耐万古霉素肠球菌临床菌株毒力基因携带情况及差异
陈敏强,汪 强,周永列,吕火烊,胡庆丰,葛玉梅
耐万古霉素肠球菌临床菌株毒力基因携带情况及差异
陈敏强1,汪 强2,周永列1,吕火烊1,胡庆丰1,葛玉梅1
目的 检测不同种类耐万古霉素肠球菌(VRE)毒力基因携带情况及其差异,为研究VRE致病机制提供基础。方法 采用微量稀释法检测浙江地区490株屎肠球菌和862株粪肠球菌临床分离菌株对万古霉素的耐药性。采用PCR检测VRE菌株ace、asa1、cylA、efaA、esp、gelE、hyl七种毒力基因。结果 10%(49/490)屎肠球菌株和0.8%(7/862)粪肠球菌株为VRE。万古霉素耐药屎肠球菌株中,5株未检出任何毒力基因,其余44株仅可检出asa1、esp、gelE和hyl基因,其中esp基因(73.5%,36/49)和hyl基因(53.1%,26/49)为优势基因,以单或双基因为主要携带模式。万古霉素耐药粪肠球菌株中可检出除hyl基因以外其他6种毒力基因,且每株菌均同时携带3~6种致病基因。结论 本地区VRE以屎肠球菌为主,耐万古霉素屎肠球菌和粪肠球菌毒力基因携带率、携带种类和模式均有明显差异。
耐万古霉素肠球菌;屎肠球菌;粪肠球菌;毒力基因
肠球菌属(Enterococcus)细菌为革兰阳性兼性厌氧球菌,广泛分布于人和动物肠道以及自然环境中[1]。由于肠球菌有较强的生存力及耐药性,近年来已成为院内感染的重要机会性病原菌,可引起皮肤、尿路感染以及更为严重的败血症、心内膜炎、脑膜炎等疾病[2-5]。根据抗生素种类不同,具有较强耐药性的肠球菌可分为高水平氨基糖苷类耐药肠球菌(high-level aminoglycoside-resistant,HLAR)和万古霉素耐药肠球菌(vancomycin-resistant enterococci,VRE)[6]。但有文献报道,VRE定值率较高的欧洲人群感染率低于VRE定值率较低的美国人群[7],提示耐药性不是肠球菌致病的唯一因素。
Shankar等(2002)首先发现,肠球菌基因组中有一个大小约为150 kb、被称为致病岛的基因片段,感染菌株的致病岛中具有多种致病基因,非感染株则否[8]。目前较为肯定的肠球菌致病基因及其产物主要有:ace基因产物辅助定植因子(accessory colonization factor)、asa1基因产物聚集性物质(aggregation substance)、cylA基因产物溶细胞素(cytolysin)、efaA基因产物心内膜炎抗原(endocarditis antigen)、esp基因产物肠球菌表面蛋白(enterococci surface protein)、gelE基因产物明胶酶(gelatinase)和hyl基因产物透明质酸酶(hayluronidase)[8-12]。此外,有文献报道不同地区人群中分离的VRE菌株携带的致病基因有差异[13-14]。因此,我们检测了肠球菌临床分离株对万古霉素的耐药性,采用PCR检测了VRE上述7种致病基因携带率,同时分析了不同VRE携带致病基因种类和模式差异。
1 材料与方法
1.1 肠球菌菌株来源 从2011年6月至2014年1月本院临床病人标本中分离肠球菌并用VITEK 2-compact型法国梅里埃(BioMérieux)全自动细菌检测分析系统及其GN细菌鉴定卡进行鉴定。
1.2 VRE菌株鉴定 参照国内临床通用的美国临床和实验室(CLSI)手册中介绍的药敏实验方法及判断标准,采用微量二倍液体稀释法检测上述肠球菌临床菌株对万古霉素(Sigma)的敏感性,若万古霉素对肠球菌最低抑菌浓度(MIC)≥8,该菌株判为VRE[15]。实验中采用北京中国食品药品检定研究院提供的金黄色葡萄球菌ATCC25923、大肠埃希菌ATCC25922和粪肠球菌ATCC29212为质控菌株。
1.3 致病基因检测 采用细菌基因组DNA提取试剂盒(Axygen)提取56株VRE(屎肠球菌49株、粪肠球菌7株)基因组DNA,紫外分光光度法测定其浓度和纯度[16]。根据文献报道引物序列(表1)[8-12],委托上海生工生物公司合成各引物。采用PCR检测上述VRE菌株基因组DNA中ace、asa1、cylA、efaA、esp、gelE和hyl基因片段。反应体积25 μl,内含:10 μL Premix Taq混合液(含反应缓冲液、dNTP及DNA聚合酶)、10 μmol/L上游和下游引物、100 ng DNA模板。反应参数:94 ℃ 4 min;94 ℃ 40 s、58 ℃ 30 s、72 ℃ 1 min,35个循环;72 ℃ 5 min。采用溴乙锭预染色的1.5%琼脂糖凝胶电泳检测各目的基因扩增片段后委托上海生工生物公司测序,采用Blast软件进行序列比对。
表1 扩增VRE致病基因引物序列
Tab.1 Sequences of primers for amplifying pathogenic genes of VRE

基因gene基因产物geneproduct引物序列(5’-3’)primersequence扩增产物(bp)amplificationproductace辅助定植因子F:GGAATGACCGAGAACGATGGC616accessorycolonizationfactorR:GCTTGATGTTGGCCTGCTTCCGasa1聚集性物质F:CACGCTATTACGAACTATGA375aggregationR:TAAGAAAGAACATCACCACGAcylA溶细胞素F:ACTCGGGGATTGATAGGC688cytolysinR:GCTGCTAAAGCTGCGCTTefaA心内膜炎抗原F:CTACTAACACGTCACGAATG499endocarditisantigenR:CGTGAGAAAGAAATGGAGGAesp肠球菌表面蛋白F:AGATTTCATCTTTGATTCTTGG510enterococcalsurfaceproteinR:AATTGATTCTTTAGCATCTGGgelE明胶酶F:TATGACAATGCTTTTTGGGAT213gelatinaseR:AGATGCACCCGAAATAATATAhyl透明质酸酶F:ACAGAAGAGCTGCAGGAAATG276hyaluronidaseR:GACTGACGTCCAAGTTTCCAA
F: upstream primer; R: downstream primer
2 结 果
2.1 VRE检出率 从217 024例临床病人标本中分离出1391株肠球菌,其中屎肠球菌(Enterococcusfaecium)490株、粪肠球菌(Enterococcusfaecalis)862株。根据药物敏感试验结果,上述490株屎肠球菌和862株粪肠球菌中分别有49株和7株为VRE。
2.2 VRE致病基因检出率 49株耐万古霉素屎肠球菌中,5株未检出致病基因,73.5%(36/49)、53.1%(26/49)和8.2%(4/49)菌株分别检出esp、hyl和gelE基因,1株检出asa1基因,但均未检出ace、cylA和efaA基因;7株耐万古霉素粪肠球菌中,100%(7/7)、85%(6/7)、85%(6/7)、71.4%(5/7)、57.1%(4/7)和42.9%(3/7)菌株分别检出efaA、asa1、gelE、cylA、esp和ace基因,但均未检出hyl基因(表2)。
表2 VRE菌株中不同致病基因检出率
Tab.2 Detection rates of different pathogenic genes of VRE isolates
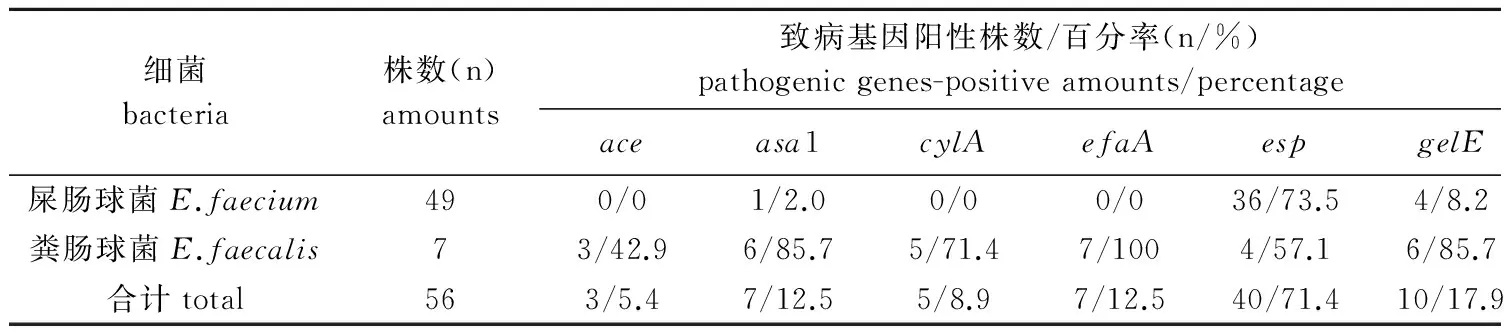
细菌bacteria株数(n)amounts致病基因阳性株数/百分率(n/%)pathogenicgenes-positiveamounts/percentageaceasa1cylAefaAespgelEhyl屎肠球菌E.faecium490/01/2.00/00/036/73.54/8.226/53.1粪肠球菌E.faecalis73/42.96/85.75/71.47/1004/57.16/85.70/0合计total563/5.47/12.55/8.97/12.540/71.410/17.926/46.4
2.3 VRE致病基因携带模式 49株耐万古霉素屎肠球菌中有5株未检出上述7种致病基因,其余44株检出1或2种致病基因,其中esp-hyl双基因(40.9%,18/44)、esp单基因(31.8%,14/44)携带率明显高于hyl单基因(18.2%,8/44)、esp-gelE双基因(6.8%,3/44)和esp-asa1双基因(2.3%,1/44)(表3)。7株粪肠球菌均携带efaA、asa1、gelE、cylA、esp或ace基因,且每株菌均同时携带3~6种致病基因(表3)。
表3 VRE菌株致病基因携带模式
Tab.3 Carrying model of pathogenic genes of VRE isolates
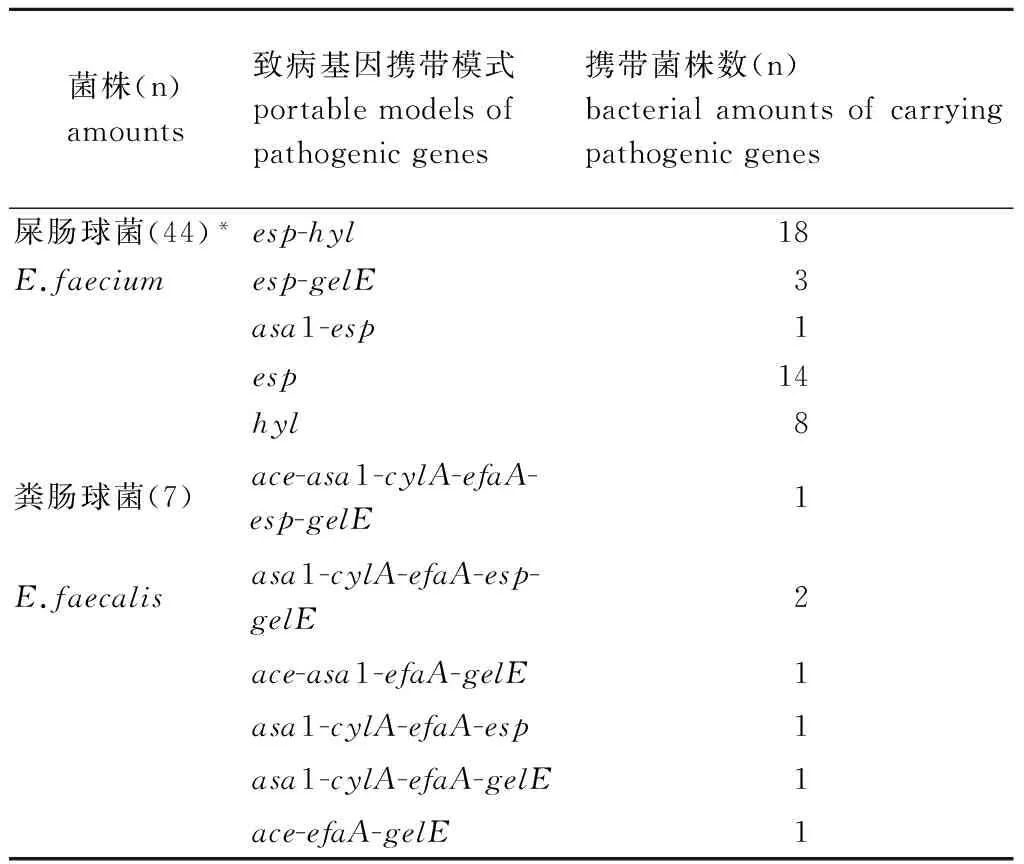
菌株(n)amounts致病基因携带模式portablemodelsofpathogenicgenes携带菌株数(n)bacterialamountsofcarryingpathogenicgenes屎肠球菌(44)*esp-hyl18E.faeciumesp-gelE3asa1-esp1esp14hyl8粪肠球菌(7)ace-asa1-cylA-efaA-esp-gelE1E.faecalisasa1-cylA-efaA-esp-gelE2ace-asa1-efaA-gelE1asa1-cylA-efaA-esp1asa1-cylA-efaA-gelE1ace-efaA-gelE1
*: another five genes were not detected
3 讨 论
国内外大量研究结果表明,肠球菌已成为主要的院内感染机会性病原菌之一,尤以粪肠球菌最为常见,其次为屎肠球菌,但屎肠球菌耐药性及获得耐药的能力均比粪肠球菌强,近年来我国其他地区临床标本中分离的粪肠球菌和屎肠球菌也不例外[4,17-18]。我们的实验结果显示,浙江地区临床病人标本中分离的1 391株肠球菌中,36.2%(490/1391)为屎肠球菌,63.8%(862/1391)为粪肠球菌。药物敏感试验结果显示,上述490株屎肠球菌和862株粪肠球菌中分别有49株和7株为VRE。上述实验结果表明,本地区人群感染的肠球菌与其他地区不同,主要以屎肠球菌为主,其次为粪肠球菌。
肠球菌不产生内、外毒素,其致病机制迄今未完全明了,但有研究证实ace基因产物辅助定植因子、asa1基因产物聚集性物质、cylA基因产物溶细胞素、efaA基因产物心内膜炎抗原、esp基因产物肠球菌表面蛋白、gelE基因产物明胶酶和hyl基因产物透明质酸酶在肠球菌侵入宿主和引起组织损伤过程中发挥了主要作用[8-12]。近年研究发现,不同种类肠球菌致病基因的检测结果差异较大[13-14]。我们的检测结果显示:49株耐万古霉素屎肠球菌中,5株未检出致病基因,其余44株也仅检出esp、hyl和gelE基因,其中esp基因(73.5%,36/49)和hyl基因(53.1%,26/49)为优势基因;7株耐万古霉素粪肠球菌虽未检出hyl基因,但均检出efaA、asa1、gelE、cylA、esp和/或ace基因。上述实验结果表明,耐万古霉素屎肠球菌和粪肠球菌致病基因携带率和种类确有明显差异。
我们对耐万古霉素屎肠球菌和粪肠球菌致病基因携带模式分析后发现,屎肠球菌以esp-hyl双基因(40.9%,18/44)、esp单基因(31.8%,14/44)为致病基因主要携带模式,但粪肠球菌每株菌均同时携带至少3种致病基因,两种VRE在致病基因携带率、携带的致病基因类型和模式上均有较大差异,与以往一些文献报道相似[12-14,19]。
[1] Yan J,Medical microbiology[M]. 3rd Edition,Beijing: The High Education Publication House,China,2016: 25-26,82-84. (in Chinese)
[2] Zhou J,Jin D,Shan LU,et al. Virulence genes and antibiotic resistance of isolated from yaks in Qinghai-Tibet Plateau,China[J]. Chin J Zoonoses,2015,31(4): 298-302. DOI: 10.3969/cjz.j.issn.1002-2694.2015.04.002
[3] Qiang H,Zhu P,Liu GY,et al. Role ofEnterococcusfaecalisendocarditis antigen EfaA on biofilm formation[J]. Chin J Zoonoses,2014,30(9): 938-940. DOI: 10.3969/cjz.j.issn.1002-2694.2014.09.013
[4] Nateghian A,Fallah F,Daghighi Z,et al. Detection of virulence genes in resistant enterococci isolated from pediatric patients at high risk for nosocomial infections[J]. Diagn Micr Infec Dis,2016,85(2): 260-262. DOI:10.1016/j.diagmicrobio.2016.03.020
[5] Britt NS,Potter EM. Clinical epidemiology of vancomycin-resistantEnterococcusgallinarumandEnterococcuscasseliflavusbloodstream infections[J]. J Glob Antimicrob Re,2016,5: 57-61. DOI: 10.1016/j.jgar.2015.12.002
[6] Agarwal J. High-level aminoglycoside resistance and β-lactamase production in enterococci at a tertiary care hospital in India[J]. Jpn J Infect Dis,2009,62(2): 158-159.
[7] Mendes RE,Castanheira M,Farrell DJ,et al. Longitudinal (2001-14) analysis of enterococci and VRE causing invasive infections in European and US hospitals,including a contemporary (2010-13) analysis of oritavancininvitropotency[J]. J Antimicrob Chemot,2016,71(12): 3453-3458. DOI:10.1093/jac/dkw319
[8] Shankar N,Baghdayan AS,Gilmore MS. Modulation of virulence within a pathogenicity island in vancomycin-resistantEnterococcusfaecalis[J]. Nature,2002,417(6890): 746-750. DOI: 10.1038/nature00802
[9] Wu LX,Wang GF,Huang WX. Experimental study on the function of the putative virulence factor hyaluronidase genehylofEnterococcusfaecium[J]. Chin J Zoonoses,2008,24(9): 799-802. DOI: 10.3969/j.issn.1002-2694.2008.09.002
[10] Shankar V,Baghdayan AS,Huycke MM,et al. Infection-derivedEnterococcusfaecalisstrains are enriched in esp,a gene encoding a novel surface protein[J]. Infect Immun,1999,67(1): 193-200. DOI: 10.1103/PhysRevE.60.1042
[11] Sava IG,Heikens E,Huebner J. Pathogenesis and immunity in enterococcal infections[J]. Clin Microbiol Infect,2010,16(6): 533-540. DOI: 10.1303/aez.2003.31
[12] Qiang H,Jiang Y,Zhu B,et al. Prokaryotic expression of the hemolysin cylL gene fromEnterococusfaecalis[J]. Chin J Zoonoses,2007,23(10): 968-970. DOI: 10.3969/j.issn.1002-2694.2007.10.003
[13] Roberta-Creti R,Monica-Imperi M,Lucia-Bertuccini L,et a1. Survey for virulence determinants amongEnterococcusfaecalisisolated from different sources[J]. J Med Microbio1,2004,53(1): 14-21. DOI:10.1099/jmm.0.05353-0
[14] Ma LY,Xu SZ. Study on virulence determinants,phenotypes and drug resistance of enterococcus from inpatients and healthy adults[J]. J Clin Exp Med,2006,58(1): 5-7. DOI: 10.3969/j.issn.1671-4695.2006.01.003
[15] Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing[S]. Twenty-fifth Informational Supplement. CLSI document M100-S25. Wayne,PA: Clinical and Laboratory Standards Institute,USA. 2015.
[16] Sambrook J,Fritsch EF,Maniatis T. Molecular cloning: A laboratory manual[M]. 2nd edition,New York: Cold Spring Harbor Laboratory Press,1989: 1.21-1.52,2.60-2.80,7.3-7.35,9.14-9.22.
[17] Hu FP,Wang X,Zhu DM,et al. Report of CHINET antimicrobial resistance surveillance program in 2015[J]. Chin J Infect Chemother,2016,16(6): 685-694. DOI: 10.16718/j. 1009-7708.2016.06.003
[18] Hu FP,Zhu DM,Wang F,et al. CHINET 2014 surveillance of bacterial resistance in China[J]. Chin J Infect Chemother,2015,(5):401-410. DOI: 10.3969/j.issn.1009-7708. 2015.05.001
[19] Bittencourt de Marques E,Suzart S. Occurrence of virulence-associated genes in clinicalEnterococcusfaecalisstrains isolated in Londrina,Brazil[J]. J Med Microbiol,2004,53(11): 1069-1073. DOI: 10.1099/jmm.0.45654-0
Distribution characteristics and diversity of virulence genes in clinical isolates of vancomycin-resistant enterococci
CHEN Min-qiang1,WANG Qiang2,ZHOU Yong-lie1,LYU Huo-yang1,HU Qing-feng1,GE Yu-mei1
(1.ZhejiangProvincialPeople’sHospital,Hangzhou310014,China; 2.XinhuaHospitalofZhejiangProvince,Hangzhou310005,China)
The aim of this study is to determine the diversity of virulence genes carried by different vancomycin-resistant enterococci (VRE),which will provides a basis for studying pathogenic mechanism of VRE. Microdilution-based drug sensitivity test was applied to detect the vancomycin resistance of 490Enterococcusfaeciumisolates and 862Enterococcusfaecalisisolates in Zhejiang area. The seven virulence genes (ace,asa1,cylA,efaA,esp,gelEandhyl) in the isolates of VRE were detected by PCR. According to the results of drug sensitivity test,10% of theE.faeciumisolates (49/490) and 0.8% of theE.faecalis(7/862) were identified as VRE. In the vancomycin-resistantE.faeciumisolates,five isolates were negative for any of the target genes and the other 44 isolates were positive forasa1,esp,gelEandhylgenes alone,in which the esp (73.5%,36/49) andhyl(53.1%,26/49) were the predominant genes and single or double virulence genes acted as the major carrying models. Except for thehylgene,the vancomycin-resistantE.faecalisisolates were positive for the other six pathogenic genes,and the isolates could carry 3-6 pathogenic genes. All the data indicate thatE.faeciumis the major species of VRE in the local area,and the carrying rate,types and models of virulence genes in the vancomycin-resistantE.faeciumandE.faecalisisolates are obviously different.
vancomycin-resistant enterococci;Enterococcusfaecium;Enterococcusfaecalis; virulence genes
Ge Yu-mei,Email: geyumei1990@hotmail.com
10.3969/j.issn.1002-2694.2017.05.007
浙江省医药卫生科技计划项目(2017KY004)资助
葛玉梅,Email:geyumei1990@hotmail.com
1.浙江省人民医院检验中心,杭州 310014; 2.浙江省新华医院,杭州 310005
R378.1
A
1002-2694(2017)05-0423-04
2016-12-07 编辑:刘岱伟
Supported by the Medical Scientific Research Foundation of Zhejiang Province (No. 2017KY004)
- 中国人兽共患病学报的其它文章
- 中国西北四省(区)结核分枝杆菌分离株一线药物耐药状况及其影响因素分析
- 美国《Emerging Infectious Diseases》2017年第3期有关人兽共患病论文摘译
- MicroRNA通用探针法的建立及布鲁氏菌病患者血浆microRNA-146a的检测
- 龟头包皮炎患者淋病奈瑟菌分离与鉴定及其耐药机制研究
- Identification of non-tuberculosis mycobacteria speciesof clinical isolates from patients clinically diagnosed with tuberculosis in Fujian Province,China
- 四对结核分枝杆菌毒素-抗毒素系统基因功能的初步研究

