Identification of non-tuberculosis mycobacteria speciesof clinical isolates from patients clinically diagnosed with tuberculosis in Fujian Province,China
LIU Hai-can,HUANG Ming-xiang,JIANG Yi,LIAN Lu-lu,,ZHAO Xiu-qin,ZHANG Li-shui,WU Yi-mou,WAN Kang-lin,
(1. State Key Laboratory for Infectious Diseases Prevention and Control,Collaborative Innovation Center forDiagnosis and Treatment of Infectious Diseases,National Institute for Communicable Disease Control and Prevention,Chinese Center for Disease Control and Prevention,Beijing 102206,China; 2. Fuzhou Pulmonary Hospital,Fuzhou 350008,China;3. Pathogenic Biology Institute,University of South China,Hengyang 421001,China)
Identification of non-tuberculosis mycobacteria speciesof clinical isolates from patients clinically diagnosed with tuberculosis in Fujian Province,China
LIU Hai-can1,HUANG Ming-xiang2,JIANG Yi1,LIAN Lu-lu1,3,ZHAO Xiu-qin1,ZHANG Li-shui2,WU Yi-mou3,WAN Kang-lin1,3
(1.StateKeyLaboratoryforInfectiousDiseasesPreventionandControl,CollaborativeInnovationCenterforDiagnosisandTreatmentofInfectiousDiseases,NationalInstituteforCommunicableDiseaseControlandPrevention,ChineseCenterforDiseaseControlandPrevention,Beijing102206,China; 2.FuzhouPulmonaryHospital,Fuzhou350008,China;3.PathogenicBiologyInstitute,UniversityofSouthChina,Hengyang421001,China)
Some species of non-tuberculosis mycobacteria (NTM) are important causes of human disease and infections. The purpose of this study was to describe the NTM species of clinical isolates from patients with tuberculosis in Fujian,China. The clinical NTM strains isolated from the patients clinically diagnosed with tuberculosis collected from 2005 to 2011 in Fuzhou Pulmonary Hospital,Fujian Province,and were identified to species level using conventional and molecular methods including multi-loci PCR,PRA-hsp65 and PRA-rpoBand sequencing forhsp65,rpoB,16SrRNAandITS. A total of 450 pre-identified NTM isolates were identified as 45 (10.00%)Mycobacteriumtuberculosisstrains and 405 (90.00%) NTM strains,which including 23 different NTM species and aGordoniabronchialisisolates. There were at least 23 NTM species as potential pathogens were found to cause pulmonary infections in human beings,and 6 NTM species andGordoniabronchialiswere firstly identified in patients with a clinical diagnosis of tuberculosis (TB) in Fujian Province. NTM species identification is very important for distinguishing between TB and NTM pulmonary diseases. And combination of several methods can increase identification accuracy and efficacy.
identification;Mycobacterium; PRA; DNA sequencing
Mycobacterium,an important cause of human disease and infections,includesMycobacterium(M.)tuberculosis,M.leprae,and non-tuberculosisMycobacterium(NTM).M.tuberculosis,which may have killed more persons than any other microbial pathogen,is a major causative agent of tuberculosis (TB)[1]. The intracellular pathogenM.lepraecauses a chronic infectious disease,leprosy,which remains an important public health problem. NTM is large group of differentMycobacteriumspecies that are ubiquitous in our living environment and can be isolated from soil,house dust,water,food and animals[2]. Some NTM species have already been recognized as the real causes of human infections. Transmission occurs by inhalation,ingestion or direct contact with a contaminated environmental source[2]. There is no evidence of human-human and animal-human transmission of NTM[2-4].
NTM affects both immune-competent and immune-compromised persons,and patients with the human immunodeficiency virus (HIV) are known to be especially vulnerable[5]. The NTM most frequently involved in disease cases includeM.aviumcomplex,M.kansasii,M.chelonae,M.abscessus,M.xenopi,M.malmoense,M.scrofulaceum,M.marinum,M.ulcerans,andM.haemophilum[6]. NTM diseases,particularly pulmonary NTM diseases with clinical signs similar to those of TB caused byM.tuberculosis,lead to a clinical dilemma with regard to diagnosis and therapy[7]. Furthermore,since differences in antimicrobial susceptibility determine treatment options,species-level identification of NTM is becoming increasingly clinically important[8]and imperative.
Traditionally,the definitive diagnosis of mycobacterial infections has been dependent on the isolation and identification of the causative agents and has required a series of specialized physiological and biochemical tests[9]. BecauseMycobacteriumspecies have different drug susceptibilities,their precise identification is crucial for the adoption of appropriate drug therapy and can ultimately influence patient outcome[10-12]. With advances in molecular microbiology and the knowledge of NTM,more species have been identified.
Combination use of the conventional biochemical testing methods with molecular biological methods such as polymerase chain reaction-restriction fragment length polymorphism (PRA) and sequencing can most accurately identifyMycobacteriaspp. strains. This study focused on identifyingMycobacteriaisolates in Fujian Province,China.
Materials and methodsMycobacterial isolates
From 2005 to 2011,450 clinical NTM strains were isolated from the clinical diagnosed TB patients’ sputum samples,and preliminarily identified with conventional Löwenstein-Jensen medium and differentiated p-nitrobenzoic acid (PNB)/2-thiophene carboxylic acid hydrazide (TCH) media in Fuzhou Pulmonary Hospital,Fujian Province,China. And then were cultured and stored at the Tuberculosis Research Laboratory,National Institute for Communicable Disease Control and Prevention (ICDC),Chinese Center for Disease Control and Prevention (China CDC).
Preliminary identification
All of the 450 selected pre-identified NTM isolates was firstly re-identified using the conventional biochemical methods--PNB and TCH testing-following standard protocols[7,13]. By which the mycobacterial isolates could be identified asM.tuberculosis,M.bovisor NTM.
Preparation of DNA
Bacterial DNA was prepared as follows. One loopful of the bacteria cultured on Löwenstein-Jensen medium was inactivated for 30 min at 85 ℃,suspended in 400 μL of TE buffer (0.01 M Tris-HCl,0.01 M ethylenediaminetetraacetic acid [pH 8.0]). Lysozyme (Sigma Chemical Co.,St. Louis,MO,USA) was added to a final concentration of 2 mg/mL and the tube was incubated for 16-20 h at 37 ℃. Bacterial DNA was prepared as described by van Soolingen et al[14]. Briefly,70 μL of 10% sodium dodecyl sulfate (SDS) and 5 μL of proteinase K (20 mg/mL) were added and the mixture was incubated for 10 min at 65 ℃. A total of 100 μL of 5 M NaCl and 100 μL of N-acetyl-N,N,N,-trimethyl-ammonium bromide were added. The tubes were incubated for 10 min at 65 ℃. An equal volume of chloroform-isoamyl alcohol (24:1,vol/vol) was added and the mixture was centrifuged for 5 min. A 0.6 volume of isopropanol was added to the supernatant to precipitate the DNA. After incubation for 30 min at 20 ℃ and centrifugation for 30 min at 14 000 × g at 4 ℃,the pellet was washed once with 70% ethanol and then dissolved in 50 μL of 1× TE buffer.
Multi-loci polymerase chain reaction (PCR)
As described by Richard C. Huard et al[15],PCR amplifications were performed in a program with an initial denaturation step of 5 min at 94 ℃ followed by 35 cycles of 1 min at 94 ℃,1 min at 60 ℃ and 1 min at 72 ℃ and ending with a final elongation step for 10 min at 72 ℃. The PCR products and a 100 bp ladder (Beijing CoWin Biotech Co.,Ltd.,Beijing,China) were visualized by 2% agarose gel electrophoresis and ethidium bromide staining. The results could distinguish theM.tuberculosiscomplex (MTBC) subspecies from NTM.
Amplification of rpoB and hsp65 DNA
A specific region of therpoBgene was amplified using primers rpoBF (5′-TCAAGGAGAAGCGCTACGA-3′) andrpoBR(5′-ATGTTGATCAGGGTCTGC-3′),resulting in a 360-bp PCR product[16-17]. Primers Tb11 (5′-ACCAACGATGGTGTGTCCAT-3′) and Tb12 (5′-CTTGTCGAACCGCATACCCT-3′) amplified a 439-bp fragment between positions 398 and 836 of the publishedhsp65 gene[18-19]. The composition of the PCR mixture (50 μL) was 25 μL 2× Taq MasterMix,2 μL of (each) primer at 10 μM,5 μL of DNA-containing supernatant and 16 μL of ddH2O. The reaction was subjected to 35 cycles of amplification (1 min at 94 ℃,1 min at 60 ℃,1 min at 72 ℃); this was followed by 10 min of extension at 72 ℃. The PCR products and a 100-bp ladder (Beijing CoWin Biotech Co.,Ltd.) were then visualized by 2% agarose gel electrophoresis and ethidium bromide staining and photographed with a ChemiDocTMXRS+ System (Bio-Rad Laboratories,Inc.,Hercules,USA).
PCR product purification
PCR products that contained partialrpoBandhsp65 DNA were purified using a Quick DNA Purification Kit (Beijing CoWin Biotech Co.,Ltd.).
PRA-hsp65[19] and PRA-rpoB[17] methods and analysis
The purified PCR products were digested with a restriction enzyme (Takara Biotechnology [Dalian] Co.,Ltd.,Dalian,China). Thehsp65 gene PCR product was digested with BstP I (60 ℃ for 2 h) and Hae III (37 ℃ for 2 h). The reactions consisted of 20 μL (15 μL of the PCR product,1 μL of restriction enzyme,1 μL of 10× buffer and 2 μL of ddH2O). For therpoBgene,the reactions contained Msp I (1 μL of Msp I,1.5 μL of DNA,2 μL of 10× T buffer and 2 μL of 0.1% bovine serum albumin) and Hae III (1 μL of Hae III,1.5 μL of DNA,2 μL of 10× M buffer and 2μL of ddH2O),which were incubated at 37 μL for 2 h. The resulting restriction fragments were separated by electrophoresis in a 4% agarose gel (LONZA MetaPhor○RAgarose; Rockland,USA) with a 20-bp ladder (Takara Biotechnology (Dalian) Co.,Ltd.) as a molecular size standard. The gels were stained with ethidium bromide and photographed with the ChemiDocTMXRS+ System (BIO-RAD). The restriction fragment sizes were estimated using Image Lab software (Bio-Rad Laboratories,Inc.,Hercules,USA). The observed PRA-hsp65 patterns were comparable to the patterns reported on PRASITE[20],in publications[18-19,21-25]or our laboratory inter-database,while the observed PRA-rpoBpatterns were comparable to the patterns reported in publications[17,23,26]or our laboratory inter-database.
Gene PCR sequencing and multi-sequence alignment
Thehsp65,16SrRNA,rpoB,ITS(16S-23Sinternal transcribed spacer) gene PCR products were sent to Beijing Tsingke Bio Tech Co.,Ltd for sequencing. The sequences were analyzed with online analysis using BLAST[27]. Multiple sequence alignment was performed using the ClustalX Program (Conway Institute,UCD Dublin,Belfield,Ireland),while the phylogenetic consensus tree was derived from DNA sequences using the neighbor-joining method with Kimura’s 2-parameter distance correction model with 1 000 bootstrap replications in the MEGA version 5.01 software package[28].
Statistical analysis
The data were recorded in Microsoft Excel (Microsoft Corporation,Redmond,USA). A certain identification was declared when consistent results were obtained between PRA-hsp65 and PRA-rpoB. When others were uncertainly identified by PRA or discordant results were obtained with the PRA and phenotypic methods,hsp65,16SrRNA,rpoBorITSgene sequencing was performed to properly identify the mycobacterial species[29-31]. Statistical analysis was performed by the χ2test with SPSS 16.0 software (SPSS Inc.,Chicago,USA).Pvalues of <0.05 were considered statistically significant.
ResultsPreliminary identification and multi-loci PCR testing
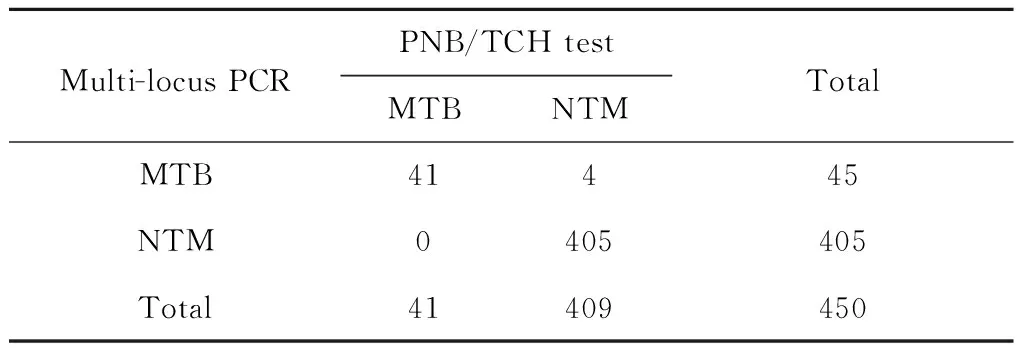
For PNB/TCH testing,the 450 pre-identified clinical NTM isolates were identified comprising 409 (90.89%) NTM and 41 (9.11%)M.tuberculosisstrains,while multi-loci PCR testing,were 405 (90.00%) NTM and 45 (10.00%)M.tuberculosisrespectively (Table 1). Although those two kinds of tests showed 4 discordant strains,we did not observe a statistically significant difference between the PNB/TCH tests and multi-loci PCR results (χ2=2.25,P=0.125).
PRA-hsp65 and PRA-rpoB results
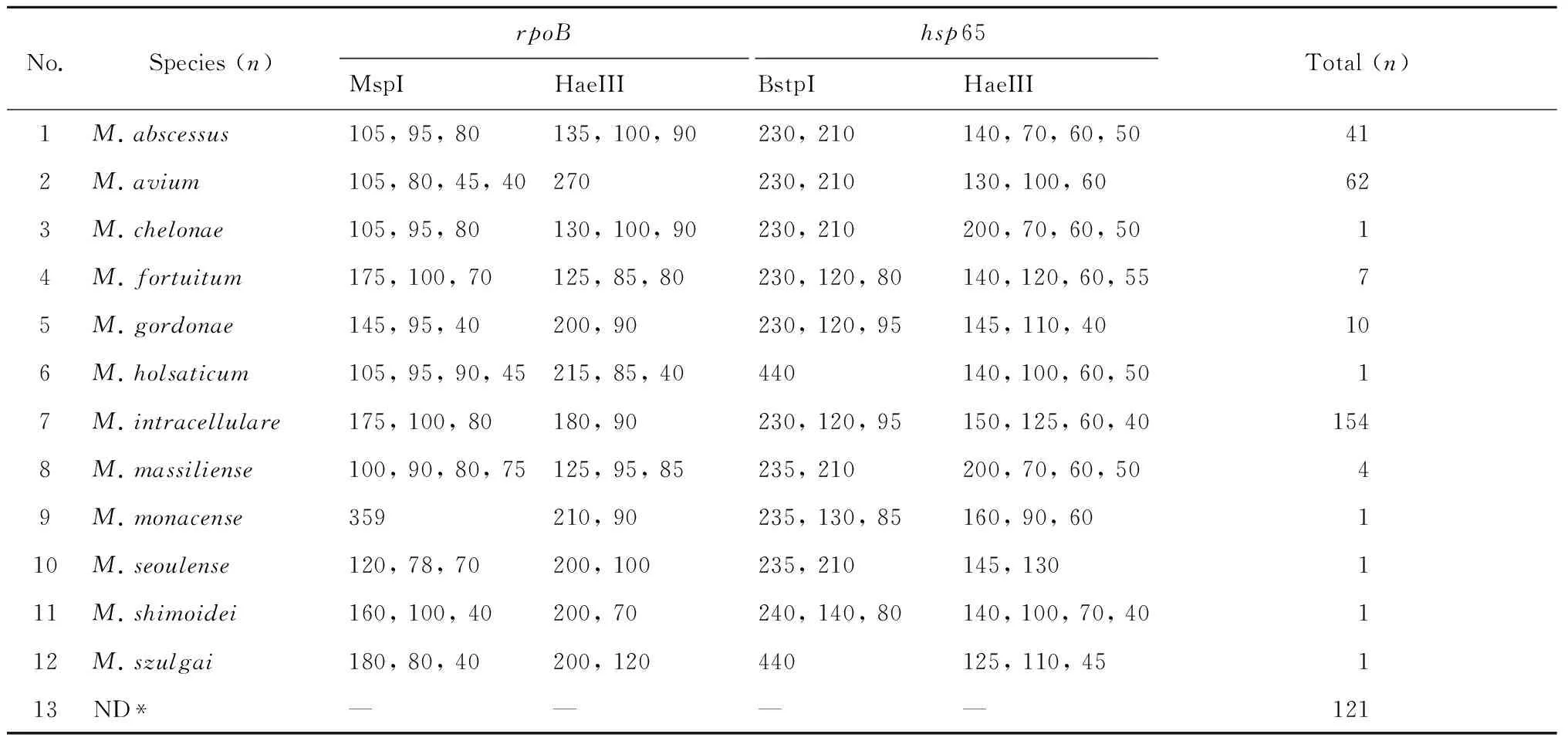
The 405 NTM strains which were identified by multi-loci PCR,were also identified by using PRA-hsp65 and PRA-rpoB. A total of 284 strains were determined to 12 NTM species (Table 2),and the rest 121 isolates could not be identified to the species level by PRA-hsp65 and PRA-rpoBsince discordant results were obtained.
Tab.1 Results of 450Mycobacterialisolates identified using PNB/TCH and multi-loci PCR tests*
Multi-locusPCRPNB/TCHtestTotalMTBNTMMTB41445NTM0405405Total41409450
Note: *,χ2=2.25;P=0.125.
Tab.2 Species identification results of the 450 NTM isolates using PRA-hsp65 and PRA-rpoB
No.Species(n)rpoBhsp65Total(n)MspIHaeIIIBstpIHaeIII1M.abscessus105,95,80135,100,90230,210140,70,60,50412M.avium105,80,45,40270230,210130,100,60623M.chelonae105,95,80130,100,90230,210200,70,60,5014M.fortuitum175,100,70125,85,80230,120,80140,120,60,5575M.gordonae145,95,40200,90230,120,95145,110,40106M.holsaticum105,95,90,45215,85,40440140,100,60,5017M.intracellulare175,100,80180,90230,120,95150,125,60,401548M.massiliense100,90,80,75125,95,85235,210200,70,60,5049M.monacense359210,90235,130,85160,90,60110M.seoulense120,78,70200,100235,210145,130111M.shimoidei160,100,40200,70240,140,80140,100,70,40112M.szulgai180,80,40200,120440125,110,45113ND*————121
Note: *ND,not determined.
Gene sequencing
Thehsp65,16SrRNA,rpoBandITSgene sequencing procedures were performed in 321 NTM strains,including 200 different NTM species strains which were selected from the 284 PRA species identified strains and all the 121 ND strains (Table 2). All the 200 NTM strains were similarly identified by the gene sequencing and PRA method. For the 121 ND strains were identified to 20 NTM species and one other genus,which including:M.intracellulare,M.avium,M.gordonae,M.massiliense,M.abscessus,M.lentiflavum,M.marseillense,M.fortuitum,M.szulgai,M.saskatchewanense,M.triplex,M.genavense,M.colombiense,M.kansasii,M.kumamotonense,M.mantenii,M.seoulense,M.septicum,M.shimoidei,M.stomatepiaeandGordoniabronchialis. In brief,in 2005 to 2012,of the 450 isolates,there were 45 strains ofM.tuberculosis,23 species of NTM and 1Gordoniagenus (Gordoniabronchialis) isolates (Table 3).
Tab.3 Frequency of the 25 differentMycobacteriumandGordoniaspecies identified from 450 clinical isolates

No. SpeciesNum(n)Ratio(%)1M.intracellulare17338.442M.avium8819.563M.abscessus5512.224M.tuberculous4510.005M.gordonae286.226M.massiliense194.227M.fortuitum92.008M.lentiflavum61.339M.marseillense51.1110M.szulgai30.6711M.saskatchewanense20.4412M.seoulense20.4413M.shimoidei20.4414M.triplex20.4415M.chelonae10.2216M.colombiense10.2217M.genavense10.2218M.holsaticum10.2219M.kansasii10.2220M.kumamotonense10.2221M.mantenii10.2222M.monacense10.2223M.septicum10.2224M.stomatepiae10.2225Gordoniabronchialis10.22Total—450100.00
Discussion
NTM species were identified in clinical specimens as early as 1885. However,their pathogenic role in human beings was long overshadowed by that ofM.tuberculosis. With recent improvements in the techniques used to isolate and identifyMycobacteriaas well as control TB,NTM have gained increasing attention from clinical and microbiological experts,because more and more NTM species have been recognized as the pathogens to cause human infection,especially in immune-compromised hosts[32]. To date,167 mycobacterial species have been recognized,and many other strains await classification[33],and approximately one third of which have been associated with diseases in humans beings[34].
Although PNB/TCH differentiation media testing is the gold standard for identifying MTB and NTM according to the Chinese clinical diagnosis of TB[35],misdiagnoses continue to occur in local hospitals. As in this study,a total of 450 strains of NTM isolates which were identified by PNB/TCH test in Fuzhou Pulmonary Hospital were re-identified using this method in ICDC,China CDC,and the results showed 41 discordant strains (Table 1),indicated that new rapid and more accurate identify methods need to be introduced to clinical diagnostic laboratories.
In this study,multi-loci PCR methods were also used to re-identify the 450 pre-identified NTM isolates,and can be identified to 405 NTM strains and 45M.tuberculosisstrains. Although the results of PNB/TCH and multi-loci PCR methods revealed 4 discordant strains,the results of these two kinds of method forMycobacteriumspecies identification did not differ significantly (Table 1). And as re-identified by combined using of gene sequencing,these 4 discordant strains also be identified as NTM isolates,so the multi-loci PCR may had higher identification accuracy than the PNB/TCH test.
All the 450 clinical Mycobacterial isolates,were identified to 45M.tuberculosisand 405 NTM isolates (details in Figure 1). And for the 405 NTM isolates which were identified to 23 NTM species,contain 404Mycobacteriumstrains and 1 strain ofGordoniabronchialis. TheM.avium-M.intracellularecomplex (M.avium,M.intracellulare,M.colombienseandM.marseillens),consisting of the main causative agents,accounted for 59.33%,which is according with the previously reports. These complex belongs to the slow-growing mycobacterium (SGM) group and usually causes pulmonary diseases. The identification of subspecies such asM.avium,M.intracellulareandM.colombienseis essential for the diagnosis and treatment of similar pulmonary infections. TheM.chelonae-M.abscessuscomplex (M.abscessus,M.chelonaeandM.massiliense),as rapidly growingMycobacteria(RGM),were also among the isolates from patients with TB in Fujian (Figure 1). With our knowledge of NTM,M.holsaticum,M.shimoidei,M.saskatchewanense,M.seoulense,M.kumamotonense,M.stomatepiaeandM.mantenii,which caused pulmonary diseases,were rarely reported in China.M.marseillense,M.saskatchewanense,M.seoulense,M.kumamotonense,M.stomatepiaeandM.manteniiwere first identified in pulmonary patients in Fujian Province,China. These NTM species should be paid more attentions to prevent its prevalence.
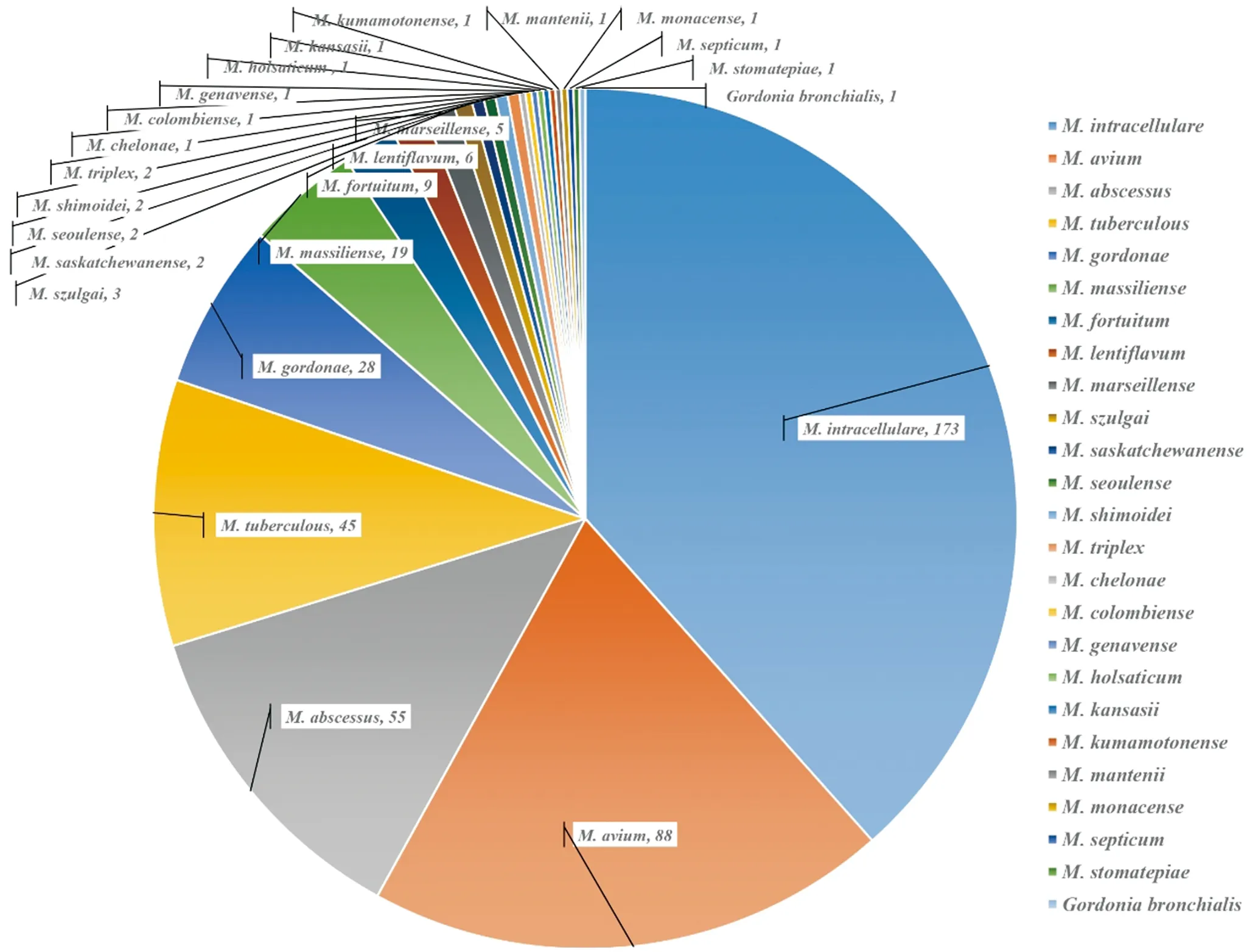
Fig.1 Species distribution of the 450 clinical mycobacterial isolates
Arnow et al investigated that endemicM.gordonaecontamination of respiratory specimens is caused byM.gordonaein hospital tap water[36]. Endemic contamination of respiratory specimens byM.gordonaeappears to be a substantial problem with an incidence as high as 14% in induced sputum samples[37]. The 18M.gordonaemay come from the hospital and laboratory tap water or laboratory buffer,which caused false-positive results in the isolates. The diversity of NTM observed here is possibly related to the geographical location of Fujian. An earlier study showed that residential soils are a likely source of pulmonary MAC infection[38]. Fujian is located in the southeast coast of mainland China,and the local subtropical climate may affect NTM recovery.
One other genus,Gordonia,was first identified in these isolates as well,but it was not differentiated using traditional identification methods such as acid-fast stains,PNB/TCH differential media and multi-locus PCR. Furthermore,althoughG.bronchialiscould be discriminated from the Mycobacteria species viahsp65 sequencing,it still has a rather close relationship with some of the mycobacteria in the phylogenetic tree. As such,the availability of an accurate and effective method is crucial to species identification. Additionally,some mycobacterium species are only identified by gene sequencing.
Due to the hyper-variability ofhsp65 gene,sequence analysis of this gene has become routine in both taxonomical studies and the identification of clinical Mycobacterium isolates[39]. TherpoBandITSgene sequencing processes are auxiliary tools. While as PRA-hsp65 and PRA-rpoBare simple,rapid and accurate to a certain extent and have been routinely used to identifyMycobacteria. Here we could use them to identify the 284 obtained clinical isolates to species level,and because of the limitations of PRA and the unknown restriction patterns,the remaining 121 isolates that were not determined (ND). Then 200 strains of the well identified NTM isolates and those ND strains were further analyzed using gene sequencing,and the results showed that PRA test and gene sequencing have well compliance,and the ND isolates also could be identified to species level. The combination use of several methods can make identification more effective.
As there has been a dramatic recent increase not only in the total number of mycobacterial species but also in the number of clinically significant species,therefore,NTM infection rates are increasing. Disseminated disease due to NTM is among the most common and severe infections in persons with advanced HIV infection. Because of some factors,particularly the prevalence of HIV/AIDS,NTM diseases will become an important public health problem. These potentially pathogenic isolates were identified as different species that will draw medical workers’ attention to control and preventMycobacteriadiseases. In China,there is not substantially more or better systematic research about NTM disease prevalence,due to China’s vast territory,larger differences exist among different areas. This study focused on the identification of NTM species and subspecies in Fujian Province,which would examine possible ways to prevent infections in at-risk patients and provide some evidence for the analysis of the diversity of NTM species and subspecies.
Acknowledgments
We thank the staff of Fuzhou Pulmonary Hospital in Fujian Province for its excellent contribution to this study.
Author contributions
For this study,Wan Kanglin conceived and designed the experiments. Huang Mingxiang,Zhao Xiuqin,and Zhang Lishui isolated and collected the clinical strains and patient information. Liu Haican,Jiang Yi and Lian Lulu performed the experiments. Liu Haican,Huang Mingxiang,Jiang Yi,Wu Yimou and Wan Kanglin analyzed the data. Wan Kanglin and Wu Yimou contributed reagents,materials and analysis tools. Liu Haican and Wan Kanglin wrote the article.
Declaration of interest
This work was funded by the project of National Key Program for Infectious Diseases of China (No. 2013ZX10006-002-001). The funders had no role in the study design,data collection and analysis,manuscript preparation process or decision to publish. The authors have read the journal’s policy and declare that no competing interests exist. Patient consent for publication was obtained.
Ethical considerations
The study was approved by the ethics committee of the National Institute for Communicable Disease Control and Prevention,Chinese Center for Disease Control and Prevention. All patients involved in the study provided written informed consent prior to participating.
[1] Daniel MT. The history of tuberculosis[J]. Respiratory Med,2006. 100(1): 1862-1870. DOI: 10.1016/j.rmed.2006.08.006
[2] Heyderman RS,Clark J. Clinical manifestations of nontuberculousMycobacteria[M]. New York: Springer,2006: 167.
[3] Griffith DE,Aksamit T,Brown-Elliott BA,et al. An official ATS/IDSA statement: diagnosis,treatment,and prevention of nontuberculous mycobacterial diseases[J]. Am J Respir Crit Care Med,2007,175(4): 367-416. DOI: 10.1164/rccm.200604-571ST
[4] Jarzembowski JA,Young MB. Nontuberculous mycobacterial infections[J]. Arch Pathol Lab Med,2008,132: 1333-1341. DOI: 10.1043/1543-2165(2008)132
[5] Lee H,Park HJ,Cho SN,et al. Species identification ofMycobacteriaby PCR-restriction fragment length polymorphism of therpoBgene[J]. J Clin Microbiol,2000,38(8): 2966-2971.
[6] Shinners D,Yeager H,Nontuberculous mycobacterial infection: clinical syndromes and diagnosis: overview[M]. 4 ed. Tuberculosis and nontuberculous mycobacterial infections,ed. D. Schlossberg,Philadelphia: W. B. Saunders Co.,1999
[7] Jing H,Wang H,Wang Y,et al,Prevalence of nontuberculousMycobacteriainfection,China,2004-2009[J]. Emerg Infect Dis,2012,18(3): 527-528. DOI: 10.3201/eid1803.110175
[8] Standards NCFCL. Susceptibility testing of mycobacteria,Nocardia,and other aerobic actinomycetes[S]. National Committee for Clinical Laboratory Standards,2003.
[9] Senna SG,Marsico AG,Vieira G,et al. Identification of nontuberculous mycobacteria isolated from clinical sterile sites in patients at a university hospital in the city of Rio de Janeiro,Brazil[J]. J Bras Pneumol,2011,37(4): 521-526. DOI: 10.1590/S1806-37132011000400015
[10] Silva C,Ueki S,Geiger D,et al.hsp65 PCR-restriction enzyme analysis (PRA) for identification ofMycobacteriain the clinical laboratory [J]. Rev Inst Med Trop Sao Paulo,2001,43(1): 25-28. DOI: 10.1590/S0036-46652001000100005
[11] Plikaytis BB,Plikaytis BD,Yakrus MA,et al. Differentiation of slowly growingMycobacteriumspecies,includingMycobacteriumtuberculosis,by gene amplification and restriction fragment length polymorphism analysis[J]. J Clin Microbiol,1992,30(7): 1815-1822.
[12] Telenti A,Marchesi F,Balz M,et al. Rapid identification ofMycobacteriato the species level by polymerase chain reaction and restriction enzyme analysis[J]. J Clin Microbiol,1993,31(2): 175-178.
[13] Kent PT,Kubica GP. Public health mycobacteriology: a guide for the level III laboratory[M]. Atlanta: US Department of Health and Human Services,Public Health Service,Centers for Disease Control. 1985: 31-46.
[14] Soolingen DV,Hermans P,Haas P,et al.,Occurrence and stability of insertion sequences inMycobacteriumtuberculosiscomplex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis[J]. J Clin Microbiol,1991,29(11): 2578-2586.
[15] Huard RC,de Oliveira Lazzarini LC,Butler WR,et al. PCR-based method to differentiate the subspecies of theMycobacteriumtuberculosiscomplex on the basis of genomic deletions[J]. J Clin Microbiol,2003,41(4): 1637-1650. DOI: 10.1128/JCM.41.4.1637-1650.2003
[16] Whang J,Lee BS,Choi GE,et al. Polymerase chain reaction-restriction fragment length polymorphism of the rpoB gene for identification ofMycobacteriumaviumsubsp. paratuberculosis and differentiation ofMycobacteriumaviumsubspecies[J]. Diagn Microbiol Infect Dis,2011,70(1): 65-71. DOI: 10.1016/j.diagmicrobio.2011.01.014
[17] Kim BJ,Lee KH,Park BN,et al.,Differentiation of mycobacterial species by PCR-restriction analysis of DNA (342 base pairs) of the RNA polymerase gene (rpoB)[J]. J Clin Microbiol,2001,39(6): 2102-2109. DOI: 10.1128/JCM.39.6.2102-2109.2001
[18] Telenti A,Marchesi F,Balz M,et al. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis[J]. J Clin Microbiol,1993,31(2): 175-178.
[19] Chimara E,Ferrazoli L,Ueky SY,et al. Reliable identification of mycobacterial species by PCR-restriction enzyme analysis (PRA)-hsp65 in a reference laboratory and elaboration of a sequence-based extended algorithm of PRA-hsp65 patterns[J]. BMC Microbiol,2008,8: 48. DOI: 10.1186/1471-2180-8-48
[20] Bodle EE,Cunningham JA,Della-Latta P,et al. Epidemiology of nontuberculousMycobacteriain atients without HIV Infection,New York City[J]. Emerg Infect Dis,2008,14(3): 390-396. DOI: 10.3201/eid1403.061143.
[21] Brunello F,Ligozzi M,Cristelli E,et al. Identification of 54 mycobacterial species by PCR-restriction fragment length polymorphism analysis of the hsp65 gene[J]. J Clin Microbiol,2001,39(8): 2799-2806. DOI: 10.1128/JCM.39.8.2799-2806.2001
[22] da Silva Rocha A,Werneck Barreto AM,Dias Campos CE,et al. Novel allelic variants ofMycobacteriaisolated in Brazil as determined by PCR-restriction enzyme analysis of hsp65[J]. J Clin Microbiol,2002,40(11): 4191-4196. DOI: 10.1128/JCM.40.11.4191-4196.2002
[23] Cheunoy W,Prammananan T,Chaiprasert A,et al. Comparative evaluation of polymerase chain reaction and restriction enzyme analysis: two amplified targets,hsp65 andrpoB,for identification of culturedMycobacteria[J]. Diagn Microbiol Infect Dis,2005,51(3): 165-171. DOI: 10.1016/j.diagmicrobio.2004.09.006
[24] Hafner B,Haag H,Geiss HK,et al. Different molecular methods for the identification of rarely isolated non-tuberculousMycobacteriaand description of newhsp65 restriction fragment length polymorphism patterns[J]. Mol Cell Probes,2004,18(1): 59-65. DOI: 10.1016/j.mcp.2003.09.003
[25] Devallois A,Goh Ks,Rastogi N. Rapid identification of mycobacteria to species level by PCR restriction fragment length polymorphism analysis of thehsp65 gene and proposition of an algorithm to differentiate 34 mycobacterial[J]. J Clin Microbiol,1997,35(11): 2969-2973.
[26] Lee H,Park HJ,Cho SN,et al. Species identification ofMycobacteriaby PCR-restriction fragment length polymorphism of the rpoB gene[J]. J Clin Microbiol,2000,38(8): 2956-2971.
[27] Reuss AM,Wiese-Posselt M,Weissmann B,et al. Incidence rate of nontuberculous mycobacterial disease in immunocompetent children: a prospective nationwide surveillance study in Germany[J]. Pediatr Infect Dis J,2009,28(7): 642-644. DOI: 10.1097/INF.0b013e3181978e8e
[28] Adékambi T,Drancourt M. Isolation ofMycobacteriumsepticumfrom the sputum of a patient suffering from hemoptoic pneumonia[J]. Res Microbiol,2006,157(5): 466-470. DOI: 10.1016/j.resmic.2005.10.006
[29] Leao SC,Martin A,Mejia GI,et al. Practical handbook for the phenotypic and genotypic identification ofMycobacteria[M]. 1 ed. 2004,Belgium: Bruges.
[30] Shin S,Kim EC,Yoon J-H. Identification of nontuberculousMycobacteriaby sequence analysis of the 16S ribosomal RNA,the heat-shock protein 65 and the RNA polymerase-subunit genes[J]. Korean J Lab Med,2006,26(3): 153-160. DOI: 10.3343/kjlm.2006.26.3.153
[31] Imperiale B,Zumárraga M,Gioffré A,et al. Disease caused by non-tuberculous mycobacteria diagnostic procedures and treatment evaluation in the North of Buenos Aires Province[J]. Revista Argentina de Microbiología,2012,44: 3-9. DOI: 10.1590/S0325-75412012000100002
[32] Jacobson K,Garcia R,Libshitz H,et al. Clinical and radiological features of pulmonary disease caused by rapidly growingMycobacteriain cancer patients[J]. Eur J Clin Microbiol Infect Dis,1998,17(9): 615-621.
[33] Euzeby JP. List of Prokaryotic names with Standing in Nomenclature-Genus Mycobacterium[EB/OL]. (2013-1-14)[2016-12-01]. http://www.bacterio.cict.fr/m/-mycobacterium.html.
[34] Zamarioli LA,Coelho AGV,Pereira CM,et al. Descriptive study of the frequency of nontuberculousMycobacteriain the Baixada Santista region of the state of São Paulo,Brazil[J]. J Bras Pneumol,2008,34(8): 590-594. DOI: 10.1590/S1806-37132008000800008
[35] Chen M,Wan K. Tuberculosis laboratory technical munual[M]. 1 ed. Beijing: Science Press,2011. (in Chinese)
[36] Arnow PM,Bakir M,Thompson K,et al. Endemic contamination of clinical specimens byMycobacteriumgordonae[J]. Clin Infect Dis,2000,31(2): 472-476. DOI: 10.1086/313940
[37] Gangadharam PR,Lockhart JA,Awe RJ,et al. Mycobacterial contamination through tap water[J]. Am Rev Respir Dis,1976,113: 894. DOI: 10.1164/arrd.1976.113.6.894
[38] Fujita K,Ito Y,Hirai T,et al. Genetic relatedness ofMycobacteriumavium-intracellulare complex isolates from patients with pulmonary MAC disease and their residential soils[J]. Clin Microbiol Infect,2013,19(6): 537-541. DOI: 10.1111/j.1469-0691.2012.03929.x
[39] Pourahmad F,Thompson KD,Adams A,et al. Comparative evaluation of polymerase chain reaction-restriction enzyme analysis (PRA) and sequencing of heat shock protein 65 (hsp65) gene for identification of aquatic mycobacteria[J]. J Microbiol Methods,2009,76(2): 128-135. DOI: 10.1016/j.mimet.2008.09.021
10.3969/j.issn.1002-2694.2017.05.002
Wan Kang-lin,Email: wankanglin@icdc.cn
2016-12-04 Editor: LIN Dan
Funded by the project of National Key Program for Infectious Diseases of China (No. 2013ZX10006-002-001). Liu Hai-can and Huang Ming-xiang contributed equally.

