Association of eleven single nucleotide polymorphisms with refractive disorders from Eskisehir, Turkey
Nadir Unlu, Ebru Erzurumluoglu Gokalp, Serap Arslan, Oguz Cilingir, Muzaffer Bilgin,Engin Yildirim, Huseyin Gursoy
1Department of Ophthalmology, Eskisehir Osmangazi University Medical Faculty, Eskisehir 26040, Turkey
2Department of Medical Genetics, Eskisehir Osmangazi University Medical Faculty, Eskisehir 26040, Turkey
3Department of Biostatistics, Eskisehir Osmangazi University Medical Faculty, Eskisehir 26040, Turkey
4Department of Pharmacology, Eskisehir Osmangazi University Medical Faculty, Eskisehir 26040, Turkey
Abstract
● KEYWORDS: refractive disorders; myopia; hyperopia;genetics; single nucleotide polymorphisms; Turkey
INTRODUCTION
Refractive errors, especially myopia is currently a global epidemic and uncorrected refractive errors are the second leading cause of blindness. Refractive errors, namely myopia, hyperopia, and astigmatism, are multifactorial ocular disorders. Their etiology is not completely understood[1].There are many environmental factors, which are responsible for myopia increase in the world. Intensive near work activity is one of the major factors defined to be related with myopia. Limited outdoor activity is the other important factor associated with myopia. Sports activities, socioeconomic status, education levels, premature birth are other risk factors claimed to be related with myopia.
There are many studies on the genetic contributions to myopic refractive error. There are more than 25 genes associated with myopia[2].MYP1, MYP2, MYP3, MYP4, MYP5, PAX6, TGIF,LUM, ARR3(arrestin 3),LRPAP1(LDL receptor related protein associated protein 1),P4HA2(prolyl 4‐hydroxylase subunit alpha 2),SLC39A5(solute carrier family 39 member 5) are the most important genes defined as candidate genes for myopia[3‐4]. Studies showed that myopia does not have a single genetic cause, but many susceptibility genes exist[5]. Results of genome‐wide association studies (GWAS) meta‐analysis support that refractive errors are caused by a light‐dependent retina‐to‐sclera signaling cascade[6]. It has also been shown that genetic factors associated with circadian rhythm and pigmentation play a role in the development of myopia and refractive error[7].
On the other hand, there is a paucity of literature on the genetic basis of hyperopia. There are a few studies which demonstrated the relation betweenMFRP(membrane frizzled‐related protein) andPRSS56(serine protease 56) genes and hyperopia[4]. The association of hepatocyte growth factor(HGF),MFN1(mitofusin 1) andVDR(vitamin D receptor)genes with myopia has been investigated in a few studies.HGFgene has been reported to be associated with low, moderate and high myopia, whileVDRandMFN1genes have been reported to be associated with low and moderate myopia[8‐10].To the best of our knowledge, there are no published studies on the genetic basis of refractive errors in Turkey. Our aim was to investigate the association of five different genes (HGF, GC,MFN1, GNB4,andVDR) with refractive errors including both myopia and hyperopia in Turkey.
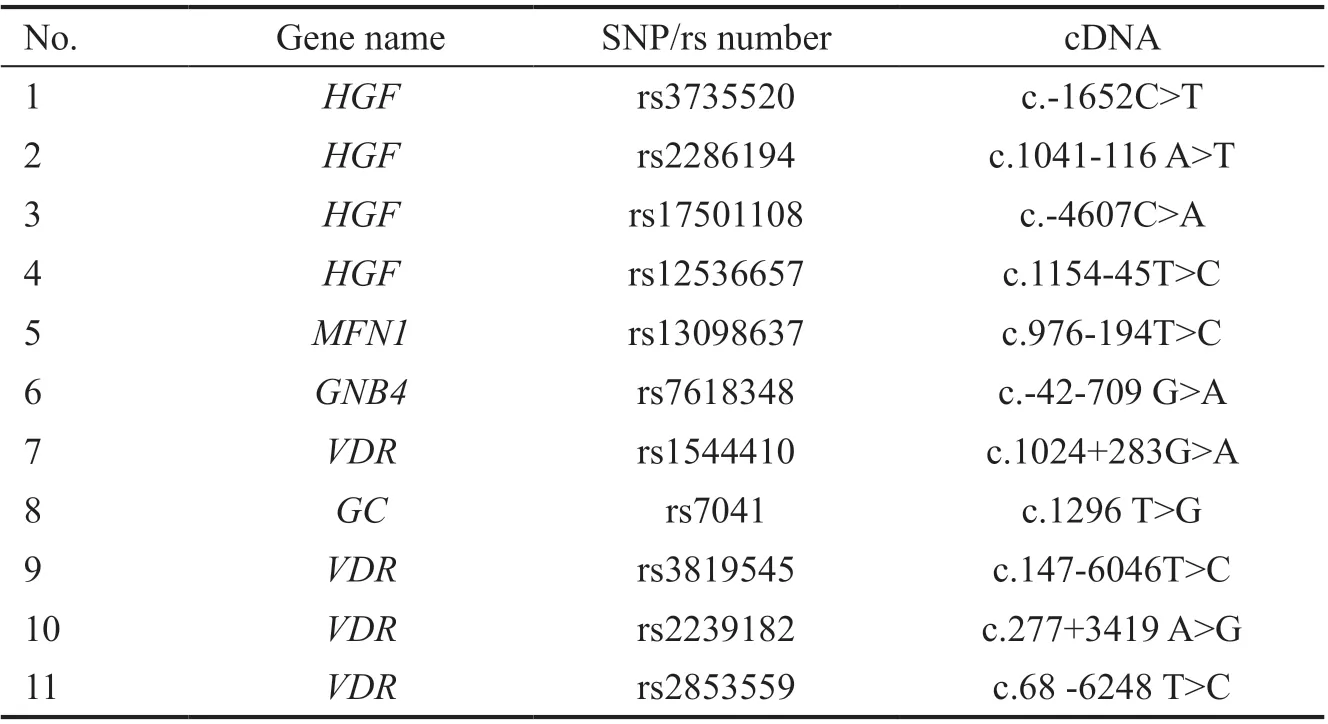
Table 1 SNPs investigated in this study
SUBJECTS AND METHODS
Ethical ApprovalThis study was conducted according to the Declaration of Helsinki guidelines and approved by the Clinical Practice Ethics Committee of Eskisehir Osmangazi University, Medical Faculty. All subjects were agreed to participate in the study and biological samples were obtained after written informed consent obtained.
ParticipantsThe effect size assumption was used to determine the preliminary sample size. Number of cases was calculated as 133. A total of 212 participants diagnosed with 91 myopia, 45 hyperopia, and 76 emmetropia by ophthalmologist at the Ophthalmology Department of Eskisehir Osmangazi University were enrolled in this study.
The autorefractometer measurements were performed under cycloplegia. Spherical equivalent (SE) was calculated by adding the spherical value and half of the cylindrical value. SE<‐0.50 diopters (D) in both eyes was considered to be myopic, while SE>0.75 D in both eyes was considered to be hypermetropic. The inclusion criteria for this study were as follows: 1) patients aged between 18 and 40; 2) 3 measurements obtained under cycloplegia. Patients meeting any of the following criteria were excluded: 1) corneal diseases; 2) lenticular diseases; 3) genetic disorders associated with myopia such as Marfan syndrome and retinitis pigmentosa; 4) previous intraocular surgical intervention; 5) nystagmus; 6) ocular trauma; or 7) patients born preterm.
Single-Nucleotide Polymorphisms GenotypingWe have selected eleven single‐nucleotide polymorphisms (SNPs;Table 1) that are associated with myopia and hyperopia or have conflicting suggestions about these SNPs in the literature.We genotyped all participants for these SNPs. Genomic DNA was isolated from peripheral lymphocytes using Magna Pure Compact LC (Roche Applied Science, Basel, Switzerland),according to the manufacturer’s recommendations.
SNPs inHGF, GC, MFN1, GNB4andVDRgenes were studied by SnapShot technique. SnapShot reactions were performed as recommended by the manufacturer. Electrophoresis of amplified PCR samples related to these polymorphism regions was performed on ABI 3130 Genetic Analyzer and the data were analyzed by using GeneMapper 4.0 Software (Applied Biosystems, Life Technologies, CA, USA).
Statistical AnalysisStatistical analysis was performed using the Statistical Package of the Social Sciences 21.0 (IBM,NY, USA). In statistical analysis of the study, continuous data are given as mean±standard deviation. Categorical data are given as percentages (%). Pearson Chi‐square analysis was used in the analysis of the created cross tables. Binary Logistic regression analysis was used to determine risk factors.Bonferroni correction adjustment was used for multiple comparison analysis statistically significant evidence of association was determined byP‐values of 0.05 or below.
RESULTS
Demographic Features of the Study GroupThe patients in this study consists of 47 female/44 male (age: 23.47±4.30)y patients with myopia, 20 female/25 male (age: 31.20±8.02)y with hyperopia and 33 female/43 male (age: 25.22±6.60)y with emmetropia.
Genotype Results
SNP analysis results in myopia and emmetropia groupsGenotypes were wild in each of cases and controls for rs2286194 polymorphism. A statistically significant difference was not observed between myopia and emmetropia groupfor rs17501108, rs2853559, rs1544410, rs13098637, rs7041,rs3735520, rs2239182, rs12536657 SNPs in terms of genotype and distribution of the allele frequencies. The genotype distribution of the rs7618348 polymorphism, which was the only statistically significant one between myopia and emmetropia group, was shown in Table 2.
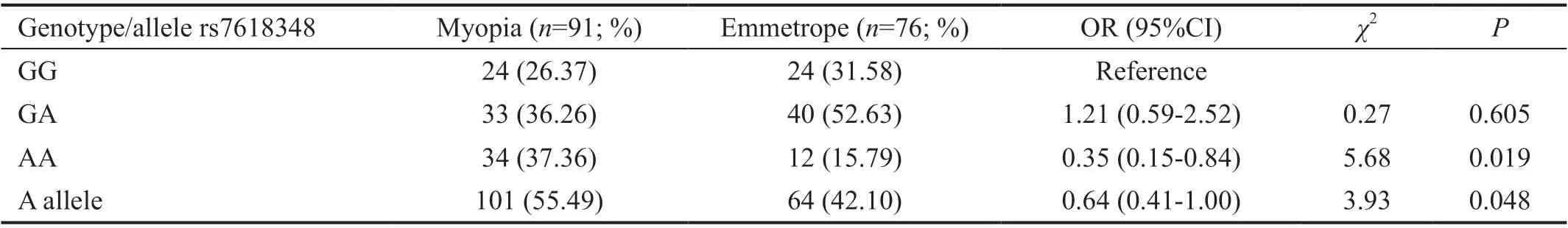
Table 2 Distribution of genotype and risk allele frequency of rs7618348 (GNB4 gene) polymorphism between myopia and emmetropia groups
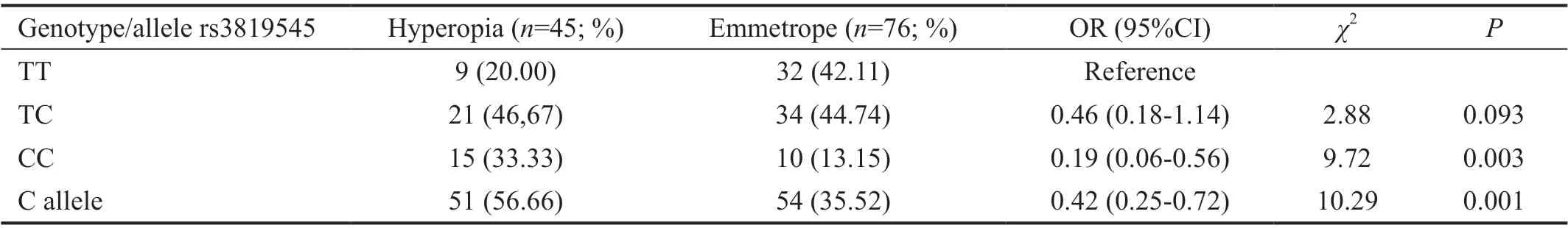
Table 3 Distribution of genotype and risk allele frequency of rs3819545 (VDR gene) polymorphism between hyperopia and emmetropia groups
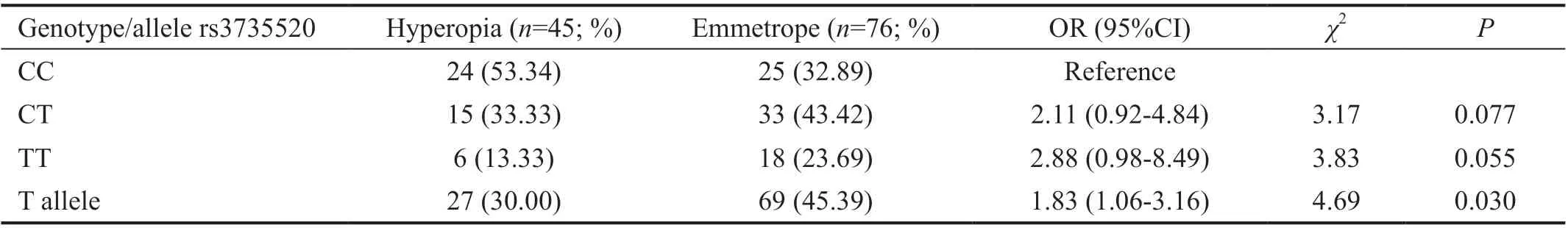
Table 4 Distribution of genotype and risk allele frequency of rs3735520 (HGF gene) polymorphism between hyperopia and emmetropia groups
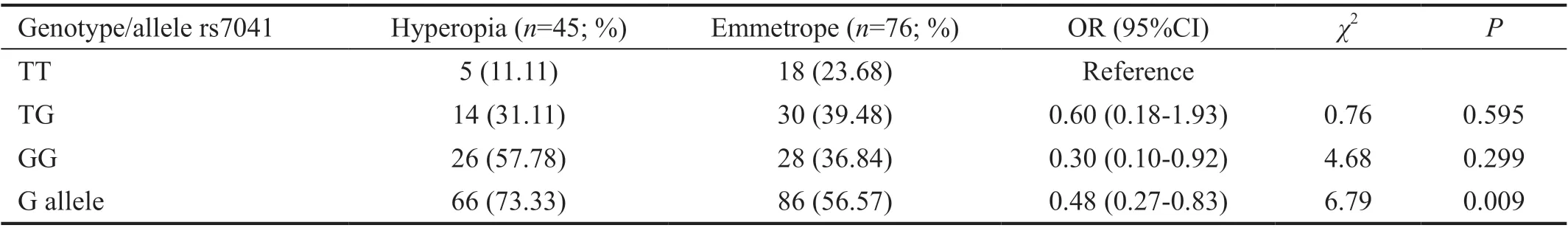
Table 5 Distribution of genotype and risk allele frequency of rs7041 (GC gene) polymorphism between hyperopia and emmetropia groups

Table 6 Distribution of genotype and risk allele frequency of rs2239182 (VDR gene) polymorphism between hyperopia and emmetropia groups
SNP analysis results in hyperopia and emmetropia groupsThere was no statistically significant difference between hyperopia and emmetropia groups for rs17501108, rs2853559,rs1544410, rs13098637, rs7041, rs3735520, rs2239182,rs12536657, rs3819545 in terms of genotype and distribution of the allele frequencies (P>0.05). The genotype distribution of the rs3819545, rs3735520, rs7041, and rs2239182 polymorphisms, which were statistically significant between hyperopia and emmetropia groups, was shown in Tables 3‐6,respectively.
DISCUSSION
rs7618348 inGNB4gene might be a risk factor for myopia.rs2239182 and rs3819545 inVDR, rs7041 in GC polymorphisms were statistically significant between the hyperopic cases and controls. Furthermore, T allele in rs3735520HGFmight be a slightly protective for the development of axial hyperopia.
Recently, more than 100 genes and 20 loci appear to be involved in refraction errors were identified with the development of next‐generation sequencing technologies,GWAS, candidate gene studies, and linkage analyses[3].
HGFgene encodes a protein binding toHGFreceptor to be involved in regulating cell growth, mobility, and morphogenesis. This protein actually has many distinct roles in like regulation of an inflammation, angiogenesis,tumorigenesis, renewing of tissues, and inhibition of apoptosis.It was first identified as a strong candidate gene in a locus related to eye weight[11].HGFand its receptor have been reported to be extensively expressed in the eye, therefore some SNPs inHGFgene play a critical role together in many ocular physiological and pathological processes. In addition, a family‐based linkage analysis studied in Chinese population found a potential locus at whereHGFis located associated with high/severe myopia[12].
Yanovitchet al[10]found a statistically significant difference in the mild‐moderate myopia group in 649 Caucasian patients compared to emmetropes, which also assert a close association between the mild‐moderate myopia and rs3735520, SNP inHGF. They also reported that the T allele inHGFrs3735520 SNP region is a risk allele for myopia in the same study.Results from this study indicate tag SNPs that are rs2286194,rs373552, and rs17501108, also called as haplotypes and rs12536657, rs2286194 in theHGFgene are associated with myopia. Vitreous chamber depth is measured longer for rs3735520 SNP in Chinese population[13]. Burdonet al[14]have found thatHGFrs3735520 SNP were associated with patients with keratoconus that this SNP was also associated withHGFserum level in genome association study. However, there is no relationship found between rs3735520 polymorphism and high myopia in a study conducted in Chinese population consisting of 1052 high myopia and 1070 controls[15]. To our knowledge, there are not any population‐based study for rs3735520 polymorphism in refractive errors except the Chinese population. On the other hand, in a study conducted in the Australian population, an association of rs2286194 with keratoconus was reported[16]. In our study data, no statistically significant difference was observed between myopia and emmetrope groups for rs3735520, rs2286194, and rs17501108 SNPs inHGF, but there was a relationship between rs3735520 and hyperopia. Although many studies reveal that rs3735520 polymorphism is associated with myopia, our data failed to support this relationship in myopia group. This risk allele was significantly higher in emmetrope group compared to hyperopia. We presumed that this allele might be a slightly protective allele for the development of axial hyperopia. These data should be supported by studies within larger cohorts.Moreover, there is a study conducted in Australian population in order to determine the effect ofHGFpolymorphisms on fracture defects, which shows that rs12536657 is associated with both mild‐moderate myopia and hyperopia[17]. In another study of 403 Spanish‐Caucasian hyperopic cases, there is no relationship found between rs12536657 and hyperopia[18].In our study, no significant difference was found between the rs12536657 SNP and both myopia and hyperopia cases compared to emmetropes. Our study supports the results obtained from the data of Barrio‐Barrioet al’s research[18], but it contradicts the findings of Veerappanet al’s[17]study, which is most probably due to population difference.
TheMFN1(mitofusin 1) gene encodes the mitochondrial membrane protein and is involved in mitochondrial mobility along with other intramembrane proteins. Considering that the eye is an organ with high energy demand, any defect in mitochondrial function is thought to lead to refractive errors[9].Only one study, which is conducted in the Chinese population,investigating the effect of rs13098637 polymorphism on refractive errors was found. In this study, an association of rs13098637 polymorphism with mild‐to‐moderate myopia was revealed. However, no significant relationship was observed in the hyperopia and myopia groups in our study.
SNPs of theGNB4gene may be associated with myopia in retinal ON‐bipolar cells since the retinal bipolar cells are involved in taking hyperopic images[19]. As a result of genome‐wide linkage studies related to myopia in dizygotic twins,Hammondet al[20]proposed that the 3q26 region could be a myopia locus for the first time in addition to the other loci they identified. In two mapping studies, including a replication study ofGNB4rs7618348 SNP, this SNP was found to be highly related to myopia, therefore they suggested that theGNB4gene may have a role in the molecular genetic etiology of myopia[19].On the other hand, no statistically significant relationship was found between myopia and rs7618348 inGNB4gene in a controlled, myopia‐related SNP study[9]. Statistical outcomes of our study givePvalue for the homozygous rs7618348 in theGNB4as 0.019, while thePvalue for the single allele change is 0.048. In particular, homozygous AA genotype has been shown to be associated with myopia in our study(P=0.019). Considering the frequency distribution of the A allele, a significant difference was found between myopia and emmetrope groups and was concluded as a risk allele.However, in order to determine its exact role and examine its relationship with myopia, it necessitates big population studies besides functional ones.
Since the vitamin‐D binds to the vitamin D‐binding protein,most of it does not circulate freely in the plasma. Thus, theGCgene, which encodes this binding protein, has been the subject of epidemiological and genetic studies in which vitamin D levels can be associated with myopia. There are few studies aimed to establish the link between SNPs in theGCgene and myopia phenotype. In one of these studies,the relationship of rs7041 variant with myopia could not be established[8]. In addition, in another Mendelian randomized study to determine the relation of low vitamin‐D level to myopic refractive defects, SNPs known to affect vitamin D level were investigated and the study group reported thatGCrs7041 polymorphism was not associated with myopia[21]. Our results support these two studies. However, as an interesting thing in our findings, the G allele was found to be significantly different between the hyperopic and emmetropic groups. In the literature, no study has been found to examine the relationship between rs7041 SNP and hyperopia. With the replication of our study data, more enlightened relationship might be established between rs7041 and hyperopia.
Many SNP regions onVDRhave been associated with many different phenotypes as it encodes a ligand‐induced transcription factor that plays a role in the transcriptional regulation of vitamin D[21]. Muttiet al[8]suggested that there was no relationship between vitamin D levels in the blood and myopia first, but then although there is no strong experimental evidence, it has been reported that there may be a relationship betweenVDRSNPs and refractive defects. In the Meta‐analysis conducted by Tanget al’s[22]study group, no association was found with myopia as a result of two‐step statistical correction forVDRSNPs rs3819545, rs2239182, and rs2853559. In our study, none of the four SNPs examined forVDR(rs1544410,rs3819545, rs2239182, rs2853559) were associated with myopia, and our results are consistent with the literature in terms of myopia. However, it was found that homozygous genotypes of rs3819545 and rs2239182 SNPs were statistically significant between hyperopia and emmetropic cases (P<0.05).No other publications investigating the relationship between the examinedVDRpolymorphisms and hyperopia have been found. In this respect, our study data is the first to suggest the relationship of these polymorphisms with hyperopia.
In our study, 212 cases were examined to determine the relationship between refractory errors and related SNPs inGC,GNB4, HGF, MFN1andVDRgenes. rs7618348 inGNB4gene might be a risk factor for myopia. rs2239182 and rs3819545 inVDR, rs7041 inGCpolymorphisms were statistically significant between the hyperopic cases and controls.Furthermore, T allel in rs3735520HGFmight be a slightly protective for the development of axial hyperopia. Our results reveal the importance of genetic predisposition to refractive errors with respect to etiology of the disease. It is known that polymorphism studies might differ because of genetic diversity among populations. Results of polymorphism studies are therefore varied particularly in where a combination of many different ethnic groups found in countries such as Turkey. In addition, it is important not to ignore the gene‐environment interactions and the effect of modifying genes when evaluating genetic risk factors. It is possible to get more exact outcomes with researches in large cohorts and populations with different ethnic backgrounds.
ACKNOWLEDGEMENTS
Foundation:Supported by Eskisehir Osmangazi University Scientific Research Projects Commission, Eskisehir, Turkey(No.2016‐1234).
Conflicts of Interest:Unlu N,None;Erzurumluoglu Gokalp E,None;Arslan S,None;Cilingir O,None;Bilgim M,None;Yildirim E,None;Gursoy H,None.
 International Journal of Ophthalmology2021年6期
International Journal of Ophthalmology2021年6期
- International Journal of Ophthalmology的其它文章
- A decrease in macular microvascular perfusion after retinal detachment repair with silicone oil
- Surgical outcomes in acute dacryocystitis patients undergoing endonasal endoscopic dacryocystorhinostomy with or without silicone tube intubation
- Dr. Father Wacław Szuniewicz, a forgotten pioneer in refractive surgery and his work in China
- Deterioration of Avellino corneal dystrophy in a Chinese family after LASlK
- A mutated CRYGD associated with congenital coralliform cataracts in two Chinese pedigrees
- Autophagy dysregulation mediates the damage of high glucose to retinal pigment epithelium cells
