Molecular identif ication of oomycete species affecting aquaculture in Bangladesh
Mohammad Nasif Sarowar,Md Jakir Hossain,Tahmina Nasrin,Tania Naznin,Zakir Hossain,Mohammad Matiur Rahman
Department of Fisheries Biology and Genetics,Bangladesh Agricultural University,Mymensingh-2202,Bangladesh
Keywords:Oomycete Saprolegnia Aphanomyces invadans Internal transcribed spacer(ITS)Phylogeny
A B S T R A C TFish mycotic disease outbreaks occur due to infections with oomycete pathogens such as Saprolegnia spp.and Aphanomyces invadans,and cause large-scale f ish production losses.Despite its negative impact on aquaculture,little is known about the diversity of oomycete pathogens.The aim of this study was to identify the diversity of pathogenic oomycetes causing infections in major aquaculture zones of Mymensingh and Jessore regions in Bangladesh.A total of 449 water and infected f ish samples were collected from 28 f ish farms in both regions of which 29 sampleswere able to grow out with mycelia on the Potato Dextrose Agar(PDA)/Glucose Peptone Yeast Agar(GPYA)plates.Sequence database searches using the rRNA Internal Transcribed Spacer(ITS)region revealed that 15 belonged to Pythium spp.,12 were Aphanomyces invadans and two corresponded to Saprolegnia parasitica.Five isolates of Pythium spp.were identif ied to the species level:one was closely related to Pythium catenulatum,four to Pythium rhizo-oryzae,rest were identif ied up to genus.The Pythium spp.were only isolated from water samples whereas A.invadans and S.parasitica were found in f ish lesions.Phylogenetic analysis revealed that a single A.invadans clone exists in the sampled area.The results obtained conf irm the existence of pathogenic oomycetes in Bangladesh f ish farms and this will pave future research on diversity,prevention and control measures.
1.Introduction
The oomycetes(phylum Heteronkontophyta)are among the most detrimental pathogens of freshwater farmed and wild f ishesall over the world(van den Berg,McLaggan,Diéguez-Uribeondo,&van West,2013).Asa group oomycete are considered asdecomposer of plantsand animalsdebrisin the aquatic ecosystem.However,some species such as
Saprolegnia parasitica and Aphanomyces invadans are often responsible for serious disease outbreaks in f ish farms and in the natural environment causing large economical losses by threatening industry production targets and long-term viability(Iberahim,Trusch,&van West,2018;Sarowar,van den Berg,McLaggan,Young,&van West,2013;van West&Beakes,2014).Fish pathogenic oomycetes are mostly undervalued as secondary pathogens(van den Berg et al.,2013)in spite of highly specialized infection mechanisms similar to many primary pathogens(Tadiso et al.,2011;Young,Cooper,Nowak,Koop,&Morrison,2008).The carbohydratesof the S.parasitica cell wall and prostaglandin E2 were shown to suppress f ish immune response during the early stages of infection(Belmonte et al.,2014).Therefore,understanding pathogen diversity and host-pathogen interactions of oomycete infections in f ish is necessary to combat the disease.
Oomycete infections are mainly spread through swimming zoospores(Beakes,Glockling,&Sekimoto,2012;van West&Beakes,2014).Because of their characteristic f ilamentous mycelial growth,oomycetes were previously grouped as Eumycota(the true fungi)but they are now classif ied in the Stramenopiles or Chromista group with the diatoms and brown algae due to the presence of bif lagellate motile zoospores(Beakes et al.,2012).Infection outbreaks are common at the onset of winter season when the water temperature decreases and favors the pathogen to infect immune compromised hosts(Bly&Clem,1992).Saprolegniosis on f ish are typically manifested by visible white or grey patches of mycelia on the body and f ins causing eventually hemodilution and death(Richards&Pickering,1979).In case of epizootic ulcerative syndrome(EUS),the early infections show petechial hemorrhagic lesions on the skin developing into severe necrotic ulcers in the muscle tissues due to penetrating hyphae of A.invadans(Chinabut,1998;Noga,Levine,Dykstra,&Hawkins,1988).The zoospore release is synchronized with environmental factors such as temperature and hostpresence(Diéguez-Uribeondo,Cerenius,&Söderhäll,1994).
Aphanomyces invadans is an important oomycete recognized as the primary pathogen responsible for EUS(Baldock et al.,2005);a detrimental f ish disease that occurs in natural waters and commercial f ish farms in Asia,Africa,parts of Australia and North America(Roberts,2012;Sosa et al.,2007).The World Organization for Animal Health(OIE)listed that this disease has been responsible for large-scale mortalities of farmed and wild f ish in more than 20 countries across four continents(Iberahim et al.,2018;Pradhan,Mohan,Shankar,&Kumar,2008).The A.invadans was f irst reported in Ayu,Plecoglossusaltivelis in Japan(Egusa&Masuda,1971)but since then it hasbeen reported from a number of f ish species including carps,snakeheads,salmonids and estuarine species(Lilley et al.,2003;Oidtmann,2012).Outbreaks of EUS in aquaculture occur regularly in Bangladesh although recent studies are rare(Khan&Lilley,2001).In Bangladesh and India,outbreaks of EUShave been responsible for large scale f ish mortalities(up to 20-50%)(Pradhan et al.,2014).However,this disease is not considered as seriously as other bacterial infections mainly due to lack of sufficient knowledge and available data.Currently,little is known about pathogen diversity and etiology of the disease that difficult the prevention and control measurements.
Comparisons of the genomes of different f ish pathogenic oomycetes such as S.parasitica,S.diclina,A.invadans revealed potential differences in the molecular and biochemical mechanisms of infection.For example,S.parasitica contains a large repertoire of proteases,kinases and disintegrins that are expressed at different stages of the infection(Jiang et al.,2013;Makkonen et al.,2016)and different Saprolegnia species use distinct infection strategies(Songe et al.,2016).In addition,several S.parasitica strains are linked to pathogenicity on specif ic life stages such as egg,alevin,juvenile and parr of Atlantic salmon(Sandoval-Sierra,Latif-Eugenin,Martín,Zaror,& Diéguez-Uribeondo,2014).Therefore,accurate identif ication of the pathogen for saprolegniosis is an essential prerequisite for disease management.Knowledge and understanding of pathogens diversity is essential to formulate sustainable prevention and control of mycotic diseases.The objective of this study wasto understand the diversity of oomycetes species causing infections in the aquaculture farms of Mymensingh and Jessore region of Bangladesh.
2.Materials and methods
2.1.Sample collection
Water samples and tissues from apparently infected f ish(Fig.1)were collected from different aquaculture farms located in Jessore and Mymensingh districts during the winter season from 15th of November 2015 to 09th of February 2016.Water temperature and p H of sampling sites were measured.All samples were collected from 28 commercial f ish farms of which 16 were from Jessore and the remaining from Mymensingh region(Table 1).The exact location of the farms are not disclosed due to the conf identiality agreement.A total of 449 samples were collected of which 248 samples(55.23%)were from Mymensingh and 201(44.77%)from Jessore.There were 399(88.86%out of 449 total samples)water samples(217 from Mymensingh and 182 from Jessore)collected from the farms and 50(11.14%)samples(31 from Mymensingh and 19 from Jessore)were tissue taken from the apparently infected f ish.Water samples(10 ml)were collected in 15 ml sterile falcon tubes.Each tube contained an autoclaved adult mosquito(Anopheles sp.)as bait.All water samples were kept at 18°C temperature for seven days to enable the colonization of the baits.For tissue samples,the infected f ish were euthanized using 1 g/L Tricaine Methanesulfonate(MS222;Sigma-Aldrich)bath until the opercula and bodily movements were ceased completely in order to comply with institutional experimental animal ethics.A small sample(around 0.5 cm3)of infected tissuesor small portion of f in(0.5 cm)wascut with a sterile scalpel and scissor,and washed with autoclaved distilled water,to reduce the unwanted microorganisms load adhered to the surface.The collected samples were placed aseptically on Potato Dextrose Agar(39 g/L PDA in d H2O and autoclaved)and GPYA plates(glucose 3 g/L,peptone 1 g/L,yeast 0.5 g/L,agar 0.5 g/L,Nacl 0.5 g/L)plates.All the collected tissue samples were incubated for a week at 18°C.The samples that showed positive growth were used for further investigation.
2.2.Laboratory culture of collected samples
The bait(mosquito)within the collected water samples were inoculated on Petri dishes(90 mm diameter)f illed with 25 mL growth medium supplemented with antibiotics(100 mg/L Vancomycin,500 mg/L Ampicillin and 20 mg/L Natamycin)to reduce bacterial growth.Two different growth media were used;Potato Dextrose Agar and GPYA.For inoculated f ish tissues on plates,a small portion of mycelia growing out from the inoculum was aseptically inoculated on new PDA and GPYA plates supplemented with antibiotics.All the plates were sealed with paraf ilm and incubated at 18°C.The isolates were reinoculated several times to eliminate bacterial contamination.
2.3.DNA extraction
A small agar plug with mycelia was aseptically placed in 20 mL Pea broth(124 g/L green pea autoclaved in dH2O).The liquid media containing inoculum were incubated at 18°Cfor 3 daysto allow mycelia to grow.DNA was extracted from the grown mycelia using the Phenol:Chloroform:Isoamyl alcohol(with the ratio of 25:24:1)method according to Zelaya-Molina,Ortega,&Dorrance,2011,quantif ied(Nanodrop,Thermo Scientif ic,USA)and stored at-20°Cfor downstream analysis.

Fig.1.Photographs showing EUSinfected f ish.A)and B)Rohu(Labeo rohita)and C)Tilapia(Oreochromisniloticus).White arrowsindicate the eroded muscles due to Aphanomyces invadans infections.
2.4.PCRamplif ication
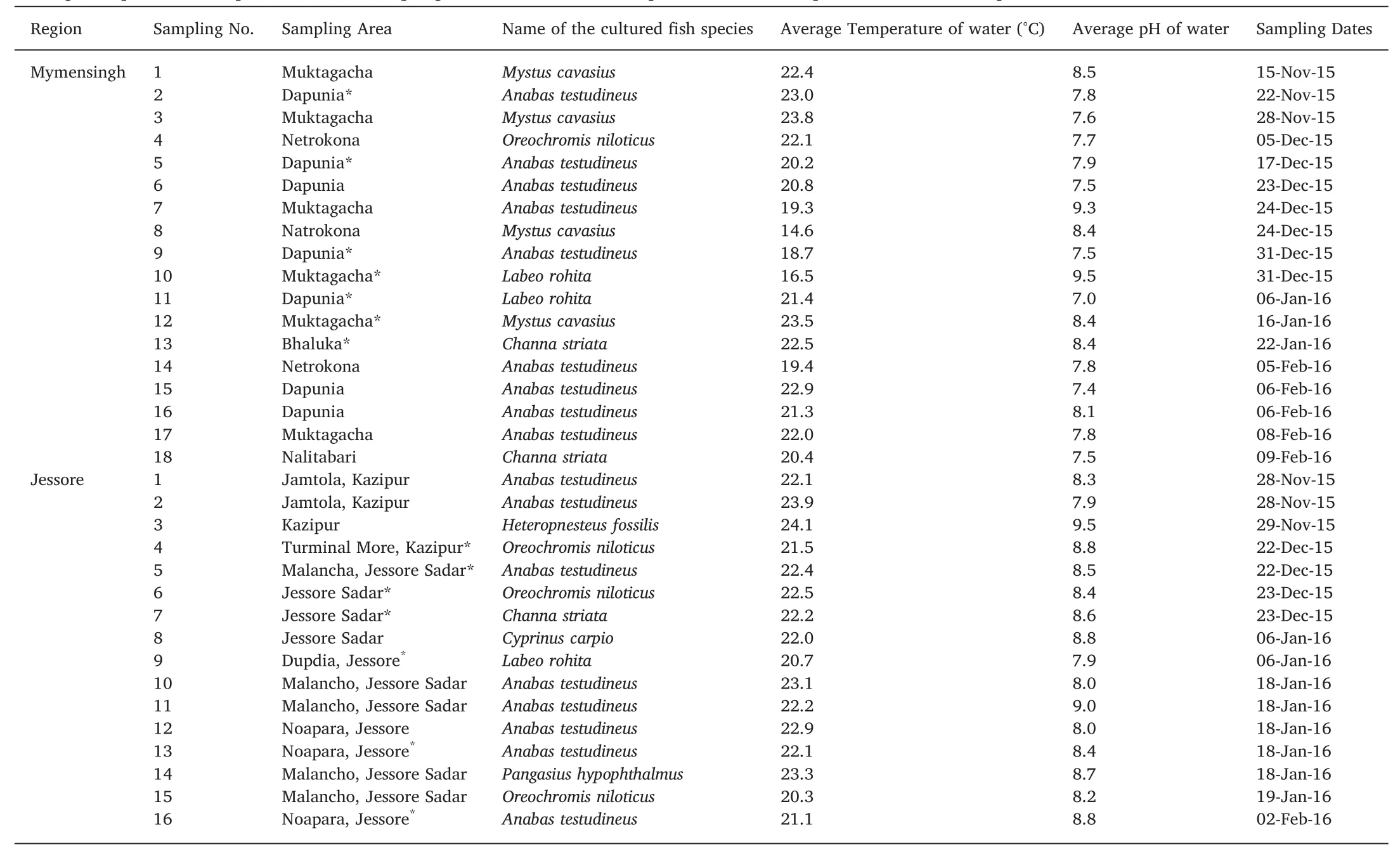
Table 1Average temperature and pH in different sampling sites at the time of sample collection and species of the f ish sampled in the farms.
PCR amplif ication of the internal transcribed spacer(ITS)region between 18S and 28Sribosomal subunits of the rDNA was performed using the primers ITS5 alt(5′TGA AAA GTCGTA ACA AGGTT 3′)and ITS 4 alt(5′TCCTCC GCT TAT TGA TAT G 3′)modif ied from(White,Bruns,Lee,&Taylor,1990).The PCR was performed for a total f inal volume of 10μL containing 5μL 2x GoTaq®G2 Colorless Master Mix(Promega®;contains DNA Ploymerase,d NTPs,MgCL2and reaction buffer),1μL of template DNA(~50 ng/μL),0.3μL of each primers(10μM)and 3.4μl of nuclease free water.The reaction was performed using a Mastercycler Gradient(Eppendorf,Germany),as follows:95°C for 5 min followed by 30 cycles(95°Cfor 30 s,57°Cfor 30 s,73°Cfor 1 min),f inal extension at 73°Cfor 7 min.PCRproducts were visualized in a 1%agarose gel electrophoresis stained with 1μLEthidium Bromide(Promega®;conc.10 mg/mL).
2.5.DNA sequencing and analysis
Sanger sequencing of the amplif ied PCR products(~750 bp-~800 bp)was done at the BIONEER,South Korea using the PCR amplif ication primers.For each amplicon,two reads(forward and reverse)were performed.The consensus sequence was searched at GenBank repository using Basic Local Alignment Search Tool(nucleotide BLAST)to conf irm.
2.6.Phylogenetic analysis
Phylogenetic analysis was carried out using the Maximum Likelihood(ML)and Bayesian inference(BI).The MEGA v7 program(Kumar,Stecher,&Tamura,2016)was used to align and construct the ML tree(Tamura&Nei,1993).The Aphanomyces and Saprolegnia analysis involved 25 and 31 nucleotide sequences(retrieved sequences available in Appendix A).The sequences were aligned with ClustalW(Thompson,Higgins,&Gibson,1994)in MEGA v7 and were manually adjusted and,positions containing gaps and missing data were eliminated.For the ML analysis the general time reversal(GTR)+Gamma(G)model was considered the best substitution model based on the lowest Bayesian information criterion(BIC)and Akaike information criterion value(AIC),and tree was built using 1000 bootstrap replications.For BI,the analysis wasperformed using MrBayes3.2.6(Ronquist et al.,2012)using the same model as the ML tree.The BI analysis included four parallel running of one and half million generations carried out separately,for each run eight Markov chain Monte Carlo(MCMC)were done having one cold and seven hot chains(temperature 0.10)for both Aphanomyces and Saprolegnia sequences.The datasets were samples every 1000thgeneration.The burn-in threshold wasanalyzed in the Tracer v1.5.0(http:/tree.bio.ed.ac.uk/software/tracer/).The f irst 25%trees were discarded for each run,the remaining trees were used for estimating posterior probabilities(pp).
3.Results
3.1.Isolation of oomycetes
A total of 29 samples(around 6.5%)out of the 449 samples that were collected during the sampling period grew positive with mycelial outgrowth.Of the 29 samples,15 were water samplesand 14 were from tissue samples.The highest temperature(23.9°C)in sampling ponds was recorded from Jamtola,Jessore on 28th November 2015 and the lowest temperature (14.6°C) was recorded from Natrokona,Mymensingh on 24th December 2015(Table 1).The mean water temperatures of the ponds from which samples were collected in Mymensingh and Jessore were 20.8±2.36°C and 22.3±1.02°C,respectively.The temperature between the two regions did not vary signif icantly during the study period.Similarly,the average water pH in Mymensingh(8.0±0.63)and Jessore(8.5±0.43)were not signif icantly different between the two regions.There was also no signif icant correlations between the positive growth from collected samples(water and apparently infected tissues)and different water temperatures or p H during the sampling period.
Fig.2.Figure showing different f ish species found to be infected by Aphanomyces invadans and Saprolegnia parasitica in the study area of Mymensingh and Jessore regions.
3.2.Molecular analysis
Of the 29 samples(9 from Mymensingh and 20 from Jessore)analyzed f ifteen were identif ied as Pythium spp.,twelve as A.invadans and two were S.paraisitca.The A.invadans and S.paraisitca were recovered from infected/moribund f ish species collected from both regions(Fig.2)while the Pythium isolates were recovered from water samples.On the basis of ITSsequences of the samples from Mymensingh region,six isolates e.g.M-FR-1,M-FR-2,M-87,M-223,M-106 and M-shol were identical to A.invadans(KC137251)(Fig.3;Table 2),and isolate M-38 was identical as S.parasitica(JN400038).Isolate M-24 and M-19 were similar(99%similarity to GenBank sequence)to Pythium catenulatum(FJ415899)and Pythium rhizo-oryzae(HQ643757).
On the other hand,ten isolates from Jessore were identif ied up to species level.Isolate J-37,J-168 and J-181 were similar(99%similarity to GenBank sequence)to Pythium rhizo-oryzae(LC150560);J-78,J-116,J-133,J-Koi,J-Rui and J-Shol isolate were identical to A.invadans(KC137251),and J-120 was identif ied as S.parasitica(JN400038).The S.parasitica isolates found in both regions shared identical sequences.In addition,ten isolates from Jessore were only identif ied up to genus level i.e.Pythium sp.(J-18,J-25,J-29,J-129,J-132,J-138,J-140,J-141,J-173 and J-175).Majority Pythium speciesfound during the study were isolated from the Jessore region(Table 2).
3.3.Phylogenetic analysis
The phylogenetic analysis of the ITSrDNA sequence collected from the Jessore and Mymensingh region with retrieved homologous sequences conf irmed that the collected isolates of M-FR-1,M-FR-2,M-87,M-223,M-106,M-shol,J-133,J-78,J-116,J-Rui,J-Koi and J-Shol are identical to the reported A.invadans(Fig.3)with high bootstrap values and posterior probabilities(pp=1,bs=100).No difference was observed between the isolatesfrom both Mymensingh and Jessore regions.Moreover,it appears that the animal pathogenic isolates of different Aphanomyces species/strains clustered apart from the plant pathogenic/saprotroph supported by bootstrap values and posterior probabilities(pp=1,bs=97;Fig.3).In Saprolegnia tree,the M-38 and J-120 isolates are identical to the reported S.parasitica(Fig.4).No difference were observed between the Mymensingh and Jessore isolates.
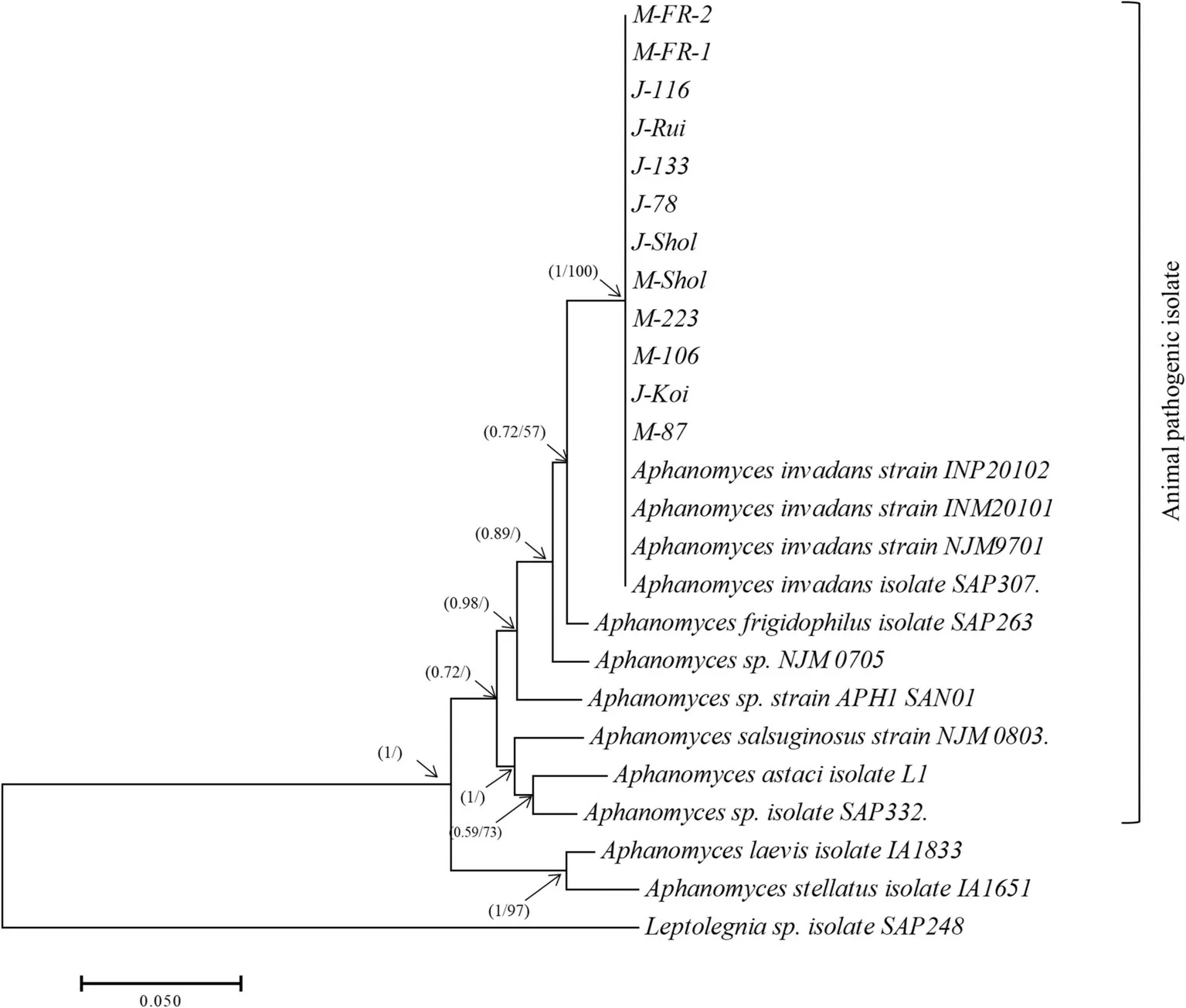
Fig.3.The f igure is a summary of the phylogenetic tree inferred by Bayesian inference and Maximum Likelihood analyses based on the internal transcribed spacer(ITS)regions of nr DNA.The analyses comprise all the Aphanomyces invadans isolates obtained in the study and closely related GenBank sequences.Isolates having J or M in the beginning of their name indicate the region where they were recovered from;MMymensingh,J-Jessore.Numbers in the branches represent the posterior probability valuesand bootstrap support(valuessmaller than 50%are not shown).
4.Discussion
Mycotic diseases of f ish,cause by oomycete pathogens,are a real threat to the sustainability of a developing aquaculture industry in Bangladesh.Molecular identif ication of oomycetes is rare in the aquaculture farms of Bangladesh and detail information and knowledge about the mycotic diseases is absent.During the span of the research period,large number of water samples were collected to unveil the diversity of the oomycetesin the study area.However,only few samples showed mycelial growth in vitro which ref lects the limitation of the traditional baiting technique to study f ish pathogenic oomycetes(Arcate,Karp,&Nelson,2006).The water sampling with bait only includes species that produced zoospores under the given conditions during sampling,leaving non-zoosporic species or species not developmentally in a state to release zoospores,to go undetected.For example,water samples collected with bait during the study did not show any A.invadans growth in laboratory conditions.Similar scenario were reported by Willoughby et al.(Willoughby,Roberts,&Chinabut,1995)and Lilley et al.(Lilley,Beakes,&Hetherington,2001)where water sampling for A.invadans zoospores were unsuccessful.In addition,the bacterial load in the collected water might have actively deterred zoospore settlement and germination on the provided bait.Using methods established to detect very low concentration of environmental DNA could helpful to overcome this problem(Thomsen&Willerslev,2015).However,the methods are expensive and require expertise compared to the traditional water sampling with baits.Moreover,the later can result in an actual live isolate that can be used for further downstream analysis.Therefore,a combination of both baiting and environmental DNA techniques is expected to result in better resolutions to study oomycete pathogen diversity.
Advances in molecular biology have helped fungal taxonomy and diagnostics of mycotic diseases in f ish.The rRNA gene internal transcribed spacer(ITS)region is a source of sequence variation among closely related species when compared to rRNA-coding genes(Belbahri et al.,2008;Windsor,Macfarlane,&Clark,2006).The ITS region is comprised by three sub-regions,ITS1 and ITS2 and the 5.8S gene in between,of which two(ITS1 and ITS2)show a higher rate of evolution and are typically species-specif ic(Bruns&Shefferson,2004;Kõljalg et al.,2005).Developments in molecular phylogeny studies for species delimitation based on DNA analysis allowed a greater resolution of taxonomically difficult oomycete species(Göker,García-Blázquez,Voglmayr,Tellería,&Martín,2009)and can uncover new species(Sandoval-Sierra,Martín,&Diéguez-Uribeondo,2014).In this study,isolation of ITSregion provided resolution down to species level which helped to depict the diversity of oomycetes in the sampled area.
The free-swimming zoospores of A.invadans initiates the infection and causes spread of the pathogen from host to host.The zoospores are released from the sporangium that is formed at the hyphal tip.The swimming zoospores attach on the host,encysts and germinate to develop f ilamentous aseptate hyphae invading and ramifying through cutaneous tissues to form ulcerative granuloma (Kiryu,Shields,Vogelbein,Kator,&Blazer,2003).However,it is difficult to identify A.invadans from water samples(Lilley et al.,2003;Willoughby et al.,1995)as it could be that the zoospores only germinate in presence of specif ic live hosts.In addition,Aphanomyces invadans are difficult to isolate from infected f ish mainly because of the fact that it is slowgrowing on agar plates compared to other saprophytic fungal species(not pathogenic)collected from infected f ish(Willoughby et al.,1995).Therefore,samplesfrom EUSinfected f ish can be easily overgrown with contaminating bacteria or other fungal species present in the water or on f ish during sample collection.Employing molecular assays to detect A.invadans directly from infected f ish(Vandersea et al.,2006)coupled to plating infected tissues on agar plates would be a more suitable approach to conf irm EUSinfections on f ish.
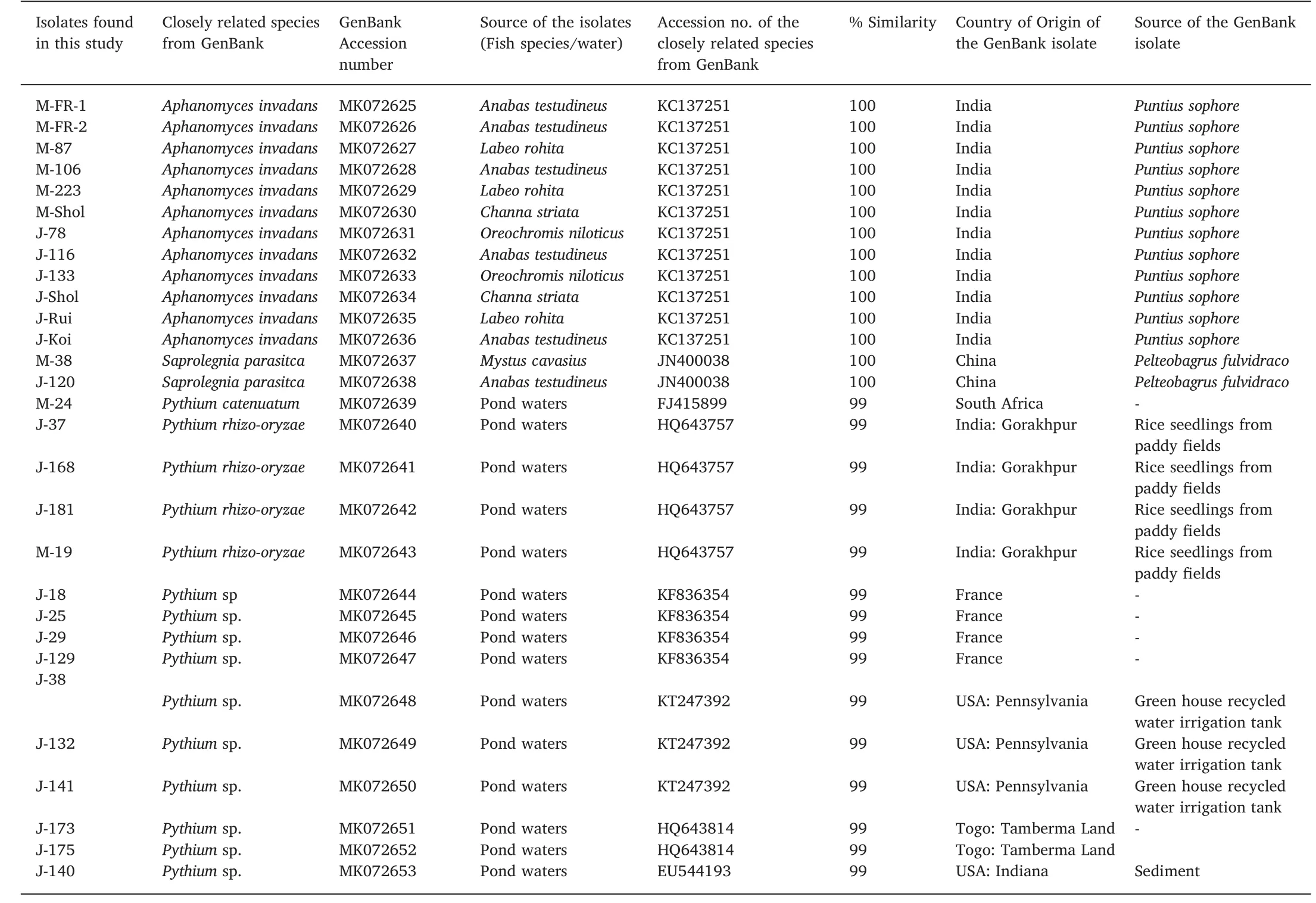
Table 2List(species,source,accession number and origin)of the isolate homologues available from GenBank.
The A.invandans isolates collected from the two geographical locations and from several different f ish species during the study appeared to be identical and are also identical to those present in the databases.Phylogenetic analysis reinforce that there is only a single clone of A.invadans broadly distributed worldwide(Lilley&Roberts,1997).Animal pathogenic Aphanomyces isolates spend energy to asexual reproduction(Diéguez-Uribeondo et al.,1994,2009)to disseminate quickly across the water column and this may explain the clonality observed in A.invadans across the different continents.Nonetheless,A.invadans appears to be the dominating oomycete f ish pathogen in the samples collected.
The phylogenetic relationship between the three lineages of the genus Aphanomyces appears to be very distinct based on ITS region.However,Diéguez-Uribeondo et al.,2009 placed few saprotroph species such as A.stellatus in the animal pathogenic clade and this could be an ongoing evolutionary trend which is not ref lected in the ITS region.Nonetheless,the Bayesian inference and maximum likelihood analysis based on ITSsequences showed that A.stellatus branches with A.laevis with high conf idence(Fig.3)grouping the animal pathogenic speciesin a separate branch.
Saprolegnia parasitica,a f ish pathogenic oomycete,was also recovered from f ish samples.The S.parasitica is one of the most devastating f ish pathogens in f ish farms(van West&Beakes,2014);and in Europe is second in economic importance to sea lice in salmonids(van den Berg et al.,2013).The genus Saprolegnia contains a number of pathogenic species to different freshwater f ish and eggs worldwide and it is responsible for at least 10%mortalities in salmon hatcheries and farms(Phillips,Anderson,Robertson,Secombes,&van West,2008).It wasa long-held belief that Saprolegnia was a minor secondary pathogen(Diéguez-Uribeondo et al.,2009),however in the recent years this view has been categorically rejected as it actively suppress host immunity during the infection process(Belmonte et al.,2014;de Bruijn et al.,2012;Minor et al.,2014;Wawra et al.,2012).In addition to S.parasitica,other speciessuch as S.diclina,S.australis,S.delica,S.ferax have been responsible for saprolegniosis in f ish and eggs,especially in salmonid aquaculture(Sandoval-Sierra et al.,2014).Saprolegnia spp.have multiple hosts and are capable of infecting different aquatic organisms such as crustaceansand amphibians(Sarowar et al.,2013).In thisstudy only two isolates were successfully obtained from infected f ish tissues.Unlike A.invadans,S.parasitica is relatively easy to grow from pathological tissues.Therefore,the lack of number of isolatessuggeststhat S.parasitica is not the most prevalent isolate in the study area.Several reports describe that different strains of S.parasitica vary signif icantly in pathogenicity(Thoen,Evensen,&Skaar,2011)and it could also be that the isolate strain was non-pathogenic hence resulted in very low infections.Nonetheless,our study provides a baseline for the diversity of animal pathogenic oomycetes in the study area.
This study has also found a number of Pythium isolates from different f ish farms that were solely recovered from water samples.The Pythium spp.are ubiquitousand present in almost every agricultural soil and they are known to be either plant pathogen or saprotrophs and to infect the root systems of various hosts reducing crop yield and quality in infected plants(Schroeder et al.,2013).The isolates found in our study are similar to P.catenulatum,P.rhizo-oryzae and a number of other Pythium sp.These were previously reported from farm soil,paddy f ield and greenhouse irrigation tanks from different parts of the world(Alcala et al.,2016).Most f ish farms and ponds studied were surrounded by crop f ields and Pythium spp.usually uses water routes to spread throughout the forests and crop f ields(Czeczuga,Mazalska,Godlewska,&Muszyńska,2005;Naznin,Hossain,Nasrin,Hossain,&Sarowar,2017).In fact,Pythium spp.has been isolated from moribund and dead f ish and crustaceans(Czeczuga,Kiziewicz,&Danilkiewicz,2002;Czeczuga,Kozłowska,&Godlewska,2002;Rahman&Sarowar,2016)but it has not been yet established if Pythium spp.are primary pathogens of f ish or of other aquatic organisms(Czeczuga et al.,2002).It might be that the traditional baiting technique resulted in the identif ication of Pythium spp.Since A.invadans zoospores are difficult to isolate from water samples and they grow slow on agar plates,Pythium spp.might have easily grown on the baits as it is prevalent in the study ponds.
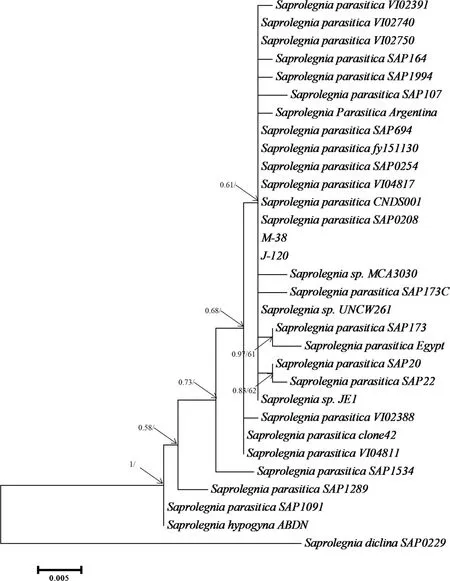
Fig.4.The f igure is a summary of the phylogenetic tree of a Bayesian inference and Maximum Likelihood analyses based on the internal transcribed spacer(ITS)regionsof nr DNA.The analysescomprise the Saprolegnia parasitica isolatesobtained in the study and closely related GenBank sequences.Isolates having Jor M in the beginning of the name indicate the region where they were recovered;M-Mymensingh,J-Jessore.Numbersin the branchesrepresent the posterior probability values and bootstrap support(values smaller than 50%not shown).
5.Summary and conclusion
The results suggests that f ish pathogenic oomycetes A.invadans and S.parasitica prevailed in f ish farms during the winter and this indicates that f ish mortality occurs due to EUSand saprolegniosis.Intense sampling could provide a clear idea on oomycetes diversity involved with f ish infections throughout the year and would be helpful to identify pathogenic isolates from f ishes that are commercially important.
Declarations of interest
None.
Acknow ledgements
The authors would like to express deepest sense of gratitude and sincere appreciation for the f inancial support of the International Foundation for Science,Sweden(IFS:A/5788-1)and COMSTECH as well as the Ministry of Science and Technology,Bangladesh(BS-57,2015-2016);TN acknowledges NST fellowship Ministry of Science and Technology,Bangladesh.The authors would also like to acknowledge the contribution of the Head of the department,Department of Fisheries Biology and Genetics,and Central Laboratory,Bangladesh Agricultural University for providing equipment and technical supports during the research.
 Aquaculture and Fisheries2019年3期
Aquaculture and Fisheries2019年3期
- Aquaculture and Fisheries的其它文章
- Waste production in aquaculture:Sources,componentsand managements in different culture systems
- The potential of Hoplias malabaricus(Characiformes:Erythrinidae),a Neotropical carnivore,for aquaculture
- Prokaryotic expression of goldf ish Tgf2 transposase with optimal codonsand its enzyme activity
- Enhancing the immune response in the sea cucumber Apostichopus japonicus by addition of Chinese herbs Houttuynia cordata Thunb as a food supplement
- Estimating blue marlin(Makaira nigricans)sustainable yield in the Indian Ocean using a data-poor approach
