implicates a close relationship between tumor occurrence and development[10-13].Additionally,dysregulated lncRNAs promote the occurrence and metastasis of malignancies[14-15].Reportedly,altered lncRNA expression can be detected in biological fluids in different cancer types[16-17],including OSCC[18-19];however,no consensus has been reached regarding the clinical value of lncRNAs in OSCC diagnosis.Therefore,we conducted a bivariate meta-analysis to summarize the diagnostic accuracy of lncRNAs for detecting OSCC.
Materials and methods
Literature search strategy and selection criteria
Two investigators searched PubMed,the Cochrane Library,Web of Science,and China National Knowledge Infrastructure (CNKI) databases for literature published until December 10,2020,to identify relevant studies by employing the following terms:“lncRNAs”or“long ncRNAs”or“lincRNAs”or“long non-coding RNAs”or“long non-translated RNAs”or“long untranslated RNAs”or“long non-protein-coding RNAs”or“long intergenic non-protein coding RNAs”or“diagnostic value”or“diagnoses”or“receiver operating characteristics”or“ROC curve”or“sensitivity”or“specificity”and“OSCC”or“oral squamous cell carcinoma”or“oral cancer”or“oral tumor.”Two investigators (WYX and LXQ) assessed study titles and abstracts and scanned full texts to eliminate irrelevant studies,using the following inclusion criteria:(1) Diagnostic data,including sensitivity,specificity,and the area under the receiver operating characteristic curve (AUC),were provided or could be calculated from the studies;(2) Studies included patients with OSCC and healthy controls;(3) Original research articles published in English or Chinese.
Data extraction and quality assessment
Two investigators (WYX,LXQ) independently reviewed and extracted the following information from each eligible study:first author,year,country,sample size,gender,mean age,tumor-node-metastasis (TNM)stage,type of sample,detection method,cut-off value,true positive (TP),false positive (FP),true negative (TN),and false negative (FN).The quality of included studies was assessed using Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2).
Statistical analysis
Statistical analyses were conducted using Stata 14.0(Stata,College Station,TX,USA).The number of subjects with a sensitivity,specificity,diagnostic odds ratio (DOR),positive likelihood ratio (PLR),negative likelihood ratio(NLR),and their corresponding 95% confidence intervals(CIs) was calculated using a bivariate meta-analysis model.Then,summary receiver operator characteristic (SROC)curve analysis and AUC were calculated to evaluate the pooled diagnostic value of lncRNAs in OSCC detection.These data were confirmed using a hierarchical summary receiver operating characteristic (HSROC) model.The heterogeneity of eligible studies induced by the threshold effect was evaluated using Spearman correlation coefficients and ROC plane analyses.Heterogeneity of non-threshold effects was tested using Cochran's Q and inconsistency index (I2) tests.For the Q test,P< 0.05 orI2> 50% indicated significant heterogeneity among identified studies.Moreover,Fagan's nomogram was used to assess relationships between prior-test probability,likelihood ratio,and post-test probability.Publication bias was assessed using Deeks' funnel plots.
Results
Study selection and characteristics of included studies
As shown in the flowchart (Fig.1),a total of eight eligible studies[20-27](981 OSCC and 585 healthy controls)evaluating 20 different lncRNAs were included in the present meta-analysis.The expression of lncRNAs was detected by reverse transcription-polymerase chain reaction (PCR) (n=7) and TaqMan quantitative PCR (n=1),while plasma (n=4) or serum (n=4) was the most common specimen assessed;in the TNM classification,four articles included patients in stages I-IV and one article included patients in stages I-III,whereas the other three failed to mention the TNM stage.According to single lncRNA expression levels,eight lncRNAs were upregulated,and data for others were unavailable.Details regarding study participants are presented in Table 1.
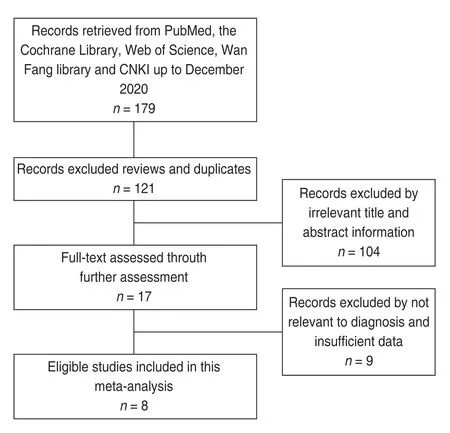
Fig.1 Flow diagram of the study selection process
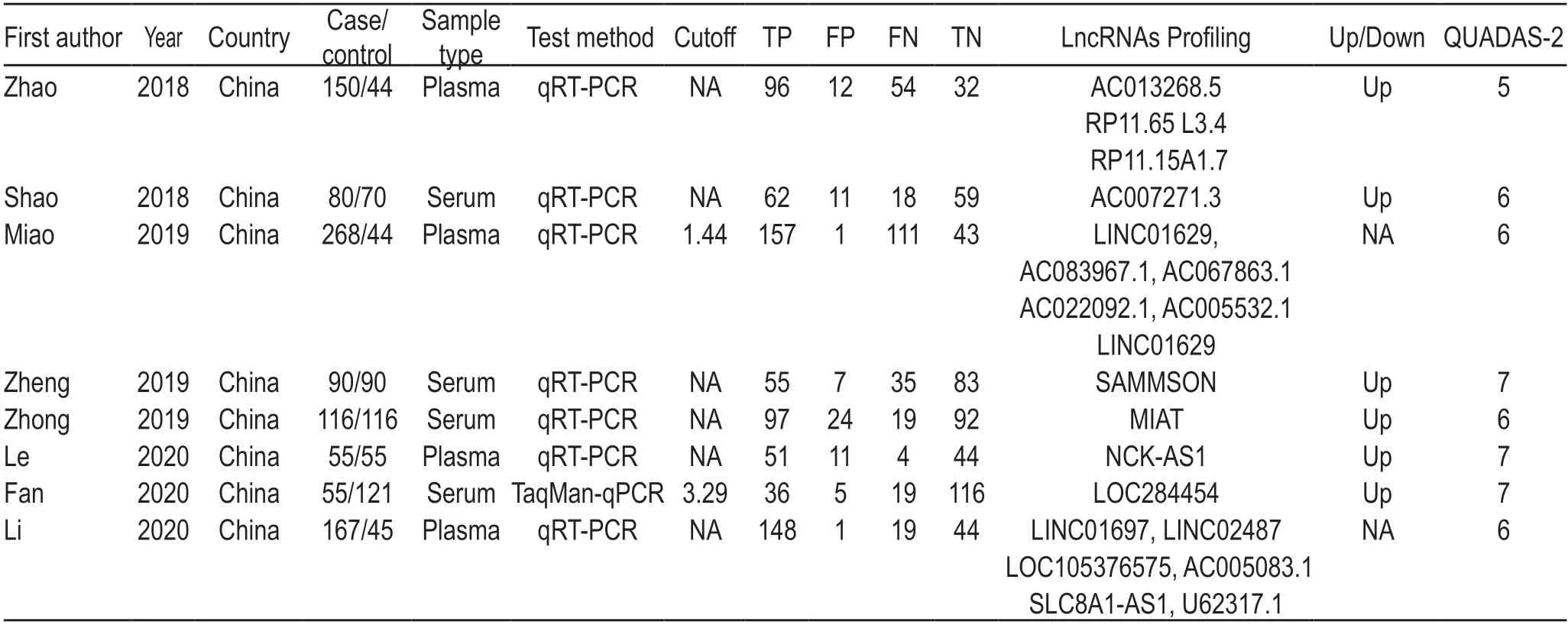
Table 1 Summary of main characteristics of included studies in this meta-analysis
Quality assessment
In the present study,the methodological quality of diagnosis was assessed using QUADAS-2.All included studies presented relatively moderate-to-high quality,with QUADAS-2 scores ranging between 5 and 7 (Table 1);this indicated the reliable foundation of our metaanalysis.
Data analysis
For the eight included studies,forest plots of lncRNA sensitivity and specificity data for diagnosing OSCC are shown in Fig.2.The pooled sensitivity and specificity of lncRNAs for diagnosing OSCC were 0.76 (95% CI,0.65-0.84) and 0.90 (95% CI,0.82-0.95),respectively.Additionally,the pooled PLR was 7.5 (95% CI,4.20-13.40),NLR was 0.27 (95% CI,0.18-0.39),and DOR was 28 (95% CI,13.00-58.00) (Figs.3 and 4).The overall SROC curve for the eight included studies is shown in Fig.5.The AUC of lncRNAs was 0.90 (95% CI,0.87-0.93),which indicates that the lncRNAs were highly accurate in differentiating patients with OSCC from controls.Next,to assess the clinical utility of the index test,Fagan's nomogram was used to predict the increasing inerrability of a positive diagnosis by using the test value,which was employed for estimating post-test probabilities.As presented in Fig.6,the pre-test probability was 20% and the positive post-test probability was increased to 65%,while the negative post-test probability was decreased to 6%.All pooled estimates indicated that lncRNAs in this analysis presented a relatively moderate-to-high accuracy in distinguishing patients with OSCC from control individuals.
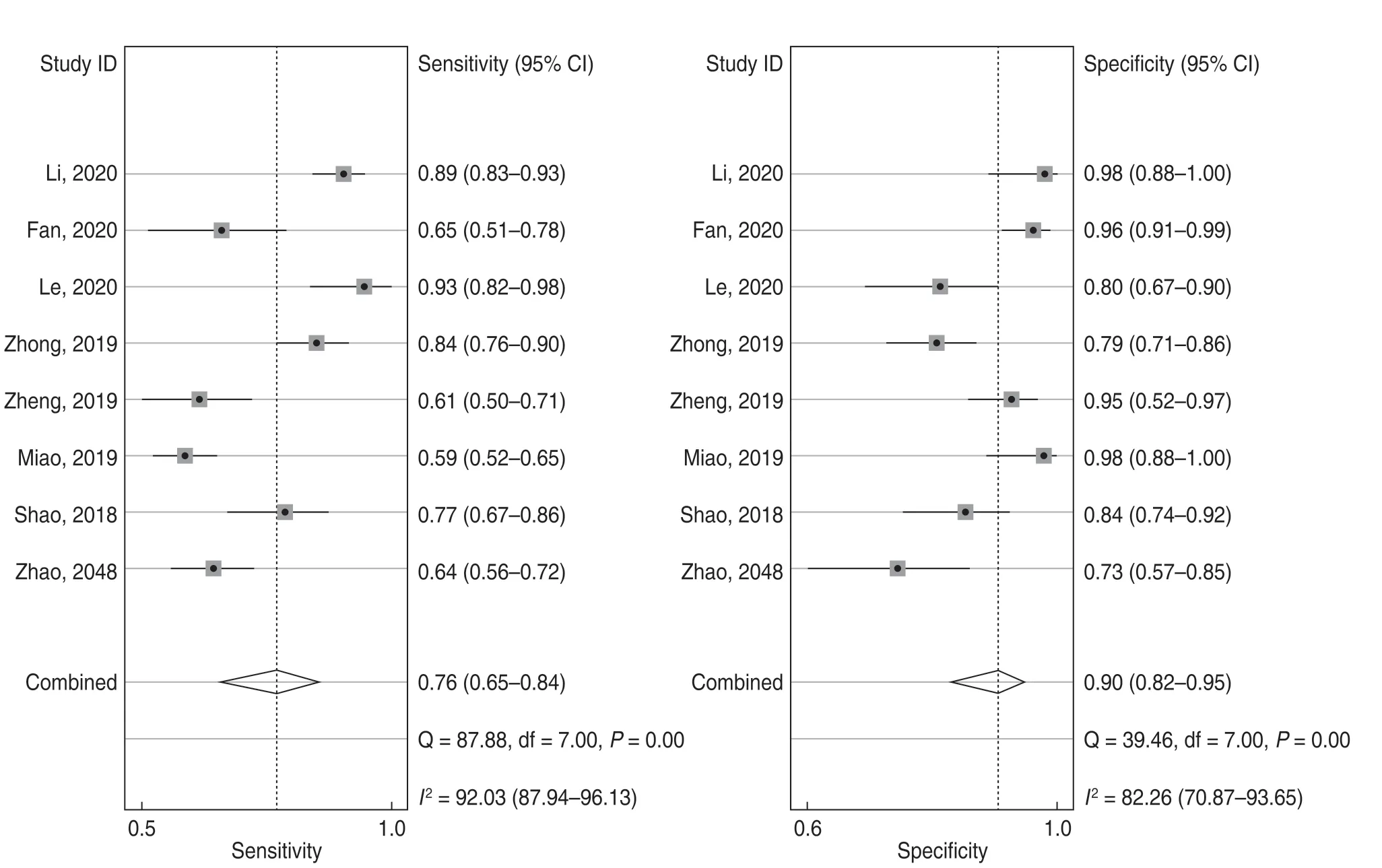
Fig.2 Forest plots of lncRNA sensitivity and specificity for the diagnosis of OSCC
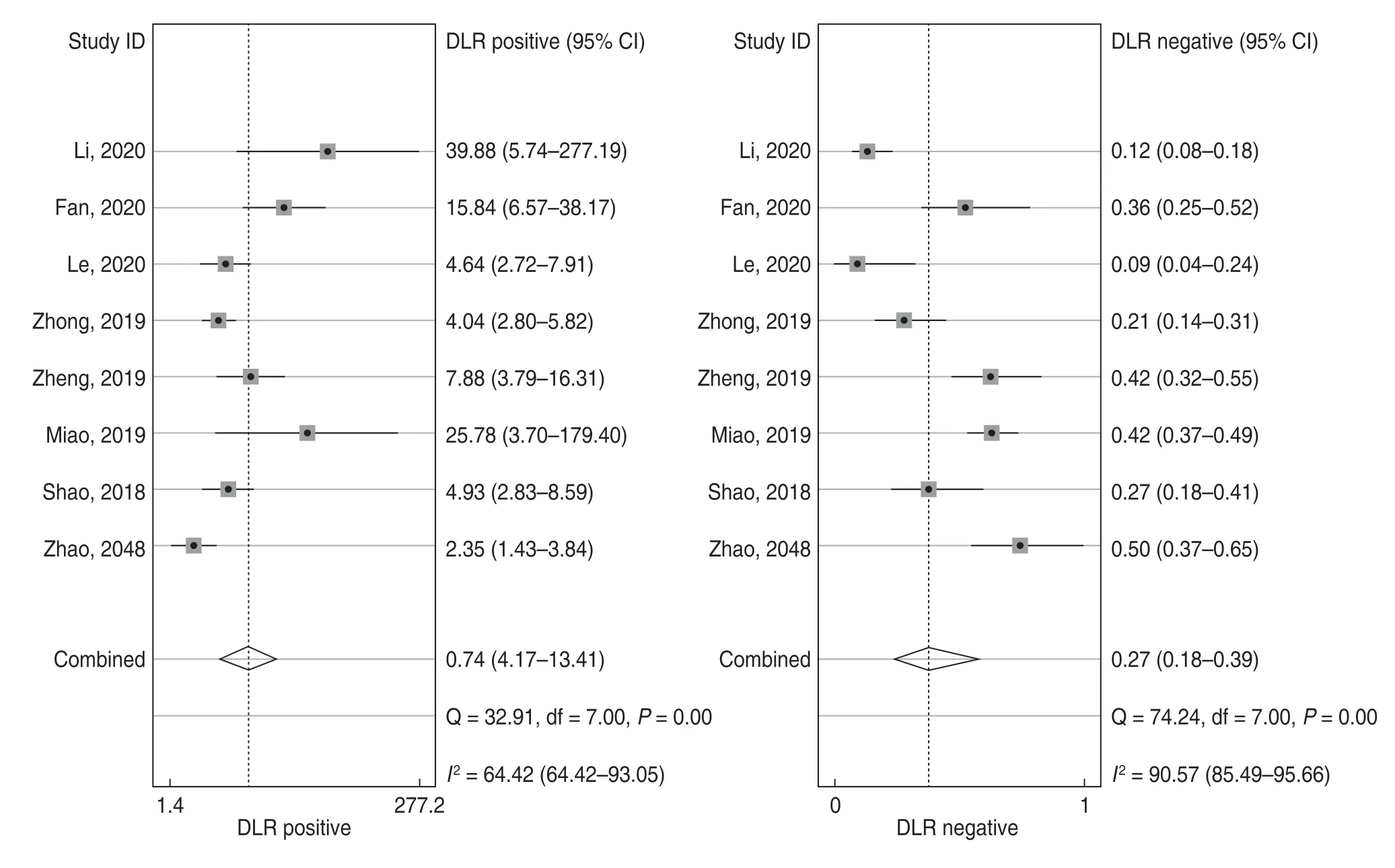
Fig.3 Forest plots of the positive likelihood ratio (PLR) and negative likelihood ratio (NLR) of lncRNAs for the diagnosis of OSCC
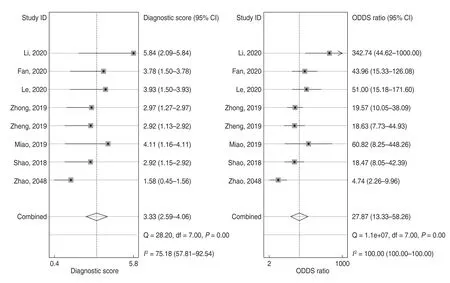
Fig.4 Forest plots of diagnostic odds ratio (DOR) of lncRNAs for the diagnosis of OSCC
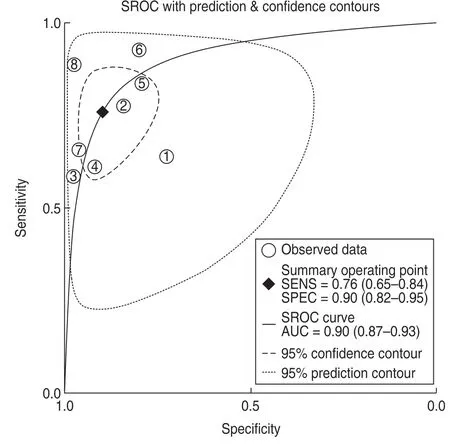
Fig.5 Summary receiver operating characteristic (SROC) curve with pooled estimates of sensitivity,specificity,and area under the ROC curve(AUC)
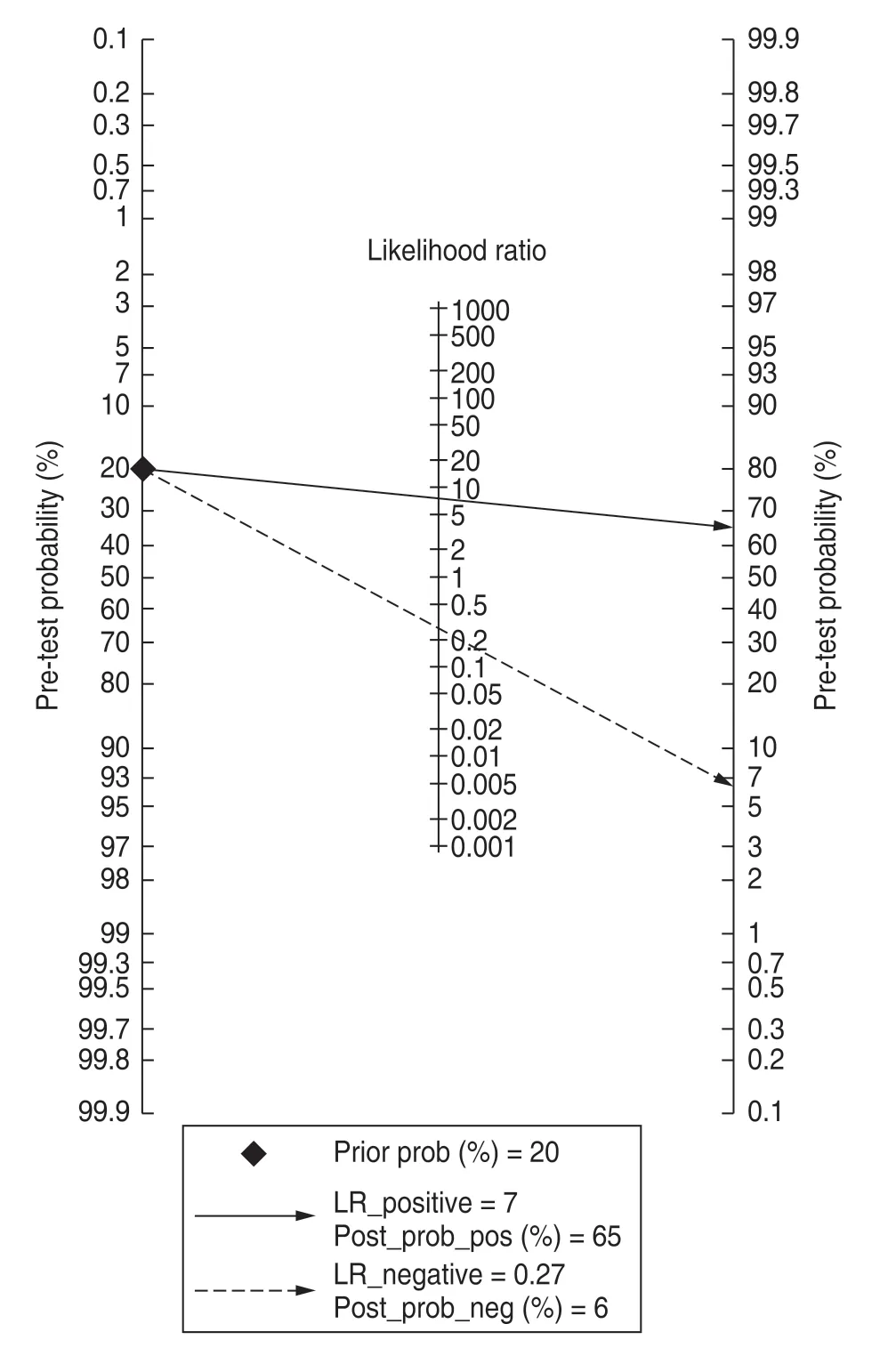
Fig.6 Fagan's nomogram assessing the post-test probabilities
Heterogeneity and publication bias
TheI2value of the heterogeneity test was 95.95%,indicating apparent heterogeneity.Nevertheless,only eight articles were included,and subgroup and metaregression analyses could not be undertaken in this meta-analysis.Moreover,we selected Deeks' funnel plot asymmetry test to assess publication bias (Fig.7),which suggested no significant publication bias (P=0.56).
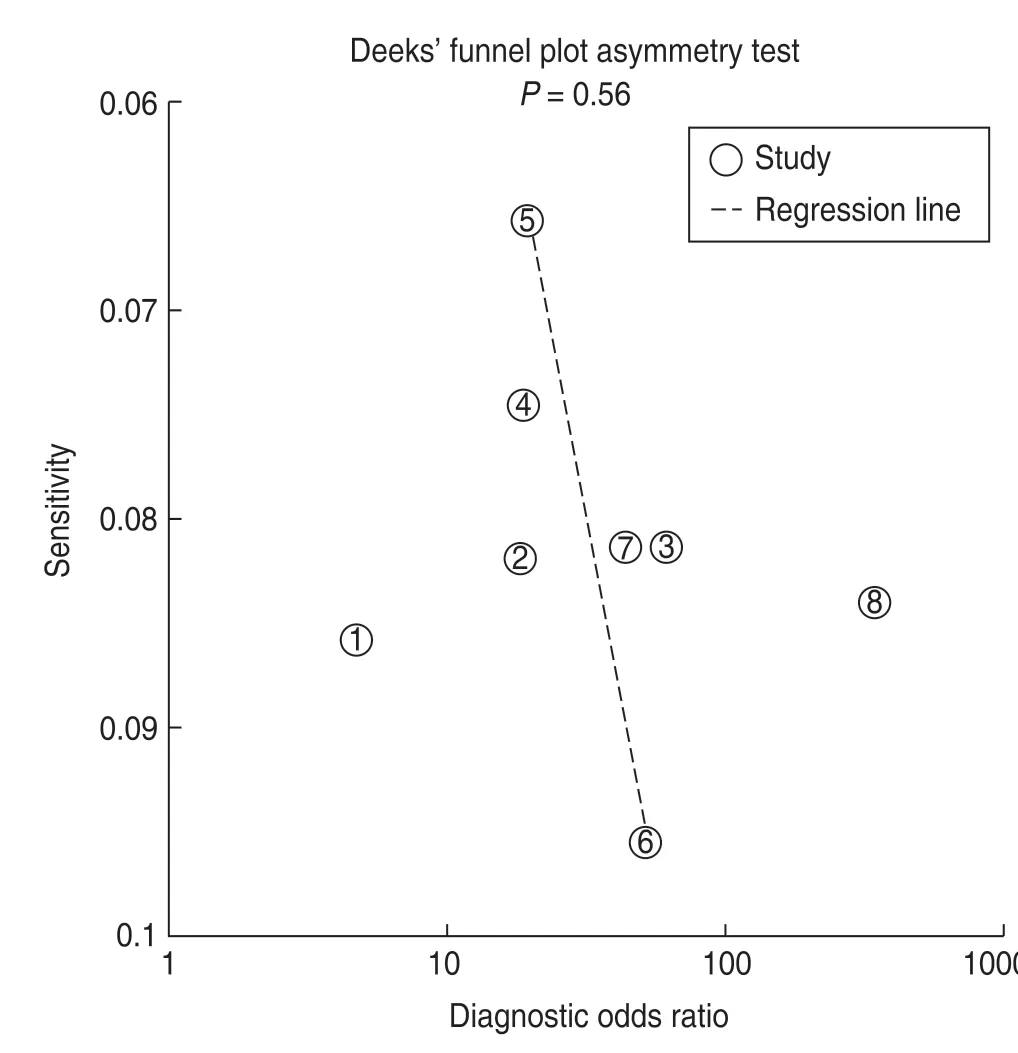
Fig.7 Graph of Deeks' funnel plot asymmetry test
Discussion
In OSCC,the initial stage is typically asymptomatic;therefore,early diagnosis remains a priority to improve survival.Most patients with OSCC are diagnosed at late stages,resulting in a poor prognosis[19,28].Therefore,to optimize the survival rate and quality of life of patients with OSCC,it is essential to explore novel,practical,and non-invasive biomarkers for early detection of OSCC.
LncRNAs exert significant effects on the occurrence and development of several human diseases.Accumulating evidence suggests that altered lncRNA expression and its mutations can promote tumor occurrence and metastasis[14-15].Furthermore,changes in lncRNA expression can be significantly associated with tumorigenesis,angiogenesis,and cell function[29].Dysregulation of lncRNAs is reportedly involved in the occurrence,metastasis,and prognosis of cancer[8,30].Moreover,given their role in cancer biology and easy detection in biological fluids,lncRNAs could be potential and predictive cancer biomarkers.Circulating lncRNAs are promising new cancer biomarkers based on their role in cancer biology,and previous studies have demonstrated their feasibility as diagnostic and prognostic tools for different types of malignant tumors[30-32].Tumor-related lncRNAs can be detected in various biological fluids,including blood[33-34].
The present review is the first evidence-based analysis to assess the diagnostic role of lncRNAs in OSCC detection.This meta-analysis included eight studies with 981 cancer cases and 585 controls,indicating that lncRNAs possess relatively high diagnostic accuracy with an overall pooled sensitivity of 0.76 (95% CI,0.65-0.84),specificity of 0.90 (95% CI=0.87-0.95),and AUC of 0.90(95% CI,0.87-0.93).Furthermore,the pooled DOR of 28 (95% CI,13.00-58.00) suggested that lncRNAs are reliable for detecting OSCC.
Additionally,PLR and NLR were used to evaluate diagnostic accuracy.In this analysis,the pooled PLR was 7.5 (95% CI,4.20-13.40),and the NLR was 0.27 (95%CI,0.18-0.38);this could be attributed to the fact that patients with OSCC have a 7.5-fold higher possibility of reporting lncRNAs positive for OSCC when compared with controls,and 27% of all individuals present a negative result.As shown in Fagan's nomogram,at a pretest probability of 20%,the positive post-test probability would increase up to 65% with a PLR of 7,while the negative post-test probability would decline to 6% with an NLR of 0.27.These results indicate the potential value of lncRNAs in OSCC detection and diagnosis.
Notably,significant heterogeneity existed among included studies in the present meta-analysis.The ROC plane showed no“shoulder arm”pattern,suggesting that the threshold effect was not a major source of heterogeneity (Fig.8).Moreover,the Spearman correlation coefficient was -0.30 withP=0.09,indicating the absence of a threshold effect.However,we could not conduct meta-regression and subgroup analyses to detect the cause of heterogeneity,owing to a lack of eligible studies.Therefore,potential factors including age distribution,TNM stage,and gender proportion were not assessed as sources of heterogeneity.
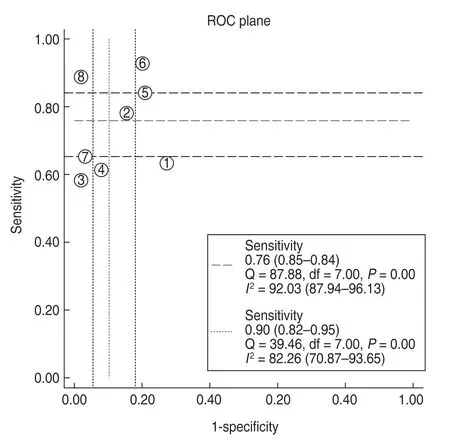
Fig.8 Receiver operating characteristics (ROC) space to assess the threshold effect of lncRNAs
However,the limitations of this study should be noted.First,our meta-analysis included a small sample size of lncRNAs in patients with OSCC.Second,the cutoff values for lncRNAs differed among included studies,which could be a potential source of heterogeneity.Third,this meta-analysis included a high proportion of Chinese populations,which may introduce unavoidable bias.Additionally,we did not determine differences in the diagnostic accuracy of lncRNAs presented in OSCCs,with respect to age distribution,TNM stage,and gender proportion.Finally,we identified the pooled lncRNA diagnostic value for OSCC but did not explore specific lncRNAs due to insufficient publications.
In conclusion,the results of this meta-analysis suggest that lncRNAs have a moderate-to-high diagnostic value in distinguishing patients with oral cancer from healthy controls,suggesting that lncRNAs can be utilized with high accuracy,particularly when performing multiplelncRNA assays.However,large-scale prospective trials,as well as different ethnic groups,are necessary to confirm and extend our findings.
Conflicts of interest
The authors indicated no potential conflicts of interest.
 Oncology and Translational Medicine2021年3期
Oncology and Translational Medicine2021年3期
- Oncology and Translational Medicine的其它文章
- Construction and validation of an immune-related lncRNA prognostic model for rectal adenocarcinomas*
- Antitumor and vascular effects of apatinib combined with chemotherapy in mice with non-small-cell lung cancer
- A study of the potential adverse effects of electrosurgical smoke on medical staff during malignant tumor surgery*
- Recombinant human vascular endostatin injection to synchronize craniospinal radiotherapy for the treatment of recurrent medulloblastoma in children:A retrospective clinical study*
- Seroprevalence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in patients with cancer and the impact of anti-tumor treatment on antibodies*
- Future of targeted therapy for gastrointestinal cancer:Claudin 18.2
