Studies on a Series of Coumarin Derivatives for Anticancer Activity against Breast Carcinoma Cell Line MCF-7 and Their Molecular Design
YANG Qi ZHANG Shu-Ping JIANG Yan
Studies on a Series of Coumarin Derivatives for Anticancer Activity against Breast Carcinoma Cell Line MCF-7 and Their Molecular Design
YANG Qi ZHANG Shu-Ping①JIANG Yan
(200093)
Breast cancer is one of the most common malignant tumors in women, and is also the focus of researchers. In this article, 3-QSAR (three-dimensional quantitative structure-activity relationship) was performed on 24 molecules which are a series of coumarin derivatives for their anticancer activity. Our team divided these compounds randomly into the training and test sets to build the CoMFA (comparative molecular field analysis)andCoMSIA (comparative molecular similarity index analysis) models. The coefficients of cross-validation2and non cross-validation2for CoMFA model were 0.684 and 0.949, and 0.579 and 0.930for the CoMSIA model, respectively. The result demonstrates that the model has strong stability and satisfactory predictability. 3contour maps suggest that the electrostatic factor has the greatest impact on activity followed by the H-bonding acceptor and hydrophilic factors. Takingthe above results into account, we designed several molecules with high anticancer activity against breast carcinoma cell line MCF-7.
3D-QSAR, CoMFA, CoMSIA, anticancer, coumarin;
1 INTRODUCTION
Breast cancer is the most common cancer among women in the world, and it is the main cause of death in both developed and developing countries[1, 2].Some progress has been made in the treatment of breast cancer[3-5], however, the incidence and mortality of breast cancer are still high[6, 7]. Coumarin is a benzopyrone derivative with anti-bacterial, anti-inflammatory, anti-oxidation and anti-tumor and other biological activities, and is distributed widely in the umbelliferae and rutaceae plants[8]. Therefore, scientists have designed and synthesized a number of coumarin derivatives with anticancer activity[9-11]. These compounds have a great effect on the treat- ment of breast cancer[12]. However, in recent years, with the development of computational chemistry, especially the emergence of quantitative structure- activity relationship (QSAR), new ideas have been provided for the design of new drug molecules[13]. 3D-QSAR is a mathematical way that quantitatively analyzes the relationship between drug chemistry information and its biological activity to find out the rule between structure and activity, and then predicts unknown compound properties based on the law and structure of the compound[14, 15].The three-dimen- sional quantitative structure-activity relationship (3D-QSAR) models, calculated via the most widely used comparative molecular field analysis (CoMFA) and comparative molecular similarity indices analy-sis (CoMSIA). The aim of CoMFA is to derive a correlation between the biological activity of a set of molecules and their 3D shapes, electrostatic and hydrogen bonding characteristics. CoMSIA is a methodology similar to CoMFA. The difference is that the factors of the steric field, electrostatic field, hydrophobic field, hydrogen-bond donor field and hydrogen-bond acceptor field were taken into account in CoMSIA. Based on the above, researchers have designed some new molecules with satisfactory results by using CoMFA and CoMSIA[16-19]. In this article, 3-QSAR model has been performed on 24 molecules which area series of coumarin derivatives for their anticancer activities by CoMFA and CoMSIA.
2 MATERIALS AND METHODS
2. 1 Preparation of data set
24 coumarin derivatives as inhibitor of MCF-7 which were derived from the work of Luo G team were randomly divided into the training sets (20 molecules) and test sets (4 molecules) in this study[12]. The structure of a series of coumarin derivatives were shown in Fig. 1. The training sets were used to construct CoMFA and CoMSIA models, and the test sets were used to validate the models. The structures of all compounds and their values of inhibitory concentrations (PIC50) are shown in Table 1.
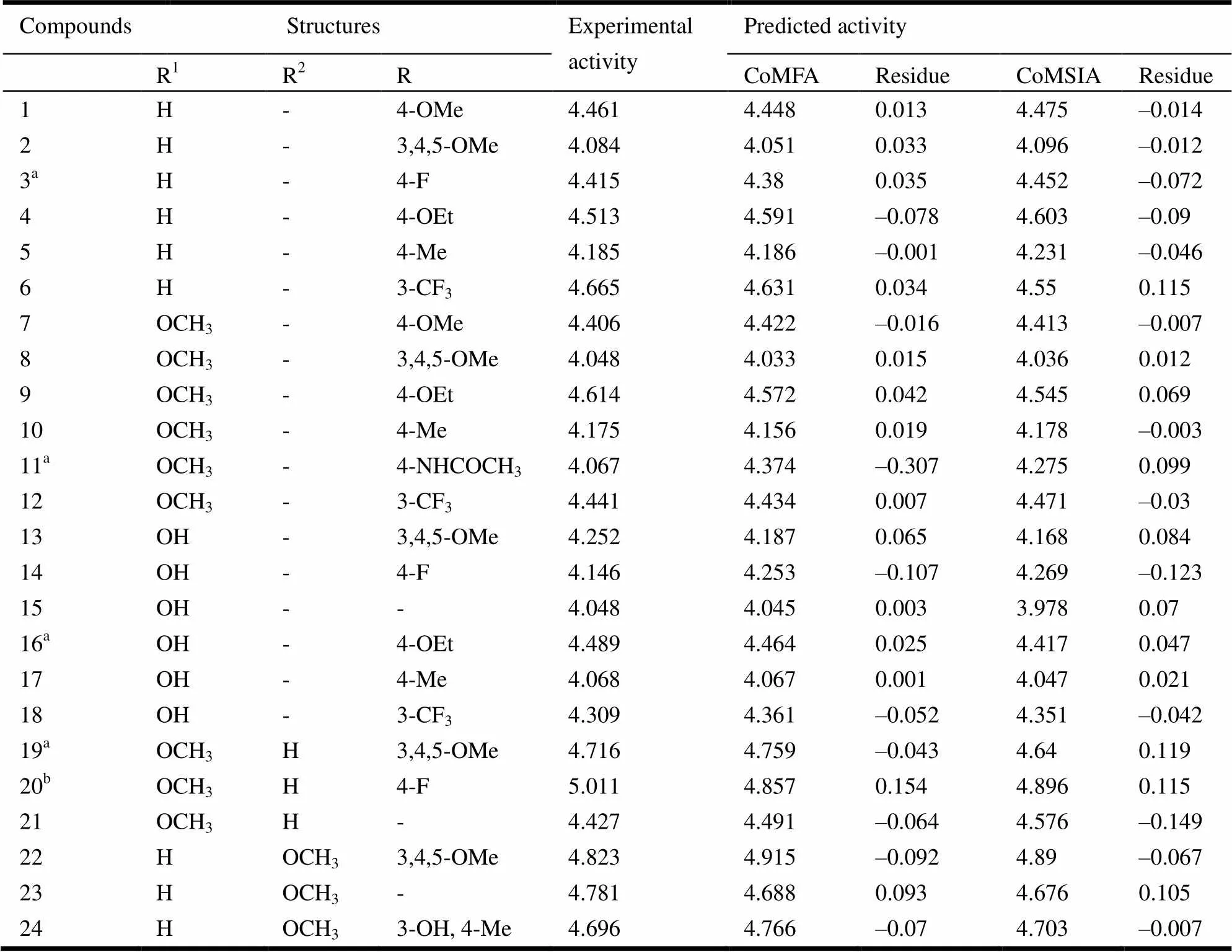
Table 1. Indoleamide Derivatives and Their Experimental and Predicted Activities
a Test set molecules, bTemplate molecule
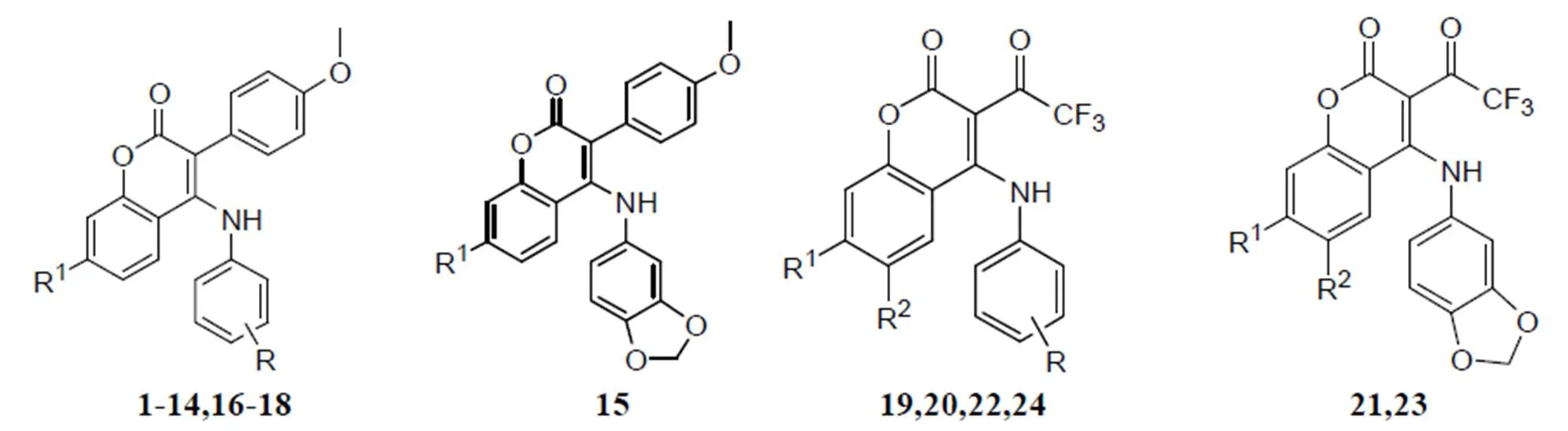
Fig. 1. Structures of a series of coumarin derivatives
2. 2 Minimization and alignment
3-QSAR model was constructed to evaluate the relationship between the structures of a series of coumarin derivatives and their anticancer activity by CoMFA and CoMSIA in SYBYL X-2.0. First of all, we constructed the initial three-dimensional struc- tures of 24 coumarin derivatives by using the sketch module in SYBYL X-2.0. Then, those coumarin derivatives were optimized. Energy minimization wasperformed using the tripos force field with the Powell conjugate gradient minimization algorithm, and the convergence criterion was 0.005 kcal/mol*A[19-21]. The maximum iteration was set to 10000. Owing to the highest inhibitory active value, compound 20 was selected as a template. Black bold atom in Fig. 2 was used as the common structure.Then, the alignment of all compounds of the training sets is shown in Fig. 3.

Fig. 2. Alignments of all molecules. Compound 20 was used as the template.
(a) Common substructure for alignment (shown in black bold atoms).(b) Alignment of all molecules
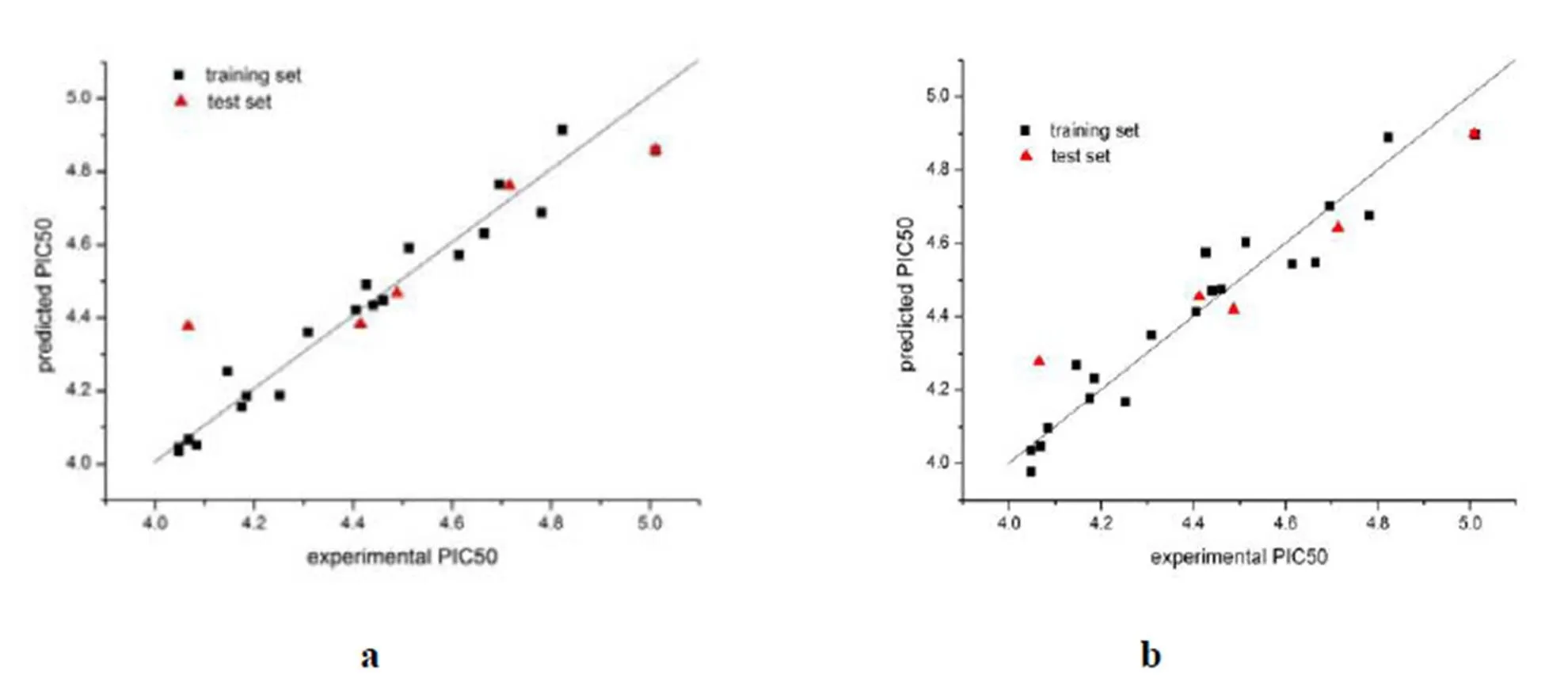
Fig. 3. Plot of experimental activity against predicted activity by the CoMFA model (a) and CoMSIA model(b)
2. 3 Establishment of the CoMFA model
The test sets which had stacked neatly were utilized to establish the CoMFA model. The size and distribution of the steric and electrostatic fields at each grid point around the overlapped molecules were calculated using the tripos force field. Meanwhile, we used the3carbon with a +1 charge as the probe atom. The rest of the parameters was the system default values[22-24]. In partial least-squares () analysis, firstly, the leave-one-out () was used for cross-validation to obtain the optimal number of components () and cross-validated correlation coefficient (2); then regression analysis was performed by non-cross validation to acquire non-cross-validated correlation coefficient (2), test value () and standard error of the estimate (cv). Finally, through analysis of the stereoscopic and steric field contour map, the favorable information was obtained that could be used to modify the existing molecules.
2. 4 Establishment of the CoMSIA model
The establishment and analysis of the CoMSIA model are basically the same as the CoMFA model. The difference is that the CoMSIA method investi- gates the interaction between the structure and activity of the compounds from five molecular fields, namely, the steric field, electrostatic field, hydro- phobic field, hydrogen-bond donor field and hydro- gen-bond acceptor field.
3 RESULTS AND DISCUSSION
3. 1 Reliability of the models
The CoMFAand CoMSIA models were establi- shed by using compounds in the training sets. Experimental and predicted PIC50values of all compounds are shown in Table1, in which their dif- ferences are also given.PLS analysis results ofCoMFA and CoMSIA areshown in Tables 1 and2, respectively. The CoMFA model got a2of 0.684 with anof 4 and2of 0.949,of 70.445, and acvof 0.073. The CoMSIA model got a2of 0.579 with anof 5 and2of 0.930,of 37.143andcv of 0.089. To validate the stability and predictive ability of the established models, 4 compounds were selected to the test sets. The predicted result for the test sets was also included inTable 1.These statis- tical parameters indicated that the CoMFA and CoMSIA modelswere dependable. Thecorrelation between the predictedandexperimental activities was depicted in Fig.3, which shows that the predicted PIC50values of the training sets compounds were in good agreement with the experimental data in a tolerable error range. Moreover, the correlation between the predicted50 values and the experi- mental data of the test sets compounds was also good.
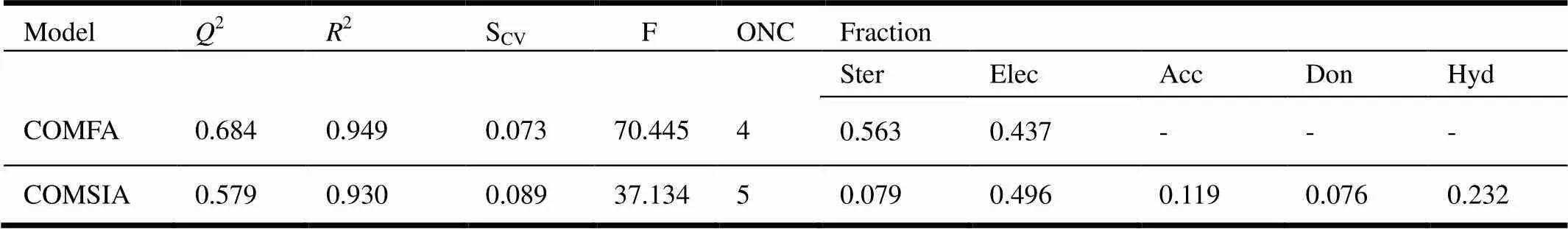
Table 2. Statistical Results of the CoMFA and CoMSIA Models
2: Cross-validated correlation coefficient.: Optimum number of components.2: Non-cross-validated correlation coefficient.cv: Standard error of the estimate.: test value.
3. 2 CoMFA and CoMSIA contour maps
The field distribution of the models could be seen in the 3-QSAR contour maps. We could modify moleculesto increase or decrease theactivity of the compound according to the 3-QSAR contour maps. Fig.4 shows thesteric and electrostaticcontour maps of CoMFA,and Fig. 5depictsthe steric,electrostatic, hydrophobic, hydrogen-bond donor and receptor contour maps of CoMSIA. We selected compound 20 as a reference structure. Allrepresented the default of 80% and20% level contributions for favored and disfavored regions, respectively.
Fig. 4a and 5a is the steric contour maps of CoMFA and CoMSIA.As shown in Fig. 4a and 5a, the steric contour maphas two colors of green and yellow. The greencontour indicated that increasing the volume of substituents in this region could enhance the activity of small molecules. The yellow contourindicated that decreasing the volume of substituents in this region could enhance the activity of small molecules. Fig. 4b and 5b is the electrostatic contour maps of CoMFA and CoMSIA.As shown in Fig. 4b and 5b, the electrostatic contour maphas two colors, blue and red. The red contour indicated that the addition of electron-donatinggroups would increase the activity. The blue contour indicated that adding electron-withdrawinggroups in this region would decrease the activity of the molecule. Fig. 5c is the hydrophobic contour map ofCoMSIA.As Fig. 5c shows, the hydrophobic contour maphas two colors of yellow and orange.The yellow contour indicatedhydrophobic groups are favored in the region. The orange contours indicated hydrophilic groups are favored in the region. Fig. 5d is the hydrogen-bond donor contour map ofCoMSIA. As shown in Fig. 5d, the hydrogen-bond donor contour maphas two colors, green and purple. The green and purple contours depicted favorable and unfavorable positions for hydrogen bond donor, respectively. Fig. 5e is the hydrogen-bond acceptor contour map ofCoMSIA.As Fig. 5e shows, the hydrophobic contour maphad two colors, magenta and red. The magenta and red con- tours depicted favorable and unfavorable positions for hydrogen-bond acceptors, respectively.
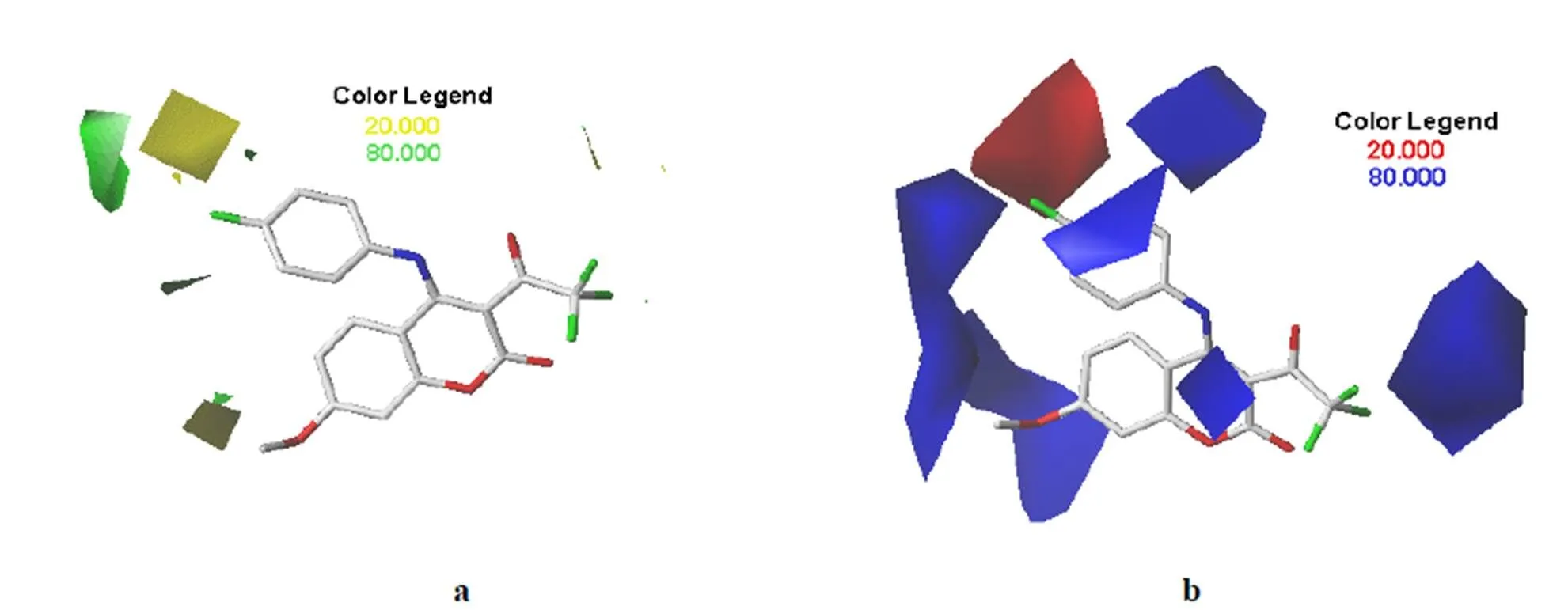
Fig. 4. Contour maps of the CoMFA models.(a) Green and yellow represent favorable and unfavorable regions for steric field. (b) Blue and red represent favorable and unfavorable regions for electrostatic field. Compound 20 was used as the template

Fig. 5. CoMSIA contour maps.(a) Favorable (green) and unfavorable (yellow) steric fields. (b) Electropositive (blue) and electronegative (red) fields. (c) Favorable (blue) and unfavorable (red) hydrophobic fields. (d) Favorable (green ) and unfavorable (purple) hydrogen-bond donor fields.(e) Favorable (magenta ) and unfavorable (red) hydrogen-bond acceptor fields. Compound 20 was used as the template
4 DESIGN OF NEWLY ANTICANCER DRUGS
It can be seen from Fig. 4b, 5b and 5c that the addition of electron-withdrawinggroups or hydro- philic groups at the R1position could increase the activity of small molecules. Based on this, we addedat the R1position. Not onlyis electron- withdrawing groups but also can form hydrogen bond with water, so we designed the compound N1 in Table 3. Compound N1 showed a good anti-cancer activity (PIC50= 4.916 and 4.931 for CoMFA and CoMSIA models, respectively). According to Fig. 4b and Fig. 5b, the addition of electron-withdrawing groups at the R2position facilitates the increase of activity of small molecules. Therefore, Compound N2 is designed by replacing a methoxy with. Compound N2 showed a good anti-cancer activity (PIC50= 4.889 and 4.994 for CoMFA and CoMSIA models, respectively). According to Fig. 5e, adding hydrogen-bond receptor at the R2position, such asand -SOH, could also increase the activity. In summary, N3~N5 molecules are designed as shown in Fig. 6 and they also have good anticancer activity.
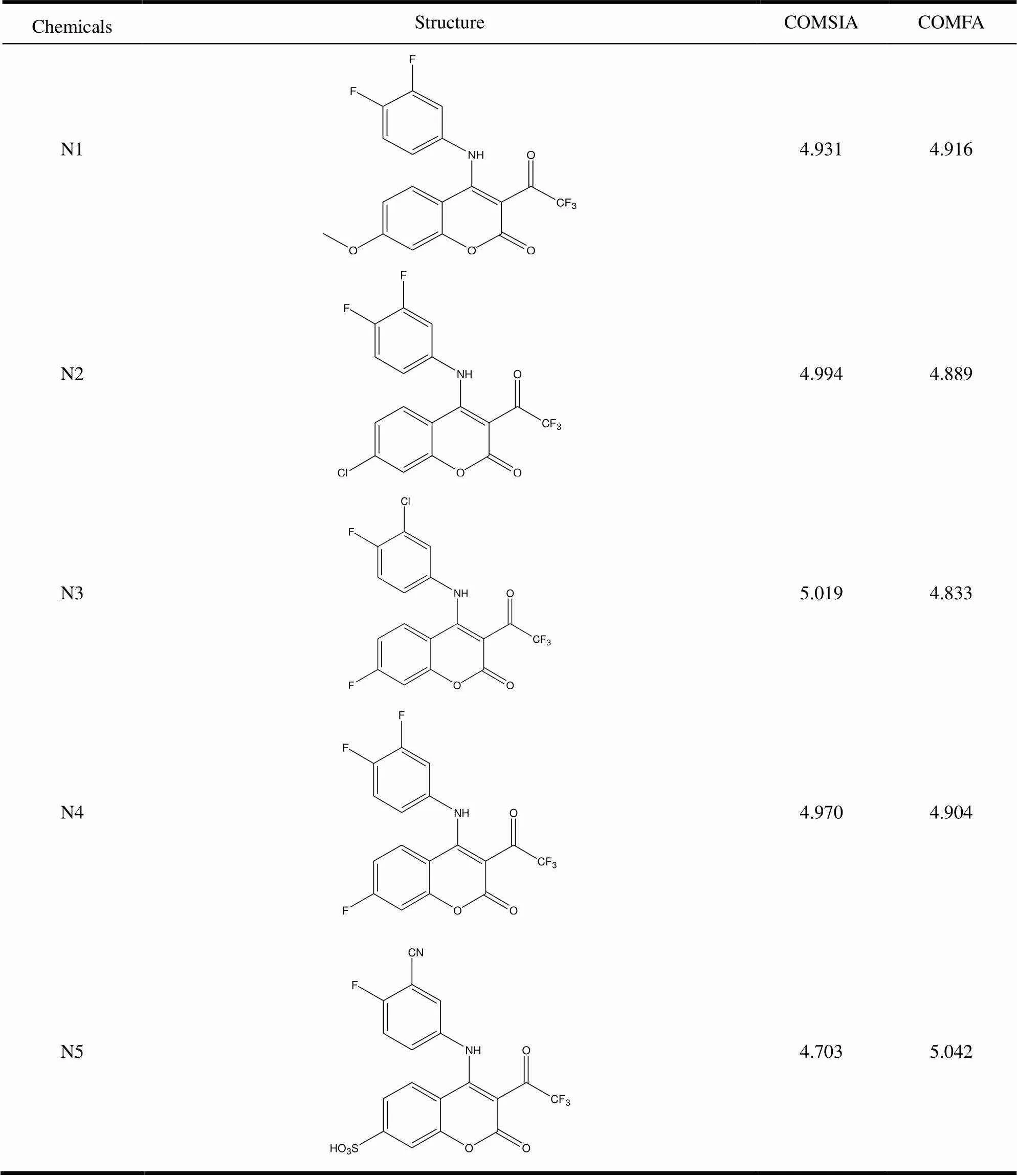
Table 3. Structure and Predicted PIC50 Values of Newly Designed Anticancer Drugs
Fig. 6. Structures of the newly anticancer drugs
5 CONCLUSION
In this article, we explored the structure-activity relationship of novel coumarin derivatives for anticancer activity against breast carcinoma cell line MCF-7 by the 3-QSAR. The cross-validation was conducted to obtain highly predictive and satis- factory CoMFA model (2= 0.684,2= 0.949) and CoMSIA model (2= 0.579,2= 0.930). Then, the contour maps ofCoMFA and CoMSIA models were generated by the SYBYL-X 2.0 software. 5coumarin derivatives with high anticancer activity were designed based on the contour maps of CoMFA and CoMSIA. Those coumarin derivatives deserve further study.
(1) Nagini, S. Breast cancer: current molecular therapeutic targets and new players.. 2017, 17, 152–163.
(2) DeSantis, C.; Ma, J.; Bryan, L.; Jemal, A. Breast cancer statistics. 2013.. 2014, 64, 52-62.
(3) Batran, R. Z.; Dawood, D. H.; El-Seginy, S. A.; Ali, M. M.; Maher, T. J.; Gugnani, K. S.; Rondon-Ortiz, A. N. New coumarin derivatives as anti-breast and anti-cervical cancer agents targeting VEGFR-2 and p38 MAPK.. 2017, 350, 1-19.
(4) Thasnim, P.; Bahulayan, D. Click-on fluorescent triazolyl coumarin peptidomimetics as inhibitors of human breast cancer cell line MCF-7.. 2017, 41, 13483-13489.
(5) Azimzadeh, M.; Rahaie, M.; Nasirizadeh, N.; Ashtari, K.; Naderi-Manesh, H. An electrochemical nanobiosensor for plasma miRNA-155, based on graphene oxide and gold nanorod, for early detection of breast cancer.2016, 77, 99-106.
(6) Harbeck, N.; Gnant, M. Breast cancer.2017, 389, 1134-1150.
(7) Fan, L.; Strasser-Weippl, K.; Li, J. J.; St Louis, J.; Finkelstein, D. M.; Yu, K. D.; Chen, W. Q.; Shao, Z. M.; Goss, P. E. Breast cancer in China.2014, 15, E279-E289.
(8) Dandriyal, J.; Singla, R. M.; Jaitak, V. Recent developments of C-4 substituted coumarin derivatives as anticancer agents.. 2016, 119, 141-168.
(9) Devji, T.; Reddy, C.; Woo, C.; Awale, S.; Kadota, S.; Carrico-Moniz, D. Pancreatic anticancer activity of a novel geranylgeranylated coumarin derivative..2011, 21, 5770-5773.
(10) Huang, X. Y.; Shan, Z. J.; Zhai, H. L.; Su, L.; Zhang, X. Y. Study on the anticancer activity of coumarin derivatives by molecular modeling.2011, 78, 651-658.
(11) Nasr, T.; Bondock, S.; Youns, M. Anticancer activity of new coumarin substituted hydrazide-hydrazone derivatives.2014, 76, 539-548.
(12) Luo, G.; Muyaba, M.; Lyu, W.; Tang, Z.; Zhao, R.; Xu, Q.; You, Q.; Xiang, H. Design, synthesis and biological evaluation of novel 3-substituted 4-anilino-coumarin derivatives as antitumor agents.2017, 27, 867-874.
(13) Tropsha, A. Best Practices for QSAR model development, validation, and exploitation.2010, 29, 476-488.
(14) Gramatica, P. Principles of QSAR models validation: internal and external.2007, 26, 694-701.
(15) Liu, P.; Long, W. Current mathematical methods used in QSAR/QSPR studies.2009, 10, 1978-1998.
(16) Bhatia, M. S.; Pakhare, K. D.; Choudhari, P. B.; Jadhav, S. D.; Dhavale, R. P.; Bhatia, N. M. Pharmacophore modeling and 3D QSAR studies of aryl amine derivatives as potential lumazine synthase inhibitors.2017, 10, S100-S104.
(17) Cao, F.; Li, X.; Ye, L.; Xie, Y.; Wang, X.; Shi, W.; Qian, X.; Zhu, Y.; Yu, H. Molecular docking, molecular dynamics simulation, and structure-based 3D-QSAR studies on the aryl hydrocarbon receptor agonistic activity of hydroxylated polychlorinated biphenyls..2013, 36, 626-635.
(18) Chen, M.; Li, P.; Hu, D.; Zeng, S.; Li, T.; Jin, L.; Xue, W.; Song, B. Synthesis, antiviral activity, 3D-QSAR, and interaction mechanisms study of novel malonate derivatives containing quinazolin-4(3H)-one moiety.2016, 26, 168-173.
(19) Dixon, S. L.; Smondyrev, A. M.; Knoll, E. H.; Rao, S. N.; Shaw, D. E.; Friesner, R. A. PHASE: a new engine for pharmacophore perception, 3D QSAR model development, and 3D database screening: 1. Methodology and preliminary results..2006, 20, 647-671.
(20) Fang, Y.; Lu, Y.; Zang, X.; Wu, T.; Qi, X.; Pan, S.; Xu, X. 3D-QSAR and docking studies of flavonoids as potent Escherichia coli inhibitors..2016, 6, 1-13.
(21) Liu, X. H.; Xu, X. Y.; Tan, C. X.; Weng, J. Q.; Xin, J. H.; Chen, J. Synthesis, crystal structure, herbicidal activities and 3D-QSAR study of some novel 1,2,4-triazolo 4,3-a pyridine derivatives.2015, 71, 292-301.
(22) Buolamwini, J. K.; Assefa, H. CoMFA and CoMSIA 3D QSAR and docking studies on conformationally-restrained cinnamoyl HIV-1 integrase inhibitors: exploration of a binding mode at the active site.2002,45, 841-852.
(23) Doytchinova, I. A.; Flower, D. R. Toward the quantitative prediction of T-cell epitopes: CoMFA and CoMSIA studies of peptides with affinity for the class I MHC molecule HLA-A*0201.2001, 44, 3572-3581.
(24) Murthy, V. S.; Kulkarni, V. M. 3D-QSAR CoMFA and CoMSIA on protein tyrosine phosphatase 1B inhibitors.2002, 10, 2267-2282.
19 March 2018;
6 June 2018
. E-mail: zhang_lucy9999@vip.126.com
10.14102/j.cnki.0254-5861.2011-2007
- 结构化学的其它文章
- Nanoclusters Au19Pd and Au19Pt Catalyzing CO Oxidation: a Density Functional Study①
- Synthesis, Single-crystal Structure and Fluorescence Property of a New Boron Compound Based on 2-(2΄-Hydroxyphenyl)-1H-benzimidazole①
- Review on Li-insertion/extraction Mechanisms of LiFePO4 Cathode Materials①
- Synthesis of a NovelImidazole Ionic Liquid Modified Mesoporous Silica SBA15 for Selective Separation and Determination of Inorganic Arsenic in Rice①
- Theoretical Investigations on the Structural and Electronic Properties of WO3Polymorphs①
- Surface Modification of Titanium Oxide by Indium for Efficient Photocatalytic Hydrogen Generation①

