Prokaryotic expression of goldf ish Tgf2 transposase with optimal codonsand its enzyme activity
Xi Zho,Ruirui Si,Mingjun He,Xiyun Jing,b,Shuming Zou
a College of Food Science and Technology,Shanghai Ocean University,Shanghai,201306,China
b Geneticsand Breeding Center for Blunt Snout Bream,Key Laboratory of Freshwater Aquatic Genetic Resources,National Demonstration Center for Experimental Fisheries Science Education,Shanghai Ocean University,Shanghai,201306,China
Keywords:Codon optimization Enzyme activity Prokaryotic expression Tgf2 transposase
A B S T R A C T Tgf2 transposase(Tgf2-TPase),a hAT transposase from goldf ish,plays an important role in f ish transgenic applications.Previously,the production of the recombinant Tgf2-TPase protein required rigorous fermentation at low temperatures(22°C)and early log phase induction(OD600=0.3-0.4)in Rosetta 1(DE3)Escherichia coli lines.In order to better expressthe Tgf2-TPase and detect its enzyme activity,83 rare codons in Tgf2-TPase were optimized and designated Tgf2-TPase83.The expression results showed that the soluble recombinant Tgf2-TPase83 was highly expressed at 30°C and was inducible at an OD600 of 0.5-0.6 in the same prokaryotic expression system.After purif ication by affinity chromatography,Tgf2-TPase83 with codon optimization had higher enzyme activity than the Tgf2-TPase control.Comparison of different preservation methods(freezedrying at-80°C,storage in 20%-glycerol,8%-sucrose,4%-mannitol),revealed storage of Tgf2-TPase83 in glycerol helped to preserve its DNase digestion activity.Furthermore,size exclusion chromatography suggested that the purif ied Tgf2-TPase83 could recognize and bind to DNA probes containing a terminal inverted repeat(TIR)and a subterminal repeat(STR)sequence of the Tgf2 transposon.Overall,the results showed that optimizing the 83 codons of Tgf2 transposase can simplify the fermentation process and improve the enzyme activity.We propose that the production of the Tgf2-Tpase83 protein in a soluble and active form could provide an alternative tool for genetic modif ication of f ish.
1.Introduction
Transposable elements(TEs),known as“jumping genes”(Panaud,2016),are DNA sequences that can move from one locus to another in the genome and are widely distributed in many species including microorganisms,plants,and animals(Janicki,Rooke,&Yang,2011).TEs can be divided into DNA and RNA transposons according to their transposition mechanism(Benjak,Forneck,&Casacuberta,2008;Kempken&Windhofer,2001).DNA transposons comprise at least 17 superfamilies such as hAT(Atikinson,2015;Rubin,Lithwick,&Levy,2001),CACTA(Zabala&Vodkin,2005),piggybac(Rad et al.,2010),Mutator(Hua-Van&Capy,2008),PIF/Harbinger(Grzebelus,Yau,&Simon,2006),Tcl/Mariner(Ivics,Hackett,Plasterk,&Izsva´k,1997),Transib(Chen&Li,2008),Merlin(Feschotte,2004),and so on.Apart from their impact on genome organization and evolution,transposable elements might serve as valuable biological tools for genetic manipulation(Abe,Suster,&Kawakami,2011;Calvi,Hong,Findley,&Gelbart,1991;Feschotte&Pritham,2007).The hAT(hobo/Ac/Tam3)is an ancient superfamily that is widespread in animals and plants(Calvi et al.,1991).However,most hAT transposons are inactive due to vertical inactivation during evolution.Tol2 and Tgf2,piscine hAT transposons,from medaka and goldf ish,respectively,are autonomous hAT transposons in vertebrates,and encode intact and active transposases(Atikinson,2015;Jiang et al.,2012;Kawakami,Shima,&Kawakami,2000).
The coding region of the goldf ish Tgf2 transposase is 2061 bp in length(Arensburger et al.,2001;Cheng,Jiang,Tian,Chen,&Zou,2014).Direct injection of Tgf2-TPase proteins into a donor harboring the Tgf2 cis-element(through co-injection into embryos),enhances insertion efficiency in aquaculture f ish species(Shibano et al.,2007;Tafalla,Estepa,&Coll,2006).However,rigorous fermentation at low temperatures(22°C)and early log phase(OD600=0.3-0.4)induction in Rosetta 1(DE3)E.coli harboring an expression construct of Tgf2 transposase are required to produce the recombinant Tgf2-TPase protein.Piscine Tgf2-TPase contains several rare codons,and optimization of expression could improve the production conditionsand increase the Tgf2-TPase activity(Liu&Li,2016;Zhao,Huo,&Li,2000).
In the present study,83 rare codons in Tgf2-TPase83were replaced synonymously taking into account the codon degeneracy and preference of E.coli.We found that the modif ied Tgf2-TPase83can be expressed under less stringent conditions and higher purity(>85%)of the protein is possible after purif ication by affinity chromatography.Furthermore,the DNase activity,DNA-binding activity,and specif icity of Tgf2-TPase83were investigated in vitro to evaluate the properties of the modif ied enzyme.Our study is aimed at reconstructing a more effective transposon system based on Tgf2-TPase83.
2.Materials and methods
2.1.Materials
Materials:Plasmid p ET28a(+),E.coli cells(DH5α),and BL21(DE3)were all produced in our laboratory.Isopropyl-β-D-thiogalactopyranoside(IPTG),kanamycin sulfate,glycerol,sucrose,mannitol,and other reagentswere provided by Sinopharm Chemical Reagent Co.(Shanghai,China).p CS2-EF1α-EGFP:6156bp,driven by the Xenopus EF1a promoter was constructed.pTgf2-EF1α-EGFP:5476bp,containing 220 bp and 185 bp of the 5′and 3'ends of the goldf ish Tgf2 transposon and driven by the Xenopus EF1a promoter was constructed.
Asthe 5′end in the transposon hasa higher binding affinity than the 3'end,four 50-bp DNA probes from the left arm of Tgf2(GenBank Accession No.HM146132,4720 bp)(as shown in Fig.3,PH(5′-CAG AGGTGTAAAAGTACTTAAGTAA TTTTACTTGATT ACTGTACTTA AGT-3′),PM1(5′-TTTTTACTTTACTTGAGTA CAATTAA AATCAATACT TTTATTTTTACTT-3′),PM2(5′-TAGAAAAAAAGTA CTTTTTACTCCTTA CAATTTTATTTACAGTCAAAAAG-3′),and PL(5′-CCTTG CTATTACCA AACCAATTGAATTGCGCTGATGCCCAGTTTAAT TTA-3)were designed for a DNA-binding assay.Oligonucleotides were synthesized,purif ied by PAGE(Sangon Ltd,Shanghai China),and annealed with their complementary sequences.
2.2.Codon optimization of Tgf2-TPase83
Tgf2-TPase(GenBank Acc.No.JN886586,2061bp)encodes 686 amino acids and consists of four open reading frames.Using the GenScript Rare Codon Analysis Tool(https://www.genscript.com/tools/rare-codon-analysis),83 rare codons in Tgf2-TPase were replaced so they complied with the codon preference in E.coli.The new cDNA,designated Tgf2-TPase83,had a codon adaption index of 0.86,a GC content of 52.96%,and a codon frequency distribution of 0.
2.3.Prokaryotic expression of Tgf2-TPase83
The 2061-bp sequence of the Tgf2-TPase83coding region wascloned into a p UC57-simple vector(TaKaRa,Dalian,China).The plasmids pUC57-simple-Tgf2-TPase83and p ET28a(+)(Merck,Shanghai,China)were digested with BamH I and Xho I(TaKaRa,Dalian,China).Linear fragments of Tgf2-TPase83cDNA and p ET28a(+)were gel-purif ied and ligated using T4 DNA ligase(TaKaRa,Dalian,China)to generate the expression vector p ET28a(+)-Tgf2-TPase83.Then,the recombinant plasmid p ET28a(+)-Tgf2-TPase83was transformed into BL21(DE3)competent cells(Merck,Shanghai,China).Positive transformants were PCR-detected and sequenced.
For protein expression,BL21 (DE3)cells harboring pET28a(+)-Tgf2-TPase83were incubated at 30°C and induced for 6 h with 1.0 mmol/L IPTG when the OD600=0.5-0.6.The cells collected by centrifugation(12,000 g,15 min,4°C)and analyzed by SDS-PAGE gel electrophoresis(Xu et al.,2015).
2.4.Purif ication of Tgf2-TPase83
The main procedures for purif ication of Tgf2-TPase83were as follows:ultrasonication for 45 min(power 100W,breaking 3s,interval 6s),followed by centrifugation(12,000 g,20 min,4°C)and then f iltration(0.22μm PESmembrane)of the supernatant.The Tgf2-TPase83f iltrate was loading onto a HisTrapTM-HP affinity column(Xu et al.,2015).The recombinant protein was purif ied by f irst equilibrating the HisTrapTM-HP column with 20 mmol/L phosphate,0.5 mol/L sodium chloride,20 mmol/L imidazole,p H 7.4,followed by binding buffer(20 mmol/L phosphate,0.5 mol/L sodium chloride,40 mmol/L imidazole,p H 7.4),and washing buffer(20 mmol/L phosphate,0.5 mol/L sodium chloride,500 mmol/Llmidazole,p H 7.4)at a f low rate of 1 mL/min using a linear gradient within 5 column volumes(Xu et al.,2015).Based on ultraviolet absorption values(OD280),the target protein was collected from the affinity column and then analyzed by SDS-PAGE(12%gel)followed by Coomassie brilliant blue staining(Zhou,Liu,&Li,2002).
2.5.Identif ication of Tgf2-TPase83
The protein band corresponding to Tgf2-TPase83on the SDS-PAGE gel was conf irmed by analysis on an ABI 4800 plus MALDI(TOF)/TOF(Applied Biosystems,Foster City,USA)and MS/MSion searches using Mascot(Matrix Science Ltd,London,UK).MALDI(TOF)/TOF analysis identif ied the recombinant Tgf2-TPase83amino acid fragment sequence using the relationship between f light time and quality(Xu et al.,2015).
The procedure for MALDI(TOF)/TOF analysis was as follows:the sample was washed twice with water and destain solution.Then it was incubated with DTT and IAM at 56°C for 1 h to denature the protein.Acetonitrile was added to the reaction mixture to dehydrate colloidal particles,so that the colloidal particlesabsorbed trypsin and the protein was digested into peptides by incubating at 37°C overnight.The reaction was stopped by adding 0.1%formic acid(FA).The mixture was analyzed using a mass spectrometer.
2.6.DNase activity determination of Tgf2-TPase83
The 125-ng p Tgf2-EF1α-EGFP plasmid was incubated with 6-ng dissolved-enzyme solution in a 20-μL volume of reaction buffer(25 mmol/LHEPES,50 mmol/LNaCl,1 mmol/LMnCl2,2 mmol/LDTT,2%glycerol,0.1 mg/mL BSA,p H 7.6)at 30°Cat various times(0,1,2,3,4,and 5 h).Reactions were stopped by adding 1%SDSand 20 mmol/L EDTA.Analysis of the reaction products was by agarose gel electrophoresis(1.2%,AGE)(Masek,Vopalensky,Suchomelova,&Pospisek,2005;Shibano et al.,2007).
2.7.Effect of different storage methods on Tgf2-TPase83 activity
Polyols and sugars can effectively increase protein stability,and four different storage methodswere tested to investigate their effectson Tgf2-TPase83activity:(1)freeze-drying to a powder;(2)20%-glycerol solution;(3)4%-mannitol solution;(4)8%-sucrose solution.The activity of the freshly purif ied enzyme was used as a control.All storage solutions containing the Tgf2-TPase83were stored at-80°C(Liao,Brown,Quader,&Martin,2002).
2.8.DNA-binding activity and specif icity assay of Tgf2-TPase83
The DNA-binding activity of the recombinant Tgf2-TPase83was evaluated by size exclusion chromatography on a Superdex 200 10/300 GL glycan chromatographic column(Hickman et al.,2005;Jiang et al.,2016;Shibano et al.,2007;Zhang,Dawson,&Finnegan,2001).Tgf2-TPase83was mixed with the probe PH,incubated for 20 min at 25°C,and then analyzed.Assessment of the change in the position of the protein elution(at OD280,OD260)after passage through the column was used to determine the DNA-binding activity of Tgf2-TPase83(Carpentier et al.,2011;Hickman et al.,2005;Jiang et al.,2016).Liquid containing the fresh enzyme was used as a blank control,and a mixture of egg white lysozyme(14.4 kDa,Sinopharm Chemical Reagent Co.,Shanghai,China)and the probe was used as a negative control.Washing buffer:0.5 mol/L NaCl phosphate buffer at a rate of 0.5 mL/min(Qu,Li,Wu,&Yang,2005).
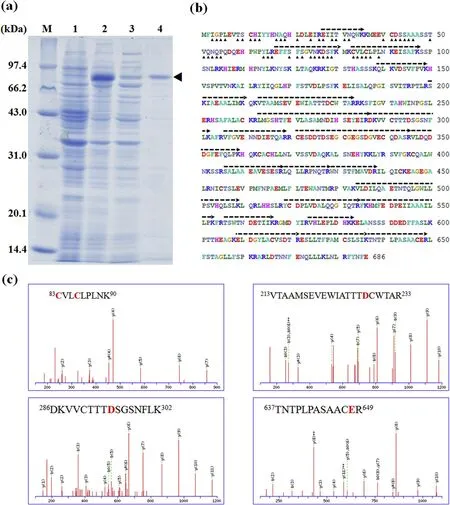
Fig.1.Purif ication and identif ication of the recombinant protein Tgf2-TPase83 with optimized genetic codons for E.coli.(a)SDS-PAGEanalysis of Tgf2-TPase83.M,standard molecular marker;1,without IPTG;2,with IPTG;3,supernatant solution after ultrasonication;4,eluate from the Ni2+-affinity column.(b)The amino acid sequence of Tgf2-TPase83.The black dotted line indicates the amino acid sequences detected by MS/MSspectra.The black triangles indicate the amino acids that were optimized.(c)MS/MSspectra of some peptide fragments(c)identif ied.C,D,E correspond to the DNA binding(C)and catalytic domain(D,E),respectively.
To assess DNA-binding specif icity(Feschotte,Osterlund,Peeler,&Wessler,2005),four different 50-bp DNA probes from the left arm of Tgf2 were designed(Fig.3).The f irst probe PHcontains six repeat sequences and was named the high repeat sequence.The second probe PM1contains three repeat sequences and was named the middle repeat sequence.The third probe PM2contains three repeat sequencesand was named as the middle repeat sequences.The last probe PLhas no repeat sequence and was named the low repeat sequence.
3.Results and discussion
3.1.Expression,purif ication and identif ication of Tgf2-TPase83
The recombinant Tgf2-TPase83isolated by Ni2+-affinity chromatography was analyzed by SDS-PAGE.As shown in Fig.1a,the recombinant protein was induced by IPTG and purif ied by affinity chromatography.Image analysis of SDS-PAGE gels showed that the recombinant protein accounted for 6.3%of the total protein(lane 2 in Fig.1a)and the purity of the recombinant protein was>85%after purif ication by affinity chromatography(lane 4 in Fig.1a).In our previous results,the recombinant protein(without codon optimization)accounted for 4.2%of the total protein and the purity of the recombinant protein was 71%(Si,Zhao,Lu,Zou,&Jiang,2017),indicating that our study using codon usage optimization for E.coli increased the expression volumes and purity of Tgf2-TPase83production.
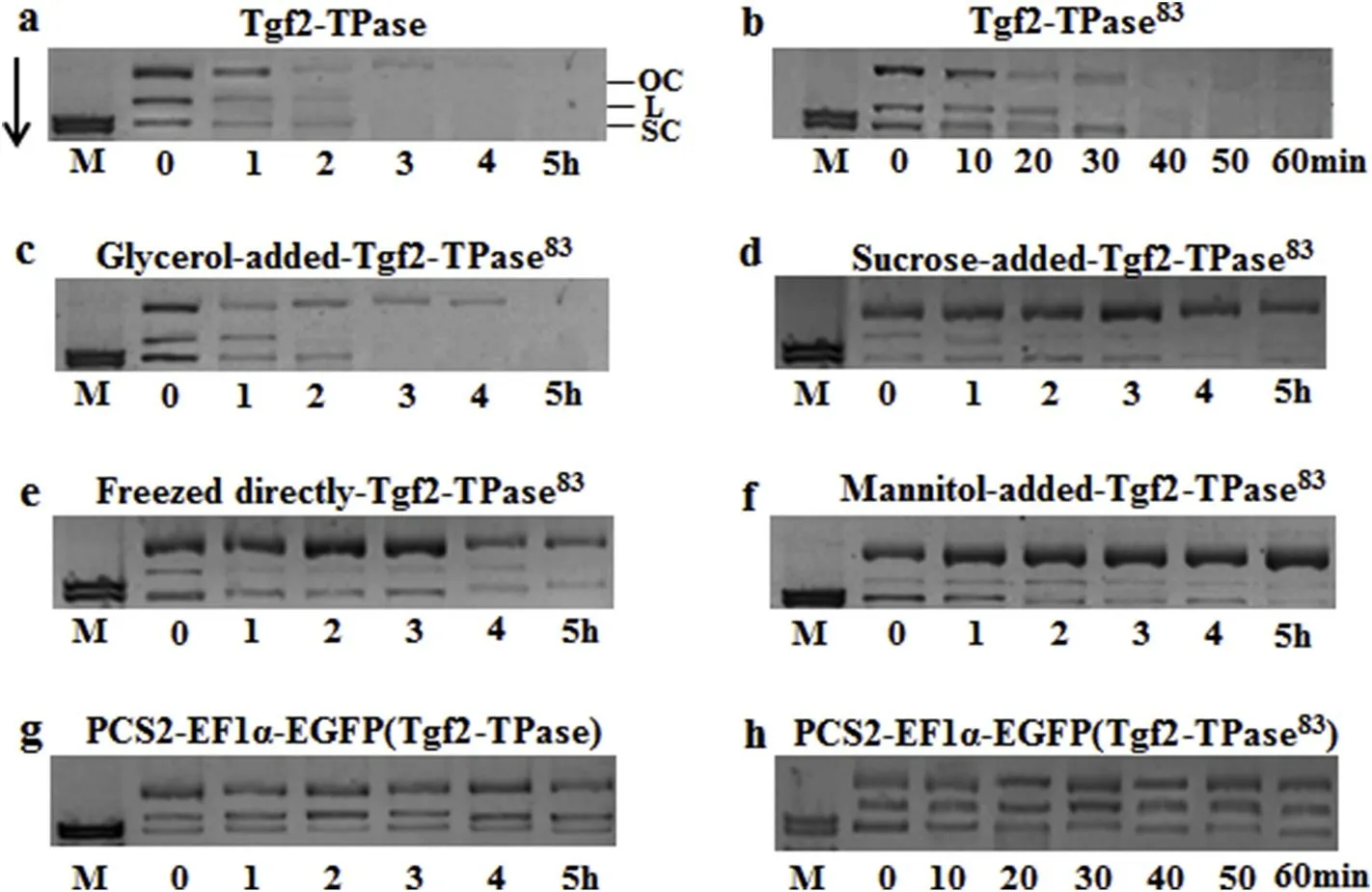
Fig.2.DNase activity analysis of Tgf2-TPase and Tgf2-TPase83 after storage under different storage conditions. The 200 ng/μL p Tgf2-EF1α-EGFP plasmid and 200 ng/μL enzyme solution were incubated at 30°C and samples removed at different time points.The products of the DNase reactions were analyzed by 1.2%agarose gel electrophoresis.Molecular weight markers are indicated on the left of the agarose gel.OC:open circular product;L:linear donor;SC:supercoiled donor.(a)The fresh Tgf2-TPase enzyme(control)was analyzed.(b)The fresh Tgf2-TPase83 enzyme was analyzed after storage under various conditions..(c)20%glycerol storage.(d)8%sucrose.(e)frozen without preserving solution.(f)4%mannitol.(g)The fresh Tgf2-TPase enzyme with PCS2-EF1α-EGFP plasmid was analyzed.(h)The fresh Tgf2-TPase83 enzyme with PCS2-EF1α-EGFP plasmid was analyzed.Arrow shows the electrophoretic direction.
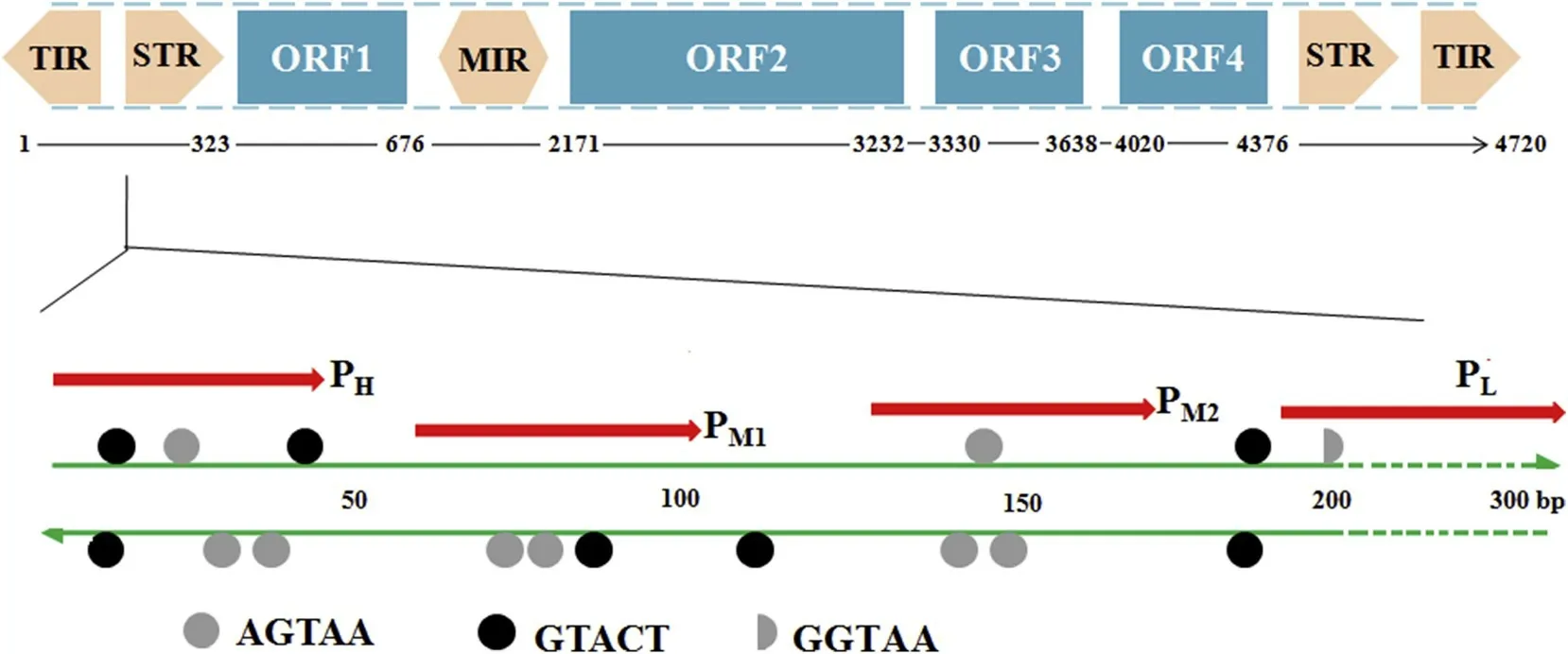
Fig.3.The location of the four DNA probes on the left end of Tgf2 transposon.PH:high repeat sequences(six repeat sequences);PM1,PM 2:middle repeat sequences(three repeat sequences);PL:low sequence sequences(no repeat sequences).
The band corresponding to the recombinant protein was cut from the SDS-PAGEgel and subject to MALDI(TOF)/TOFanalysis.After MS/MS ion searching using Mascot,at least 21 peptide fragments were detected that matched the putative transposase from goldf ish,(AFC96942.1)as depicted in Fig.1b.Several oligopeptides are displayed in Fig.1c.The peptide fragment(83-90 amino acid residues)included two conserved cysteines(C83 and C86)located in the Nterminal BED zinc f inger domain involved in DNA binding of the Tgf2 transposase(Yant,Park,Huang,Mikkelsen,&Kay,2004).A catalytic triad and residuesof the DDE(D228,D295 and E648)in the C-terminus RNase-H domain was also identif ied in the fragments(213-233 amino acid residues,286-302 amino acid residues,637-649 amino acid residues)(Michel,O'Brochta,&Atkinson,2003).Therefore,the recombinant Tgf2-TPase83protein was successfully expressed in BL21(DE3)and MALDI-TOF conf irmed the protein was goldf ish Tgf2 transposase(Xu et al.,2015).
In this study we optimized rare codons and developed an effective system for prokaryote expression of goldf ish Tgf2-TPase83.The expression plasmid pET28a(+)-Tgf2-TPase83in BL21(DE3)E.coli host cells successfully produced a recombinant.
Tgf2-TPase83protein of~80 kDa.Successful expression and purif ication of Tgf2-TPase83wasconf irmed by MALDI(TOF)/TOFanalysis.
3.2.DNase activity of Tgf2-TPase83
Asa hAT element,Tgf2 transposase catalyzes DNA transposition in a cut-and-paste mode as follows:(1)pairing of transposases by dimerization or oligomerization,(2)DNA-specif ic binding of the transposase to the transposon end,(3)cleavage of DNA strands at the ends of the transposon,and(4)capture of a new locus and integration into target DNA.As DNA cleavage is a crucial step in transposition,its DNase activity was chosen to evaluate transposition activity(Carpentier et al.,2011;Cheng et al.,2014;Jiang et al.,2012).
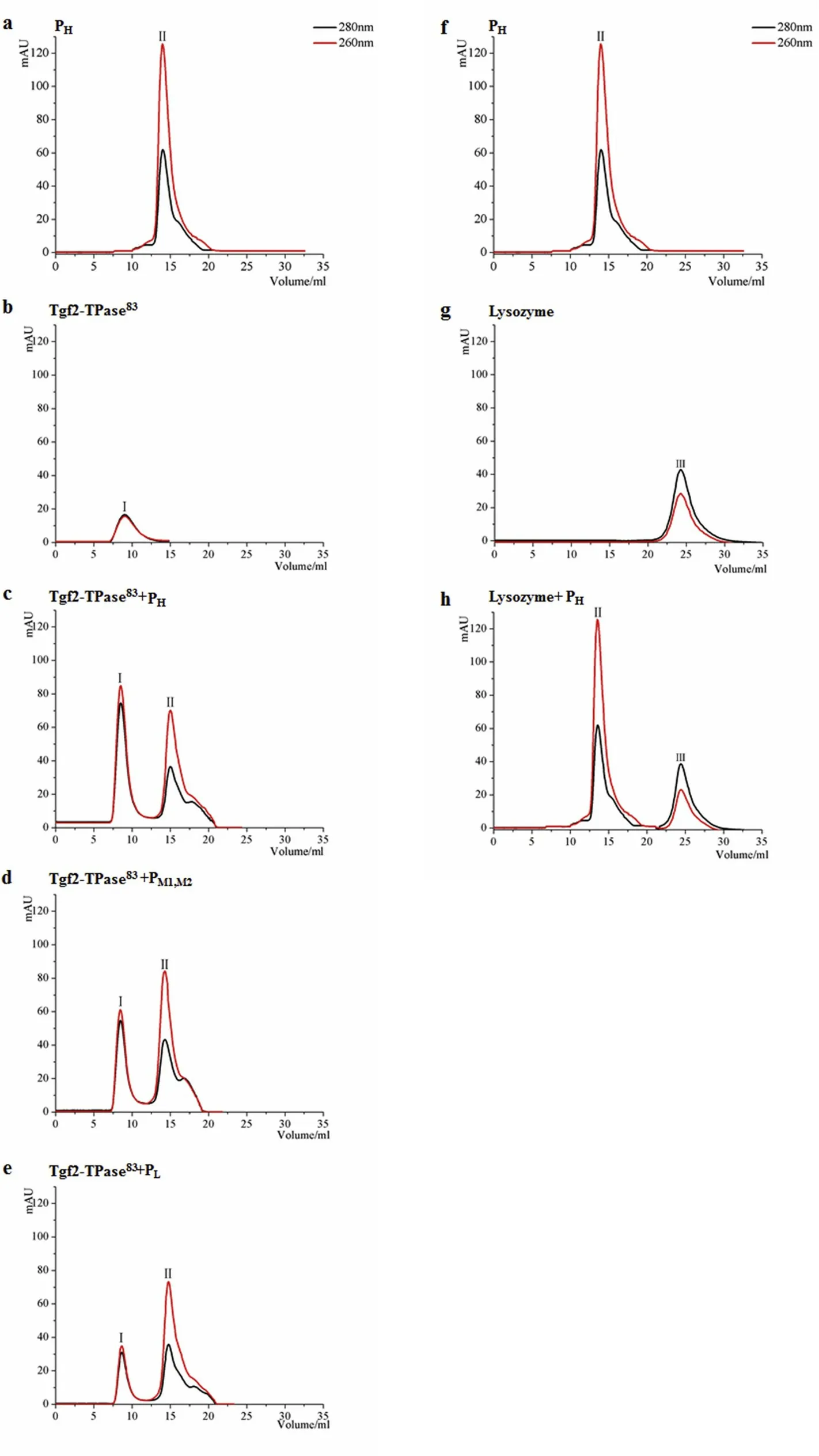
Fig.4.DNA-binding activity and specif icity of Tgf2-TPase83 analyzed by size exclusion chromatography.(a,f)Elution prof iles of the PH double-stranded DNA probe alone.(b)Elution prof iles of Tgf2-TPase83 alone.(c)A mixture of Tgf2-TPase83 and PH.(d)A mixture of Tgf2-TPase83 and PM1or PM2.(e)A mixture of Tgf2-TPase83 and PL.(g)Elution prof iles of lysozyme alone.(h)A mixture of lysozyme and PH.Based on the elution time,peak I corresponds to the Tgf2-TPase83 and peak II corresponds to probe PH,peak III presents the lysozyme.
Comparison of the DNase activity of Tgf2-TPase and Tgf2-TPase83was established by incubation of the proteins with the plasmid pTgf2-EF1α-EGFPcontaining the left and right arms of Tgf2 in buffer at 30°C.The disappearance of reactants across time was visualized by agarose gel electrophoresis(Fig.2).Cleavage of plasmids by Tgf2-TPase83occurred within 60 min,and nearly 100%of the DNA including open circular,linear,and supercoiled forms were completely digested within 40 min(Fig.2b).However,cleavage of the plasmid p Tgf2-EF1α-EGFP by Tgf2-TPase took up to 5 h although nearly 100%of the DNA(circular,linear,and supercoiled)was completely digested after 4 h incubation(Fig.2a).The Tgf2-TPase83with codon optimization had higher enzyme activity than the Tgf2-TPase control.Freshly-made Tgf2-TPase and Tgf2-TPase83incubated with the plasmid p CS2-EF1α-EGFP without the left and right arms of Tgf2 had no DNase activity(Fig.2g and h).
Four common methods for protein preservation were investigated using recombinant Tgf2-TPase83,and the results are displayed in Fig.2c-f.In the 20%-glycerol treatment,nearly 100%of the DNA,including supercoiled and linear forms,was completely digested in less than 3 h,whereas the open circular form required just over 4 h for complete digestion(Fig.2c).The preserved Tgf2-TPase83enzyme in 20%glycerol had a decreased activity that was similar to the control(Tgf2-TPase).Of the various concentrations of glycerol tested(10%,20%,30%,40%,50%),20%glycerol was the best choice for preservation of Tgf2-TPase83activity(data not shown).Therefore,all the above results suggest that adding 20%glycerol to recombinant Tgf2-TPase83at-80°Cwas helpful for protein stability.In previous studies,DNase activity was widely used to evaluate protein activity such as mariner,pacmmer and Tol2(Carpentier et al.,2011;Delauriere et al.,2009;Shibano et al.,2007).We found that DNase activity was more stable when the recombinant Tgf2-TPase83was stored in 20%glycerol.
3.3.Tgf2-TPase83 DNA-binding activity and specif icity to Tgf2 transposon
The goldf ish Tgf2 transposon is 4720 bp in length and consists of four open reading frames,TIR(terminal inverted repeat),STR(subterminal repeat)and MIR(middle inverted repeat)(Fig.3).Among hAT family members,there is signif icant diversity of sequence,number,spacing,and orientation of small repeat sequences.Tgf2,together with Tol2,appears to contain more small repeats,whereas Herms carries the lowest number(Michel et al.,2003).Subsequent analysisof the TIRand STR regions using gel retardation assays with a combination of recombinant Tgf2-TPase83transposase and DNA oligomers containing repeats of 5′AGTAA3′,5′GTACT3′and 5′GGTAA3′was carried out in the present study.Four groups of probes were designed,PH,PM1,PM2,and PL,denoting incorporation of high,medium,and low numbers of small repeats,as shown in Fig.3.To conf irm the in vitro DNA-binding activity of Tgf2-TPase83,the size exclusion chromatography elution prof iles of Tgf2-TPase83were investigated,as shown in Fig.4.Each probe exhibited a UV prof ile characteristic of nucleic acids(peak II,~15 min)with an OD260higher than the OD280(Fig.4a,f),whereasthe OD260and OD280value of Tgf2-TPase83(peak I,~9 min)was characteristic of protein(Fig.4b).
When Tgf2-TPase83and PHwere mixed and incubated at 25°C for 20 min,and then eluted on size exclusion chromatography,the peak of OD characteristic of the proteins was almost lost(peak I(Fig.4c)and a shift to a peak with OD characteristics of nucleic acids was evident.The modif ication in the elution prof ile on size exclusion chromatography when Tgf2-TPase83was mixed with PHrevealsits DNA binding activity,which was identical to the Hermes transposases(Hickman et al.,2014).When Tgf2-TPase83and PM1,PM2,or PLwere mixed,the results were similar to that obtained with PH(Fig.4d and e).These data indicate Tgf2-TPase83has DNA-binding activity with various repeat sequences at the left end of Tgf2.Furthermore,our data suggested that Tgf2-TPase83had different DNA-binding activities with PH,PM1,PM2,and PL.The transposon terminal TIR and STR sequences both appeared essential for transposase DNA-binding,and the upstream repeat sequences seemed to have greater binding affinity than the downstream repeat sequences,probably due to the higher number of small repeats,or more specif ic binding information(Fig.3).That is,the more repeats in the transposon terminal regions,the more sensitive the DNA-binding activity of Tgf2-TPase83(Fig.4c-e)(Feschotte et al.,2005;Hickman et al.,2014;Yant et al.,2004;Zhang et al.,2001).Our detailed research on the relationship of TIR,STR,and transposase provides insight into transposition,which may facilitate future technological application.
Lysozyme(~14.4 kDa)was used as a negative control to establish the specif icity of goldf ish Tgf2-TPase83binding to DNA.The DNA probe PHwas mixed with lysozyme and incubated as indicated above.The elution prof ile is shown in Fig.4h.No obvious shift in UV absorption was observed,compared to peak II in Fig.4f and peak III(~25 min,OD280>OD260)in Fig.4g.The results indicate that lysozyme has no DNA-binding activity.In conclusion,our results suggest that recombinant Tgf2-TPase83had DNA-binding activity to the left arm of Tgf2.
The goldf ish Tgf2 transposon was identif ied and is the second autonomous transposon isolated in f ish after the medaka Tol2 transposon(Cheng et al.,2014;Zou,Du,Yuan,&Jiang,2010).The Tgf2-TPase83was expressed in E.coli and tested for its ability to recognize and bind 50 bp DNA sequencescorresponding to the TIRand STRof itsrespective transposon.The experiments in the present study revealed that the Tgf2-TPase83binds to TIR and STRsequences from its own transposon.Furthermore,this DNA binding was specif ic to Tgf2-TPase83and was similar to the previously reported rice marine-like transposases(Feschotte et al.,2005).
4.Conclusions
In summary,Tgf2-TPase codons were optimized for E.coli,and expressed and the results revealed that optimizing 83 rare codons improved the yield and purif ication of goldf ish Tgf2-TPase83.Furthermore,20%glycerol improved stability of Tgf2-TPase83on storage.The purif ied goldf ish Tgf2-TPase83had better DNA binding activity,and we propose it may be a helpful tool for the development of f ish transgenesis in the future.
Acknow ledgements
This work was supported by the National Science Foundation of China (31572220,31272633,31201760);the National High Technology Research and Development Program of China(863 Program)(2011AA100403);and the Shanghai University Knowledge Service Platform(ZF1206).
 Aquaculture and Fisheries2019年3期
Aquaculture and Fisheries2019年3期
- Aquaculture and Fisheries的其它文章
- Waste production in aquaculture:Sources,componentsand managements in different culture systems
- The potential of Hoplias malabaricus(Characiformes:Erythrinidae),a Neotropical carnivore,for aquaculture
- Molecular identif ication of oomycete species affecting aquaculture in Bangladesh
- Enhancing the immune response in the sea cucumber Apostichopus japonicus by addition of Chinese herbs Houttuynia cordata Thunb as a food supplement
- Estimating blue marlin(Makaira nigricans)sustainable yield in the Indian Ocean using a data-poor approach
