Cyclic adenosine monophosphate signal pathway is involved in regulation of triacylglycerol biosynthesis following nitrogen deprivation in Chlamydomonas reinhardtii*
MIAO Rongli HUANG Kaiyao
1 Key Laboratory of Algal Biology, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan 430072, China
2 University of Chinese Academy of Sciences, Beijing 100049, China
Abstract The unicellular green alga, Chlamydomonas reinhardtii is a model organism for studying various biological processes, such as photosynthesis, f lagellar motility, and lipid metabolism. To f ind some novel genes regulating the lipid metabolism under various stress conditions, the paromomycin resistance gene aphVIII was transferred into the genome of C. reinhardtii to establish a mutant library. Two genes mutated in two of the TAG-reduced mutants (Cre06.g278111 in M2 mutant, Cre06.g278110 in M6 mutants) were neighboring in the genome, and their expression levels were down-regulated in their corresponding mutants in parallel with their reduced TAG levels following N deprivation. The proteins encoded by these two genes(KCN11 by Cre06.g278111, ACYC3 by Cre06.g278110) contained a conversed cyclic mononucleotide phosphate (cNMP) binding protein and an adenylate domain, respectively. Since cNMP binding protein and adenylate domain have been known as important components of cyclic adenosine monophosphate (cAMP)signaling pathway, suggesting that these two genes might aff ect cellular TAG biosynthesis through cAMP signal pathway.
Key word: Chlamydomonas reinhardtii; mutant; lipid droplet (LD); triacylglycerol (TAG); cyclic adenosine monophosphate (cAMP)
1 INTRODUCTION
As a classical organism model,Chlamydomonas reinhardtiihas many powerful advantages for screening specif ic mutants. Its nuclear (Merchant et al., 2007), mitochondrial and chloroplast (Maul et al.,2002) genomes have all been sequenced and can be transformed (Boynton et al., 1988; Kindle, 1990;Randolph-Anderson et al., 1993). There are numerous selection markers and inducible expression vectors(Harris et al., 2009; Gonzalez-Ballester et al., 2011),which allow screening and identif ication of mutants,rapidly.
Under stress conditions such as nitrogen deprivation, high light etc., many microalgae accumulate large amount of TAG as forms of lipid droplets (LDs) (Hu et al., 2008; Belotti et al., 2013).As an organelle for TAG storage, LDs were found to be more than just storing energy, and with the discovery of perilipin, a phosphoprotein associated with LDs (Greenberg et al., 1991), researchers are paying more and more attention to the physiological functions of LDs. Nguyen et al. (2011) have found a major lipid droplet protein (MLDP) in the proteomic prof iling of LDs isolated fromC.reinhardtiiwhich could aff ect the size and degradation of LDs(Moellering and Benning, 2010; Tsai et al., 2014).Subsequently, major lipid droplet proteins were also found inHaematococcuspluvialisandNannochloropsis(Peled et al., 2011; Vieler et al.,2012). Interestingly, lipid-related proteins such as perilipin, diacylglycerol acyltransferase DGAT and phospholipase exist on the surface of LDs, which also aff ect LD formation, degradation and stability(Chapman et al., 2012; Pol et al., 2014). In addition,some environmentally responsive molecules and genes also aff ect the lipid metabolism in algae and plants.
There have been many reports that genes involved in lipid metabolism have anti-stress function in higher plants.Sensitivetofreezing2(SFR2) inArabidopsiscan use the MGDG as the galactosyl donor for synthesizing DGDG and TriGDG to protect chloroplasts against damage caused by low temperature (Moellering and Benning, 2011; Roston et al., 2014; Barnes et al., 2016). Homologs of SFR2 have also existed in all high plants, and they can defense against the damage of salt and drought in tomato and freezing inArabidopsisthrough chloroplast membrane lipids remodeling (Moellering et al., 2010; Wang et al., 2016). In addition, the overexpression of endogenous diacylglycerol acyltransferase1 (AtDGAT1) inArabidopsiscould increase its resistance to freezing stress, and led TAG accumulation (Arisz et al., 2018). Some genes related to lipid metabolism also have function of anti-biotic stress in microalgae. Genes involved in fatty acid desaturation such asCiFAD9,CiFAD3inChlamydomonassp. ICE-L in Arctic had the function of protecting cells against freezing and salt stress by regulating fatty acid desaturases (An et al., 2013b),and the temperature regulated the transcriptional expression ofCiAD9,CiFAD1, andCiFAD2to modulates the fatty acid prof ile (An et al., 2013a). In addition, disruption of plastidgalactoglycerolipid degradation1(PGD1) increased ROS stress in photosynthetic system (PSI) during TAG accumulation(Li et al., 2012).
Although there have been some studies on genes related to both abiotic stress and lipid metabolism, the molecular mechanism of TAG accumulation in microalgae under stress conditions is not clear. To clarify this problem, we established a mutant library and screened mutants with reduced TAG contents to f ind some genes involved in both abiotic stress and lipid metabolism.
2 MATERIAL AND METHOD
2.1 Strains and growth conditions
CC4348 was used as a parental strain, it is a cw15 cell-wall-def icient strain in the BAFJ5 background(Zabawinski et al., 2001), and it was obtained from theChlamydomonasResource Center. M2 and M6 were screened from our insertional mutant library. All strains were grown in Tris-acetate-phosphate (TAP)medium (Harris et al., 2009) with 7.5 mmol/L NH4Cl at 25°C under continuous illumination of 60 μmol/(m2·s), 120 r/min at 25°C in liquid cultures or on solid plates containing 1.5% agar. To induce LD formation,cells were collected at a density of 6×106cells/mL by centrifugation at 1 500×gfor 5 min, then washed with N-free TAP medium (TAP-N), and resuspended in the same volume of TAP-N medium under constant light(approximately 60 μmol/(m2·s)) at 25°C for 2 days.
2.2 Construction of a mutant library
TheaphVIIIgene fromStreptomycesrimosus(Sizova et al., 2001) was amplif ied and transferred into theChlamydomonasgenome with glass beads as a reported method (Kindle, 1990). After transformation and 4 h of recovery, cells were spread onto solid TAP plates containing 10 μg/mL paromomycin. Transgenic lines were picked to new solid TAP plates for duplicates. The primers used to amplifyaphVIIIare listed in Table 1.
2.3 Cells staining
To observe LDs in vivo, cells were stained with a f inal concentration of 0.5 μg/mL Nile Red (Sigma-Aldrich) and incubated in the dark for 10 min at room temperature (Chen et al., 2009). Stained cells were visualized using a confocal microscope (Zeiss LSM710) equipped with specif ic excitation/emission f ilters: chlorophyll autof luorescence, ex 488 nm/em 660-680 nm; Nile Red f luorescence, ex 488 nm/em 560-610 nm or analyzed using a Multi-Mode Microplate reader (Filter Max F5; Molecular Devices)with f ilters: chlorophyll autof luorescence, ex 488 nm/em 670 nm; Nile Red f luorescence, ex 488 nm/em 580 nm.
2.4 Lipid extraction and analysis
Total lipids were extracted according to the Bligh and Dyer method (Bligh and Dyer, 1959). Brief ly,8×108cells or 50 mg dried algal samples were extracted in 1.5 mL of chloroform/methanol (2:1, v/v)and centrifuged at 3 000×g, 25°C for 15 min for three times. All the upper layers were collected, 5 mL of 50 mmol/L phosphate buff er solution were added to the supernatant and centrifuged at 3 000×g, 25°C for 5 min. The extracts were collected to pre-weighted vials and dried under an N2steam. For fatty acid composition analysis, the total lipids were processed for fatty acid methyl esterif ication (FAME) and quantif ied by gas chromatography-mass spectrometry(GC-MS) as previously described (Rossak et al.,1997). To separate TAGs, the organic phase was spotted on a silica thin layer chromatography (TLC)plate using petroleum ether/diethyl ether/acetic acid(70:30:1, v/v/v) as the solvent system (Ghosal et al.,2007). To visualize TAGs, the developed TLC plates were air dried and exposed to iodine vapor at 37°C for 5 min (Yang et al., 2015). For each plate, 20-μg glyceryl trioleate was used as the TAG marker and the TAG bands was quantif ied by gray semi-quantitative analysis using ImageJ (ver. 1.45, NIH) (Tsihlis et al.,2010). In each independent experiment, the loading samples on each TLC plate were the same (20 μL of the total lipids extracted from cells with same dry weight or same number).

Table 1 Information of primers used in this study
2.5 Cloning of mutated genes
The mutated genes were cloned by restriction enzyme site-directed amplication PCR (RESDAPCR) (González-Ballester et al., 2005). Four degenerate primers were designed based on the random distribution of abundant restriction sites(AluI,PstI,SacII, andTaqI) in the genome ofC.reinhardtii. After two rounds of PCR procedures using the degenerate primers combined with four specif ic primers of theaphVIIIinsertion, four sets of PCR products were obtained upstream and downstream of the insertion sequence, the bands with a length of 750-2 000 bp were selected for sequencing.All the primers are listed in Table 1.
2.6 RNA isolation and quantitative real-time PCR
RNA was isolated from algal samples usingTransZolup Plus RNA Kit (Transgen Biotech)according to the operating manual. RNA was then reverse transcribed into f irst-strand cDNA using a High-capacity cDNA Reverse Transcription Kit with RNase Inhibitor (Invitrogen). Gene transcription was measured using SYBR Green PCR Master Mix(Applied Biosystems) and the ABI 7900 HT Real-Time PCR System (Life Technologies). The expression levels of target genes were normalized usingRACK1as the internal control (Moellering and Benning, 2010). The expression level ofRACK1was calculated by the delta-delta method (Livak and Schmittgen, 2001). Primers used for real-time PCR are shown in Table 1.
2.7 Statistical analysis
All experiments were performed with at least three biological repeats to ensure reproducibility. Values are presented as means±SD. Statistical analyses were performed using the GraphPad Prism 5, the signif icance of the diff erences was tested using a oneway ANOVA orTtest. *P<0.05; **P<0.01;***P<0.001; ns is no signif icant diff erence.
3 RESULT
3.1 Isolation of TAG-reduced mutants
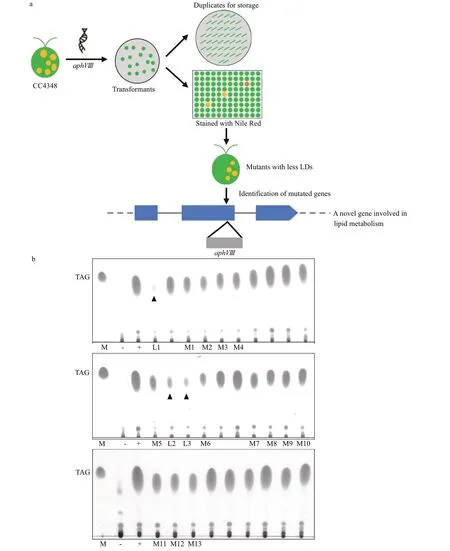
Fig.1 Screen mutants lacking lipid droplets (LDs) in C. reinhardtii
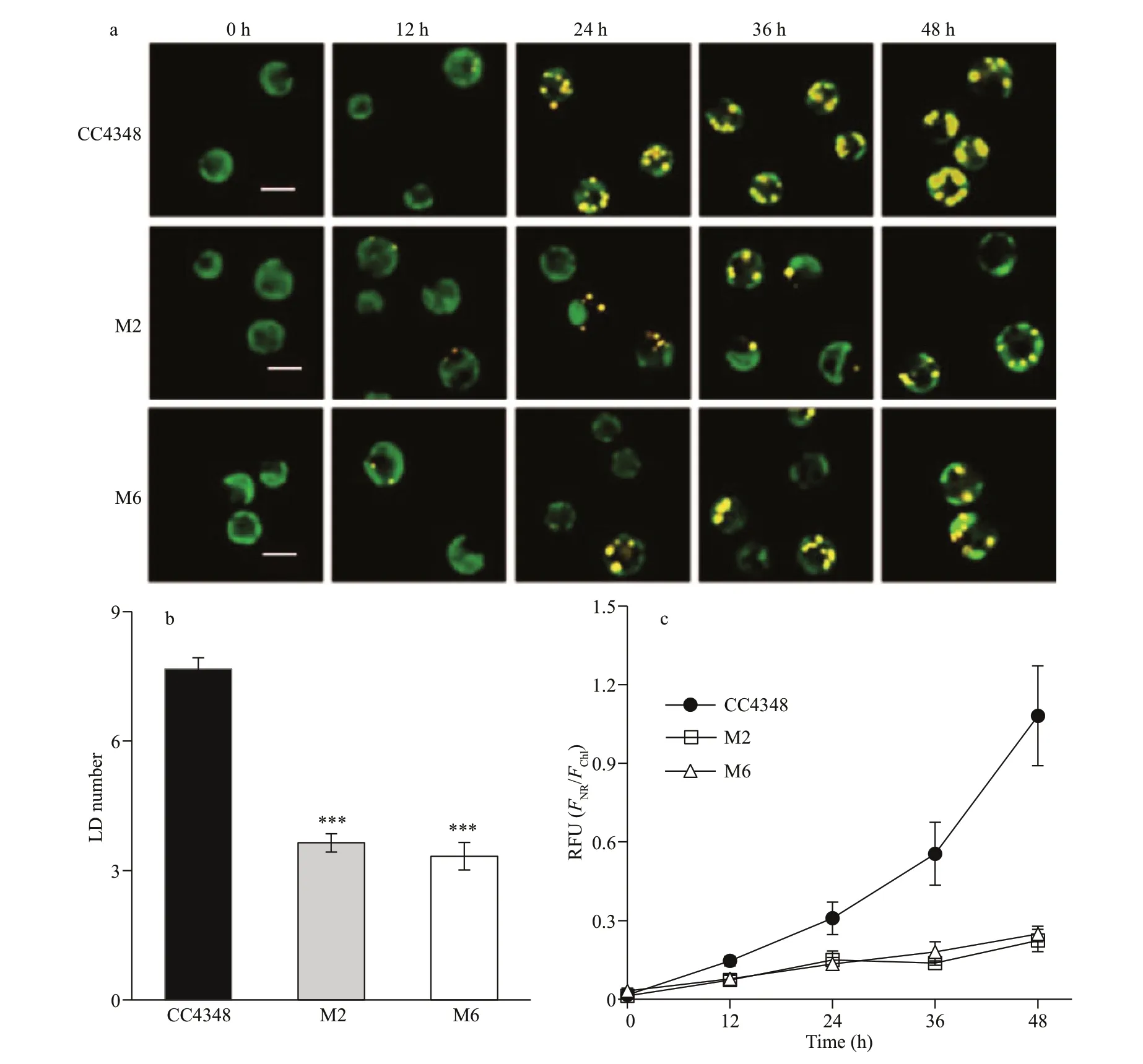
Fig.2 Fluorescence analysis of the lipids in CC4348 and mutants M2, M6 following N deprivation
The main steps of screening mutants with low lipid content are shown in Fig.1a. Firstly, the resistance gene of paromomycin (aphVIII) was transferred into the nuclear genome ofC.reinhardtii, and a series of random insertional transformants were obtained by antibiotic plates. Next, all transformants were kept duplicates and transferred to 96-well plates containing TAP-N medium for 1 day, and the cells in 96-well plates were stained by Nile Red (NR) to measure their f luorescence intensity by MD using chloroplast (Chl)autof luorescence as standard. In the capacity of 10 560 transform mutant library, 120 mutants with weaker NR f luorescence intensity were initially isolated. Some mutants were died in the process of nitrogen-def icient culture (indicated by black triangles in Fig.1b), and others showed unstable phenotypes.After quantitative TLC conf irmation, 13 mutants with stable phenotypes were f inally obtained (Fig.1b). This study focused on analysis of the phenotypes and mutated genes in two of these mutants, M2 and M6,as the genes mutated in M2 and M6 occupy neighboring positions in the genome.
3.2 Nile Red f luorescence intensity in M2 and M6 mutants
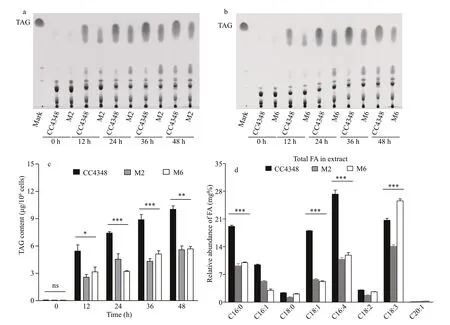
Fig.3 Analysis of the lipid phenotypes of CC4348 and mutants M2, M6 by TLC or GC-MS following N deprivation
During the course of N deprivation, the wild type cells of CC4348, and mutants M2 and M6 were stained by Nile Red and observed under the confocal microscope (Fig.2a). After 24 h of N deprivation,there were only about 3-4 LDs in each cell of M2 and M6, but about 7-8 LDs in the parental strain CC4348(Fig.2b). After 48 h of N def iciency, the LDs in CC4348 began to fuse and almost occupied the entire cell, whereas those in M2 and M6 only showed an increase in LD number but smaller LD size than CC4348 (Fig.2a). It is suggested that the TAG content in M2 and M6 reduced as a result of their inability of LD fusing. Subsequently, the relative f luorescence intensity of Nile Red-stained LDs (FNR/FChl) in M2,M6, and CC4348 cells were measured, and theFNR/FChlin M2 and M6 was only 48.4% and 43.6% of that in CC4348 after 24 h of N deprivation,respectively, then decreased to 21% and 23% that in CC4348 after 48 h of N deprivation, respectively(Fig.2c). This result indicated that it was both intuitive and rapid to analyze the lipids in vivo using the specif ic lipophilic dye staining (Chen et al., 2009;Cooper et al., 2010), avoiding the cumbersome steps of lipids extraction. Therefore, it is the preferred method to screen mutants at the early stage.
3.3 TAG contents and fatty acids in M2 and M6 mutants
To conf irm the phenotypes of M2 and M6 mutants,their total lipids in same cell number (8×108cell/sample) or same cell dry weight (50 mg/sample) were extracted, and their TAG contents and fatty acids composition were analyzed by TLC and GC-MS,respectively (Fig.3). In order to compare the TAG content of CC4348 and mutants M2, M6, their TAGs were analyzed by TLC and visualized by iodine vapor fumigation. The plaque areas indicating TAG levels of the three strains all increased with the prolongation of N def iciency time, but the TAG plaque areas of CC4348 were much bigger than those of M2 and M6 at the same duration of N deprivation (Fig.3a, b).Semi-quantitative analysis of their TAG plaque areas found that the TAG content in M2 and M6 were only 61.2% and 43.4% of that in CC4348 cells at 24 h of N def iciency, 55.7% and 56.6% of that in CC4348 at 48 h, respectively (Fig.3c). These results showed that the TAG content in M2 and M6 did reduced.
To detect changes in the fatty acid composition of M2 and M6, their total lipids were extracted and the amount and composition were analyzed after N deprivation for 24 h (Fig.3d). The result showed that there was no diff erence in the composition of major fatty acids between M2/M6 and CC4348, but the relative fraction of fatty acids C16:0, C16:4. C18:1 was reduced signif icantly. It is worth noting that the fatty acid composition of C18:3 in M2 is decreased signif icantly, but it is increased in M6, compared to that in CC4348. It suggested that the lipids synthesis in M2 and M6 were initially inhibited, in the process of synthesizing of long-chain fatty acids from acetyl-CoA. The fatty acids C16:0 and C16:4 are usually used as precursors for fatty acids C18:3, which can also be formed from higher-saturation fatty acids C18:1 catalyzed by fatty acid desaturases (Wakil et al., 1983). It was speculated that this might be one of reasons that the proportion of C16:0, C16:4, and C18:1 in M2 and M6 mutants decreased signif icantly but C18:3 increased obviously.
3.4 Location of the genes mutated in M2 and M6 in the genome
To identify theaphVIIIinsertion site, RESDAPCR (González-Ballester et al., 2005) was employed to amplify the f lanking sequence. Four random primers combined with four specif ic primers annealing to the positive strand of the paromomycin resistance gene (aphVIII) generated a series of products (Fig.4a,b) after two rounds of PCR, and products with a length ranging from 750 bp to 2 000 bp were sent for sequencing. It was found that the insertion site in M2 located in the f irst exon of a gene on chromosome 6(locus Cre06.g278111) (Fig.4c) encoding a potassium ion channel protein (KCN11) and this protein contained a conversed cNMP binding domain (Xu et al., 2016). Its homologous protein sequences in four higher plants were blasted on phytozome v12.1(https://phytozome.jgi.doe.gov/pz/portal.html), and the sequences with the highest similarity were selected. They were potassium transport 2/3 inArabidopsisthaliana, potassium channel AKT2 inOryzamarina, potassium channel protein ZMK2 inZeamaysand putative potassium channel AKT2/3 inRicinuscommunis, respectively. Then, their conserved domains were scanned on the Prosite (https://prosite.expasy.org/prosite.html), and the conversed cNMP binding domain was hit on the sequences of all these four homologous proteins (Fig.4d). Alignment of their sequences of the cNMP motif showed a high similarity (Fig.4e), which indicated that the function of KCN11 inChlamydomonasmight have the similar function to homologous proteins in higher plants.
With the same method, we found that the insertion site in M6 was located in the 3′UTR of a gene also on chromosome 6 (locus Cre06.g278110), and the position of this gene was adjacent to the gene mutated in M2 in the genome (Fig.4c). Scanning the protein sequence encoded by this gene showed that it was rich in leucine repeats and contained an adenylate cyclase (class3) domain (AcyC3) (Fig.4d). Adenylate cyclase is the major component of cAMP signaling pathway and activated by the α-subunit of the heterotrimeric G proteins to synthesize cAMP(Lengeler et al., 2000), which indicates thatACYC3might aff ected the TAG accumulation by cAMP signal pathway.
Insertions into the f irst exon ofKCN11and 3'UTR ofACYC3were expected to aff ect genes expression of them. After 24 h of N def iciency, the transcript levels ofKCN11in M2 andACYC3in M6 were quantif ied by real-time PCR, respectively, and it showed that both transcriptional expression level of them decreased signif icantly (Fig.4f & g). Down-regulation of the expression ofKCN11gene in M2 led to a decrease in its TAG content, which was consistent with the previously report that the mRNA expression ofKCN11increased in the wild-type strain following N deprivation (Miller et al., 2010). The downregulation of expression ofKCN11andACYC3following N deprivation in parallel with the decrease of TAG accumulation suggested that the products of these two genes might play a role in TAG biosynthesis.
4 DISCUSSION
4.1 Mutants with abnormal TAG content
In our study, two methods were used to detect neutral lipid content, one was Nile Red staining and other was TLC method, and the results detected by these two methods were consistent before 36 h of N deprivation, but inconsistent at 48 h of N deprivation.The TAG contents in M2 and M6 were 55%-57% of that in CC4348 when detected by TLC, but only 21%-23% of that in CC4348 when detected by Nile Red staining, which might be the inconsistent rate of chloroplast degradation in M2, M6 mutants and CC4348 during LD formation. Due to more number and larger size of LDs in CC4348, its chloroplasts degraded faster than the mutants. When the f luorescence intensity of Nile Red was normalized by the autof luorescence intensity of the chloroplast, it might result in a higher detection value in CC4348 and a lower detection value in the mutants at the later stage of LD formation (after 36 h of N def iciency). It indicated that it is best to screen mutants with abnormal lipid content within 36 h of N deprivation using Nile Red staining.
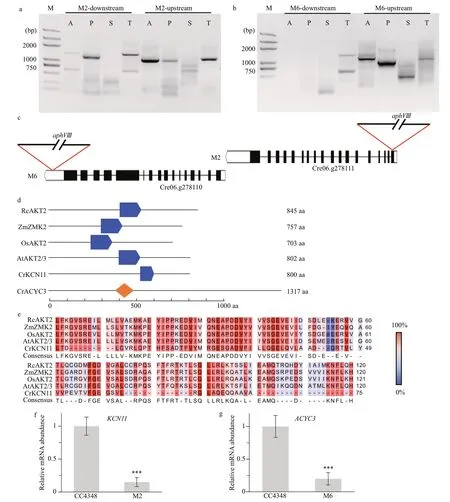
Fig.4 Clone and identif ication of the mutated gene in M2 and M6 mutants
4.2 cAMP signal pathway involved in the TAG accumulation
cAMP signaling pathways, have been known as one of conserved signal pathways to play the key roles in many eukaryotic organisms upon external environmental stimuli (Choi and Xu, 2010). Potassium transport 2/3(AKT2), the homologous protein of CrKCN11 inArabidopsisthaliana, can be regulated by a variety of cytokines such as Ca2+, pH (Latz et al.,2007). It was critical for cellular responses to various environmental or endogenous stimuli, such as the defense of virus infection (Ascencio-Ibanez et al.,2008). It has been reported that the levels of cAMP increase in response to biotic and abiotic stress and inf luence calcium (Ca2+) inf lux subsequently(Alqurashi et al., 2016). KCN11 inChlamydomonasalso has been reported that could respond to K+and some other biotic and abiotic stimuli (Xu et al., 2016).Therefore, we speculated thatKCN11could response to N def iciency stress and aff ect the TAG accumulation in M2 cells indirectly.
Although there was no sequence similar to the sequence of the gene mutated in M6 in higher plants,the protein encoded by this gene was rich in leucine repeats. It has been reported that the protein rich in leucine repeats inArabidopsisthalianacan protect itself from infection ofP.syringaeinfection (Kobe and Kajava, 2001). In addition, the protein rich in leucine repeats in rice also has been reported to be resistant to disease (Wang et al., 1999), which suggested that CrACYC3might belong to the antistress gene family. In fact, genes involved in both lipid metabolism and disease resistance signal transduction did exist not only in plants but also in microalgae, such assuppressoroffattyaciddesaturase def iciency1(SDF1) inArabidopsis, which could defense against bacterial infections (Nandi et al.,2004). Furthermore, omega-3 fatty acid desaturase inChlamydomonassp. ICE-L, also could protect cells from damage of freezing and high salinity stress in Antarctic (Zhang et al., 2011), which suggested that genes against diff erent abiotic stresses were closely related to TAG synthesis.
A conserved cNMP domain also called cAMP /cGMP binding domain and an adenylate cyclase domain existed on the protein of CrKCN11 and CrACYC3, respectively (Fig.4f). Adenylate cyclase is one of the major components of cAMP signaling pathway, and cAMP -binding protein is one of the cAMP targets, suggesting they are both important for the activation of cAMP signaling pathway during the physiological processes inChlamydomonasincluding photosynthesis and abiotic stress responses(Donaldson et al., 2016, Chatukuta et al., 2018).According to Choi et al. (2015), cAMP signaling pathways are positively associated with lipid biosynthesis of microalgae. In mammals, induction of cAMP signal in both white and brown adipose tissues promoted lipolysis (Ravnskjaer et al., 2015). For example, stimulating β-adrenaline increase the level of cAMP and promote lipid degradation (Kolditz and Langin, 2010). In addition to its function on lipolysis,cAMP signal also plays an important role in adipocyte diff erentiation, such as diff erentiating f ibroblasts precursors into mature adipocytes (Siersbæk et al.,2014), and turning the white adipose tissue to a more phenotype of brown adipose tissue (Dempersmier et al., 2015); and regulation of lipid metabolism, such as hydrolyzing the triglycerides stored in adipocyte LDs to glycerol and fatty acids (Frayn, 2002). In our study,the expression level ofKCN11andACYC3decreased in M2 and M6 with the reduction of cellular TAG levels, respectively. It conf irmed our hypothesis that cAMP signal pathway might be involved in the TAG accumulation ofChlamydomonasfollowing N deprivation.
5 CONCLUSION
Our study found that the relative f luorescence intensityFNR/FChlcould accurately indicate intracellular TAG content within 36 h of N def iciency.The TAG contents decreased with the down-regulation of mRNA expression levels ofKCN11andACYC3 in M2 and M6, respectively. Protein domain analysis showed that both KCN11 and ACYC3 belonged to the cAMP signaling pathway and aff ected the TAG accumulation ofChlamydomonas.
6 DADA AVAILABILITY STATEMENT
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
 Journal of Oceanology and Limnology2020年2期
Journal of Oceanology and Limnology2020年2期
- Journal of Oceanology and Limnology的其它文章
- Contribution of surface wave-induced vertical mixing to heat content in global upper ocean*
- Upper ocean response to typhoon Kujira (2015) in the South China Sea by multiple means of observation*
- Inf luence of simulating deep-sea environmental factors on cathodic performance of seawater battery*
- Adsorption characteristics of chitooligosaccharides onto activated charcoal in aqueous solutions*
- Eff ects of hypoxia on survival, behavior, and metabolism of Zhikong scallop Chlamys farreri Jones et Preston 1904*
- Distinct inf luence of trimethylamine N-oxide and high hydrostatic pressure on community structure and culturable deep-sea bacteria*
