拟斯卑尔脱山羊草的FISH核型分析
董磊,董晴,张文利,胡晓龙,王洪刚,王玉海
(1枣庄学院,山东枣庄 277160;2山东农业大学农学院/山东农业大学作物生物学国家重点实验室,山东泰安 271018)
拟斯卑尔脱山羊草的FISH核型分析
董磊1,2,董晴1,张文利1,胡晓龙1,王洪刚2,王玉海1
(1枣庄学院,山东枣庄 277160;2山东农业大学农学院/山东农业大学作物生物学国家重点实验室,山东泰安 271018)
【目的】建立拟斯卑尔脱山羊草的FISH核型,分析明确不同来源拟斯卑尔脱山羊草的FISH核型特点,比较不同拟斯卑尔脱山羊草及其与普通小麦的FISH核型差异。【方法】以荧光标记的寡核苷酸Oligo-pTa535和Oligo-pSc119.2为探针,利用荧光原位杂交(fluorescence in situ hybridization,FISH)技术分析pTa535和pSc119.2在不同拟斯卑尔脱山羊草、四倍体小麦和普通小麦染色体上的杂交信号分布特点;以禾本科植物着丝粒专化寡核苷酸Oligo-CCS1为探针,明确拟斯卑尔脱山羊草的着丝粒位置,测量拟斯卑尔脱山羊草染色体相关参数;通过FISH核型比较明确不同拟斯卑尔脱山羊草及其与小麦核型的多态性差异。【结果】Oligo-pTa535主要分布在小麦的D和A组染色体上,在小麦的B组染色体上仅有零星分布,在5份拟斯卑尔脱山羊草的染色体中未显示Oligo-pTa535杂交信号。Oligo-pSc119.2杂交信号主要分布在小麦的B组染色体上,在小麦的A、D组染色体中分布较少,但在5份拟斯卑尔脱山羊草染色体上均有广泛分布。根据Oligo-pTa535和Oligo-pSc119.2杂交信号在小麦染色体上的分布特点,可以将小麦的不同染色体相互区分开来。Oligo-pSc119.2杂交信号在不同倍性、不同品种的小麦B组染色体上的分布特点基本相似,而不同来源拟斯卑尔脱山羊草的Oligo-pSc119.2的FISH核型差异较大,甚至在同一细胞内的2条同源染色体上Oligo-pSc119.2杂交信号的分布也具有明显差异。不同来源的拟斯卑尔脱山羊草与小麦B染色体组的FISH核型存在明显差异。PI542238的7对染色体均为中间着丝粒染色体,核型公式为2n=14=14m。其余4份拟斯卑尔脱山羊草的4S染色体均为近中着丝粒染色体,其余染色体均为中间着丝粒染色体,核型公式皆为2n=14=12m+2sm。【结论】拟斯卑尔脱山羊草染色体上含有丰富的与pSc119.2高度同源的重复序列,不含有与pTa535高度同源的重复序列。不同来源的拟斯卑尔脱山羊草之间以及同一来源的拟斯卑尔脱山羊草的个体间甚至同一个体内的同源染色体间在 pSc119.2的分布上均具有遗传多样性。以Oligo-pSc119.2为探针建立的拟斯卑尔脱山羊草染色体FISH核型与小麦B组染色体的核型具有显著差异。利用荧光标记的Oligo-pTa535和Oligo-pSc119.2为探针进行FISH分析,可以准确区分拟斯卑尔脱山羊草的不同染色体,并能将拟斯卑尔脱山羊草与小麦的染色体区分开来。
普通小麦;拟斯卑尔脱山羊草;核型分析;FISH;寡核苷酸
0 引言
【研究意义】小麦近缘物种是进行小麦品种遗传改良的重要基因资源[1-3]。拟斯卑尔脱山羊草(Aegilops speltoides,SS,2n=14)是山羊草属拟斯卑尔脱山羊草组中的重要物种,是普通小麦B染色体组最可能的供体种[4-6]。拟斯卑尔脱山羊草遗传变异丰富,在小麦的遗传改良中具有重要利用价值,它的许多抗病(如Lr28、Lr35、Lr36、Lr47、Lr51、Sr32、Sr39、Pm12、Pm32和Pm53等)、抗虫(如Gb5与抗Hessianfly基因)和耐盐基因已被转入普通小麦[7-14]。拟斯卑尔脱山羊草在小麦的籽粒品质性状改良和耐热育种中也具有重要价值[15-16]。植物染色体核型分析是研究植物染色体数量及结构变异、形态结构特征和物种起源与进化关系的重要方法,在植物育种特别是植物远缘杂交育种中亦具有重要应用价值[4,17-19]。利用FISH技术,对不同来源拟斯卑尔脱山羊草的染色体核型进行分析,并与小麦的B染色体组核型进行比较,可为拟斯卑尔脱山羊草基因组的遗传多样性以及拟斯卑尔脱山羊草染色体组与小麦B染色体组差异研究提供参考依据。【前人研究进展】前人对山羊草多个物种的染色体核型进行了分析。TEOH等[20]研究发现不同来源的拟斯卑尔脱山羊草的 C-分带核型表现出明显的多态性,并建立了4个不同居群的标准核型及相应的核型模式图。FRIEBE等[17]研究建立了不同来源的8份单芒山羊草、8份无芒山羊草和10份顶芒山羊草的标准核型,发现3种山羊草的C-分带带型在种内均表现出较高的多态性。FRIEBE等[21]对分别来自前苏联、土耳其和希腊的19份尾状山羊草的C-分带带型进行了分析,结果表明,同一来源的尾状山羊草带型的一致性较高,而不同来源的尾状山羊草之间的带型差异较大。此后,FRIEBE等[22-23]又分别建立了西尔斯山羊草和小伞山羊草的C-带核型图及其模式图。上述多个山羊草物种的染色体核型图均是基于 C-带技术建立的,信号及染色体背景颜色均为黑色,反差小,视觉效果较差。近年来,在植物染色体核型分析中越来越多的研究采用某些克隆重复序列或寡核苷酸序列为探针,利用FISH技术建立FISH核型图。特别是同时利用多个探针建立的核型图,探针信号与染色体背景反差大、条带丰富、视觉效果明显好于C-带核型,有效提高了染色体核型分析的效果。BADAEVA等[24]以克隆重复序列pAs1和pSc119.2为探针,建立了4套单芒山羊草的标准核型。MIRZAGHADERI等[25]分别利用寡核苷酸pSc119.2和(CTT)10、pSc119.2和pTa535作为探针,建立了尾状山羊草、小伞山羊草、柱穗山羊草和钩刺山羊草的标准FISH核型。TANG等[26]开发了寡核苷酸序列pTa535和pSc119.2,前者主要分布于普通小麦的A、D组染色体上,在B组染色体上无分布或分布较少,后者主要分布于小麦B组染色体上,在A组和D组染色体上仅有零星分布。进一步支持了小麦A-D基因组之间的同源性远高于A-B和D-B之间同源性的研究结果[27]。将荧光标记的Oligo-pTa535和Oligo-pSc119.2用作探针进行小麦染色体FISH分析,根据两种探针所产生的 FISH核型可有效区分小麦的21对染色体,因此,在小麦的遗传育种研究中得到广泛应用[28-35]。DELGADO等[36]发现pTa535在大麦染色体上也有较多分布,根据 pTa535所产生的带型可将大麦的7对染色体区分开。CUADRADO等[37]证明 pSc119.2在黑麦染色体上也有大量分布,并可用于区分黑麦的染色体。【本研究切入点】关于pTa535和pSc119.2在拟斯卑尔脱山羊草染色体上的分布特点以及利用这两种寡核苷酸序列构建拟斯卑尔脱山羊草FISH核型的研究尚未见报道。【拟解决的关键问题】本研究利用pTa535和pSc119.2两种寡核苷酸重复序列为探针,对5种不同来源的拟斯卑尔脱山羊草进行FISH核型分析,建立FISH核型图和模式图,比较分析不同来源拟斯卑尔脱山羊草的染色体核型差异;并以四倍体小麦和普通小麦为对照,比较拟斯卑尔脱山羊草S组染色体FISH核型与小麦B组染色体的核型差异。
1 材料与方法
1.1 植物材料
5份拟斯卑尔脱山羊草(PI369594、PI422448、PI542244、PI542238和PI542264,SS,2n=14),由中国科学院遗传与发育生物学研究所王道文博士实验室提供。PI542244主要分布于土耳其东南部的乌尔法地区,采集于土耳其哈兰遗址与乌尔法市交界处东北7 km处,具体位置为北纬36.95°,东经38.92°,海拔350 m。其主要性状为:一年生,叶片无毛,每个小穗的最外侧两小花的外稃顶端各有一个长芒。PI542238主要分布于土耳其东南部的迪亚巴克尔地区,采集于土耳其区域农业研究站,具体位置为北纬37.97°,东经40.33°,海拔820 m。其主要性状为:一年生,叶片无毛,侧生小穗外稃无芒。PI542264主要分布于土耳其的乌尔法地区,采集于土耳其乌尔法市中心以东48 km处,具体位置为北纬37.20°,东经39.23°,海拔625 m。其主要性状为:一年生,穗粗壮,侧生小穗无芒,穗长约20 cm,顶部小穗具两根长芒。PI369594和PI422448的具体来源和地理分布不详,前者的主要性状为一年生,叶片无毛,侧生小穗含3—4朵小花,外稃具有细长的芒;后者的主要性状为一年生,叶片有毛,穗细长微弯,每小穗含3—5朵小花,除顶端小穗外稃具长芒外,侧生小穗外稃无芒,每个小穗的护颖及顶端小穗的芒上具有较密集的毛状刺。栽培二粒小麦DM4(AABB,2n=28)引自中国农业科学院作物科学研究所;普通小麦品种烟农 15和中国春(AABBDD,2n=42)由山东农业大学小麦染色体工程育种实验室繁殖保存。
1.2 方法
染色体制片和原位杂交参照KATO等方法[38]进行,拟斯卑尔脱山羊草核型制作参照MOLNÁR等[39]的标准进行。以Tamra(6-carboxytetramethyl -rhodamine)标记的禾本科着丝粒专化寡核苷酸CCS1为探针,分析明确拟斯卑尔脱山羊草的着丝粒位置;每个材料随机选取5个细胞轮廓清楚、染色体数目完整、分散良好、形态清晰的根尖细胞,利用 Micromeasure 3.3软件测量并计算染色体的相对长度和臂比。四倍体小麦和普通小麦的核型制作参照TANG等[26]的标准进行。每份山羊草随机选取25粒种子、四倍体小麦DM4和2个普通小麦品种均随机选取 3粒种子,利用 Tamra(6-carboxytetramethylrhodamine)标记的寡核苷酸pTa535和6-FAM(6-carboxy -fluorescein)标记的寡核苷酸pSc119.2为探针,对其进行FISH核型分析。pTa535和pSc119.2 2种重复序列最初分别来源于普通小麦和黑麦[26]。CCS1、pTa535和pSc119.2 3种寡核苷酸由上海英维捷基公司合成。
2 结果
2.1 拟斯卑尔脱山羊草和小麦的FISH分析
同时利用CCS1、pTa535和pSc119.2对5种不同来源的拟斯卑尔脱山羊草、栽培二粒小麦 DM4和 2个普通小麦品种的根尖细胞进行了原位杂交分析(图1),CCS1在5份拟斯卑尔脱山羊草所有染色体的着丝粒部位均显示了明显的红色杂交信号。pTa535在5份拟斯卑尔脱山羊草的根尖细胞染色体上未显示明显的杂交信号,而pSc119.2在5份拟斯卑尔脱山羊草所有染色体上均显示了丰富的绿色杂交信号,这些信号主要分布于拟斯卑尔脱山羊草染色体的两端,在染色体的中部几乎没有信号(图1-A—图1-E)。与拟斯卑尔脱山羊草相似,pSc119.2在DM4的B组染色体上也显示了丰富的杂交信号,这些信号主要分布在染色体长臂的端部、近端部或中部及短臂的端部,在着丝粒附近未见分布。在DM4的A组染色体中,除了4A长臂末端及 5A短臂末端产生了少量杂交信号外,A组其他染色体未发现pSc119.2的杂交信号。pTa535的信号主要分布在DM4的A组染色体上,在B组的7对染色体上未显示明显的信号。除了5B染色体的着丝粒信号较弱外,CCS1在DM4的其他染色体的着丝粒部位均显示了强烈的红色荧光信号(图1-F)。
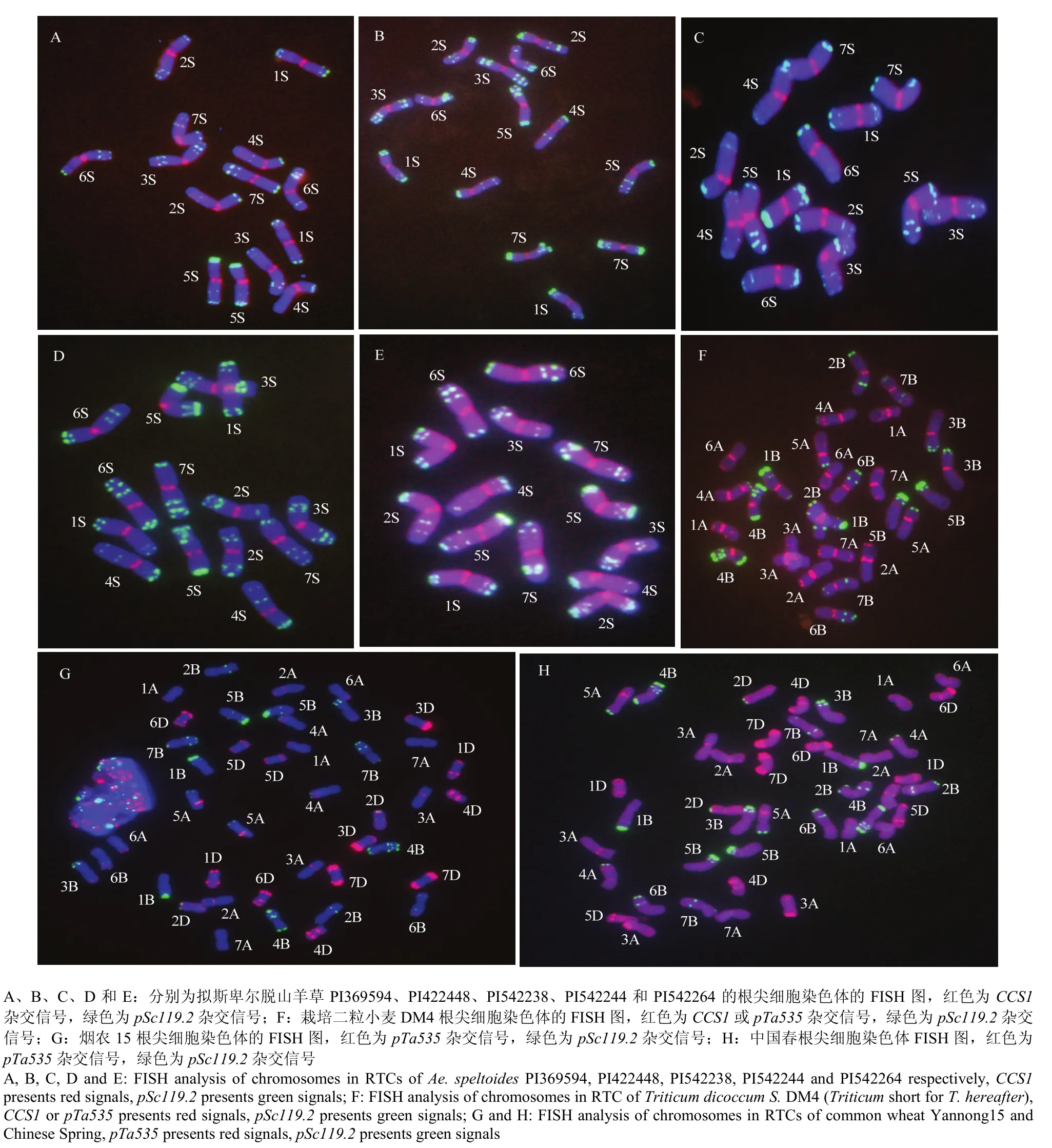
图1 拟斯卑尔脱山羊草、栽培二粒小麦DM4、烟农15和中国春的染色体FISH分析Fig. 1 FISH patterns of the chromosomes in Ae. speltoides, T. dicoccum S. DM4, T. aestivum Yannong 15 and Chinese Spring
在2个普通小麦中,pTa535的红色杂交信号在D组染色体上的分布最广泛,其次是A组染色体,而在B组染色体上几乎没有分布或分布极少。与DM4相似,pSc119.2的绿色杂交信号在普通小麦A组中的4A长臂末端及5A短臂末端或长臂中部有少量分布,在2D短臂末端也有少量分布。pSc119.2的杂交信号在B组染色体上的分布最丰富,且在每对染色体上分布的位置、数量均有明显差异(图1-G、图1-H)。
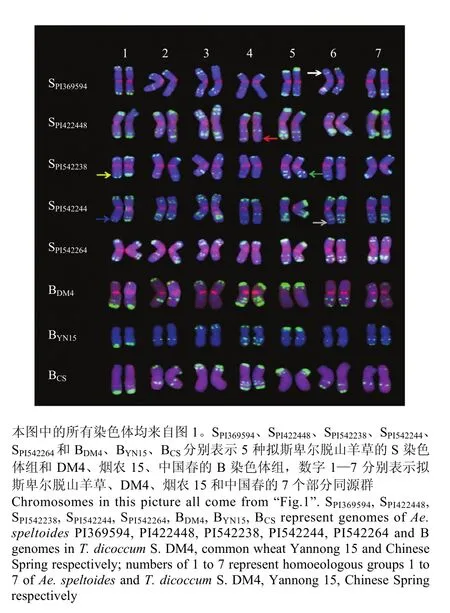
图2 拟斯卑尔脱山羊草与小麦B组FISH 核型比较Fig. 2 Karyotype comparison between Ae. speltoides and B genomes of wheat
以四倍体小麦DM4、普通小麦品种烟农15和中国春的B组染色体核型为对照,对5份拟斯卑尔脱山羊草的FISH核型进行了比对(图2)。结果表明,在同一个部分同源群内,DM4的FISH带型与普通小麦相似,而不同来源的拟斯卑尔脱山羊草染色体的FISH带型均与小麦中对应的 B组染色体存在明显差异。不同拟斯卑尔脱山羊草相同部分同源群内的染色体,其FISH带型也具有明显差异,甚至同一个细胞内的一对同源染色体的FISH带型也不完全相同。例如,PI369594的2条6S染色体短臂末端的随体信号差异明显,一个随体信号出现在随体与短臂的交界处,另一个随体信号则出现在随体末端(白色箭头所示);PI422448的2条4S染色体,其中一条染色体长臂近末端显示pSc119.2荧光信号,而其同源染色体的对应部位没有杂交信号(红色箭头所示);PI542238的2条 1S染色体中,一条染色体长臂末端仅有较弱的荧光信号,而另一条染色体的对应部位的荧光信号较强,5S的2条染色体中,一条染色体长臂上3处显示荧光信号,而另一条染色体的长臂上仅有2处显示荧光信号(黄色和绿色箭头所示);PI542244的2条1S染色体在长臂末端存在明显信号差异(蓝色箭头所示),其2条6S染色体的长臂带型也存在差异,其中一条染色体的长臂有2处显示pSc119.2的杂交信号,而另一条染色体的长臂上有3处显示杂交信号(灰色箭头所示)。但是,无论在四倍体小麦DM4还是在2个普通小麦品种中,每一对同源染色体的pSc119.2荧光带型都完全一致。
2.2 拟斯卑尔脱山羊草的FISH核型图
在5份拟斯卑尔脱山羊草中分别随机选取25粒种子萌发、采集根尖进行染色体制片,获得良好中期细胞分裂相的种子数分别为 8粒(PI369594)、8粒(PI422448)、9粒(PI542244)、9粒(PI542238)和7粒(PI542264)。为了更直观准确地描述pSc119.2杂交信号在5份拟斯卑尔脱山羊草7对染色体的分布特点,进一步建立了它们的 FISH核型模式图,对同一份拟斯卑尔脱山羊草不同籽粒来源的相同序号染色体存在的差异带型及频率进行了统计,并将不同带型的频率标注到相应染色体的FISH模式图上(图3)。
2.3 拟斯卑尔脱山羊草的染色体核型参数
对5份拟斯卑尔脱山羊草染色体的短臂和长臂长度等参数进行了测定(表1)。PI542238的7对染色体均为中间着丝粒染色体,其核型公式为2n=14=14m。其余4份山羊草的7对染色体,除4S为近中着丝粒染色体外,其他染色体均为中间着丝粒染色体,核型公式皆为2n=14=12m+2sm。
3 讨论
建立植物 FISH核型图,不仅在植物的分类及进化研究中具有重要意义[40-41],在植物育种特别是植物远缘杂交育种中也具有重要价值。LINC等[42]以克隆重复序列pSc119.2和Afa family为探针建立了二倍体长穗偃麦草的 FISH核型,不仅能区分二倍体长穗偃麦草的7对染色体,而且也能将偃麦草与普通小麦的染色体完全区分开,并根据核型特征进一步对普通小麦-二倍体长穗偃麦草一套附加系及11个普通小麦-二倍体长穗偃麦草双端体附加系进行了鉴定,成功地识
别出其中的偃麦草染色体或染色体臂。DELGADO等[36]证明,依据寡核苷酸Oligo-pTa535和Oligo-pAs1在野生大麦染色体上的分布特点,可以将7对大麦染色体与小麦染色体区分开。本研究结果表明,通过FISH核型分析可以在染色体基组的水平上准确区分拟斯卑尔脱山羊草和小麦的染色体,建立的拟斯卑尔脱山羊草的 FISH核型,揭示了不同来源拟斯卑尔脱山羊草及其与小麦B染色体组的多样性差异,它在区分拟斯卑尔脱山羊草不同染色体及鉴定小麦遗传背景中的拟斯卑尔脱山羊草染色体或染色体片段中具有利用价值。
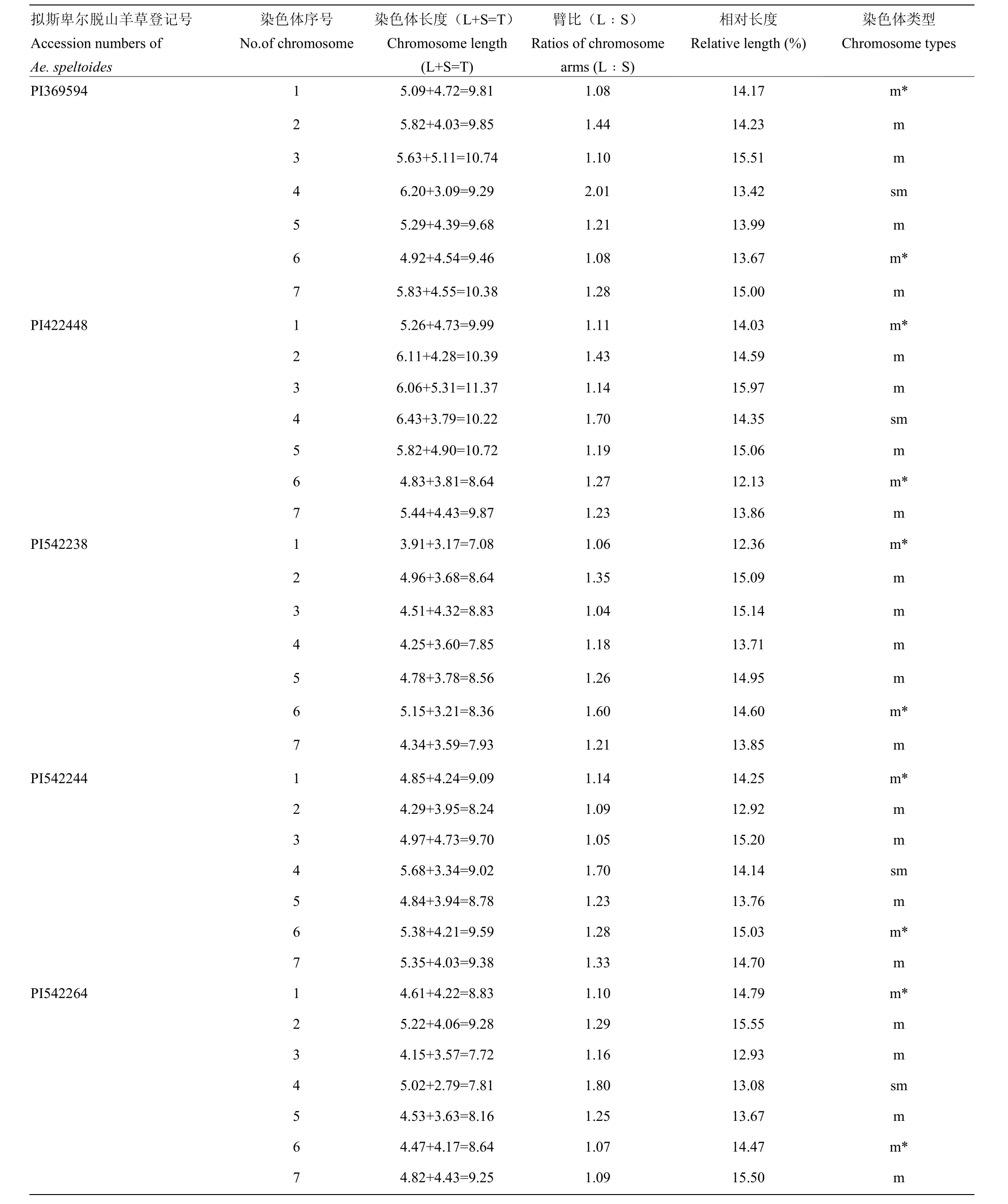
表1 拟斯卑尔脱山羊草的染色体参数Table 1 Chromosome parameters of Ae. speltoides used in this study
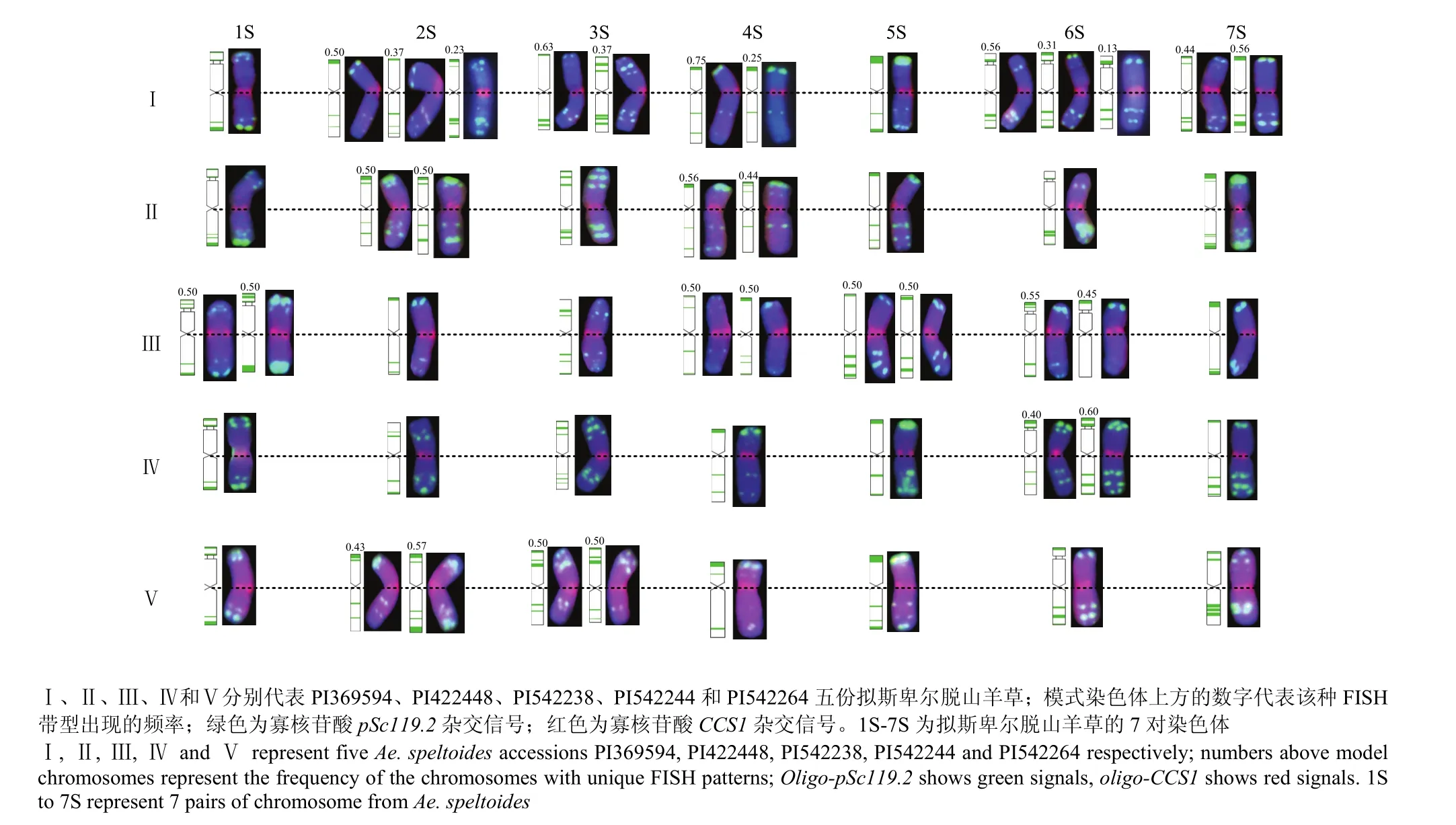
图3 拟斯卑尔脱山羊草的染色体FISH核型模式图Fig. 3 FISH idiogram of the chromosomes of five accessions of Ae. speltoides
关于小麦属B染色体组的起源问题,至今尚未找到与其完全相同的供体。颜济等[43]综合分析了目前多方面研究结果认为,小麦的B染色体组来源于拟斯卑尔脱山羊草S染色体组的可能性很大,S染色体组通过组间染色体的局部代换、重组及基因突变等重要变化而演化形成B染色体组。通过对拟斯卑尔脱山羊草S染色体组FISH核型与四倍体小麦DM4及2个普通小麦品种 B染色体组 FISH核型的比较分析发现,5份拟斯卑尔脱山羊草的pSc119.2杂交信号分布与四倍体小麦DM4和2个普通小麦品种的B组染色体确实存在明显差异。这表明,拟斯卑尔脱山羊草的染色体组与小麦的B染色体组确实存在较大的遗传分化。这些结果对于深入研究小麦B染色体组的起源与进化及其与拟斯卑尔脱山羊草染色体组的遗传关系具有参考价值。
由 U.S. National Plant Germplasm System网站( https://npgsweb.ars-grin.gov/gringlobal/view2.aspx?d v=web_site_taxon_accessionlist¶ms=:taxonomyid= 1547;:siteid=19)信息可知,本研究的5份拟斯卑尔脱山羊草中,除PI369594和PI422448等2份拟斯卑尔脱山羊草的来源和和分布不详外,其他3份山羊草均来自土耳其。其中,PI542244和PI542264主要分布于土耳其的乌尔法地区,PI542238主要分布于土耳其的迪亚巴克尔地区,而乌尔法和迪亚巴克尔均位于土耳其的东南部,地理位置相近。另外,PI542244、PI542238和PI542264的性状差异明显。这说明,拟斯卑尔脱山羊草在土耳其东南部地区(特别是乌尔法和迪亚巴克尔)可能有着广泛分布,且遗传变异丰富,加之拟斯卑尔脱山羊草具有异花授粉习性[44],具有遗传差异的拟斯卑尔脱山羊草混生在一起,相互之间容易发生天然杂交,这可能是造成不同拟斯卑尔脱山羊草的FISH核型表现差异的原因。
4 结论
拟斯卑尔脱山羊草染色体上含有丰富的与pSc119.2高度同源的重复序列,不含有与pTa535高度同源的重复序列。不同来源拟斯卑尔脱山羊草之间以及同一来源的拟斯卑尔脱山羊草的单株间甚至单株内的同源染色体间在pSc119.2的分布上均具有遗传多样性。以Oligo-pSc119.2建立的拟斯卑尔脱山羊草染色体FISH核型与小麦B组染色体的核型具有显著差异。利用荧光标记的Oligo-pTa535和Oligo-pSc119.2为探针进行 FISH分析,可以准确区分拟斯卑尔脱山羊草的不同染色体,并能将拟斯卑尔脱山羊草与小麦的染色体区分开来。
[1] 周阳, 何中虎, 张改生, 夏兰琴, 陈新民, 高永超, 井赵斌, 于广军. 1BL/1RS易位系在我国小麦育种中的应用. 作物学报, 2004, 30(6): 531-535.
ZHOU Y, HE Z H, ZHANG G S, XIA L Q, CHEN X M, GAO Y C, JING Z B, YU G J. Utilization of 1BL/1RS translocation in wheat breeding in China. Acta Agrinomica Sinica, 2004, 30(6): 531-535. (in Chinese)
[2] 张丽, 张怀渝, 任正隆, 罗培高. 小麦-黑麦 1BL/1RS易位系在小麦遗传改良中的应用现状及前景分析. 分子植物育种, 2010, 8(14): 1-8.
ZHANG L, ZHANG H Y, REN Z L, LUO P G. The progress and prospect of 1BL/RS translocation line in wheat genetic improvement. Molecular Plant Breeding, 2010, 8(14): 1-8. (in Chinese)
[3] 钟冠昌, 穆素梅, 张正斌. 麦类远缘杂交. 北京: 科学出版社, 2002: 92-97.
ZHONG G C, MU S M, ZHANG Z B. Wide Hybridization in Triticeae. Beijing: China Science Press, 2002: 92-97. (in Chinese)
[4] MAESTRA B, NARANJO T. Homoeologous relationships of Aegilops speltoides chromosomes to bread wheat. Theoretical and Applied Genetics, 1998, 97: 181-186.
[5] SALSE J, CHAGUÉ V, BOLOT S, MAGDELENAT G, HUNEAU C, PONT C, BELCRAM H, COULOUX A, GARDAIS S, EVRARD A, SEGURENS B, CHARLES M, RAVEL C, SAMAIN S, CHARMET G, BOUDET N, CHALHOUB B. New insights into the origin of the B genome of hexaploid wheat: Evolutionary relationships at the SPA genomic region with the S genome of the diploid relative Aegilops speltoides. BMC Genomics, 2008, 9: 555.
[6] PETERSEN G, SEBERG O, YDE M, BERTHELSEN K. Phylogenetic relationships of Triticum and Aegilops and evidence for the origin of the A, B, and D genomes of common wheat (Triticum aestivum). Molecular Phylogenetics and Evolution, 2006, 39(1): 70-82.
[7] MAGO R, VERLIN D, ZHANG P, BANSAL U, BARIANA H, JIN Y, ELLIS J, HOXHA S, DUNDAS I. Development of wheat-Aegilops speltoides recombinants and simple PCR-based markers for Sr32 and a new stem rust resistance gene on the 2S#1 chromosome. Theoretical and Applied Genetics, 2013, 126: 2943-2955.
[8] JIA J, DEVOS K M, CHAO S, MILLER T E, READER S M, GALE M D. RFLP-based maps of the homoeologous group-6 chromosomes of wheat and application in the tagging of Pm12, a powdery mildew resistance gene transferred from Aegilops speltoides to wheat. Theoretical and Applied Genetics, 1996, 92: 559-565.
[9] NAIK S, GILL K S, PRAKASA RAO V S, GUPTA V S, TAMHANKAR S A, PUJAR S, GILL B S, RANJEKAR P K. Identification of a STS marker linked to the Aegilops speltoidesderived leaf rust resistance gene Lr28 in wheat. Theoretical and Applied Genetics, 1998, 97: 535-540.
[10] PETERSEN S, LYERLY J H, WORTHINGTON M L, PARKS W R, COWGER C, MARSHALL D S, BROWN-GUEDIRA G, MURPHY J P. Mapping of powdery mildew resistance gene Pm53 introgressed from Aegilops speltoides into soft red winter wheat. Theoretical and Applied Genetics, 2015, 128: 303-312.
[11] YU G T, KLINDWORTH D L, FRIESEN T L, FARIS J D, ZHONG S B, RASMUSSEN J B, XU S S. Development of a diagnostic co-dominant marker for stem rust resistance gene Sr47 introgressed from Aegilops speltoides into durum wheat. Theoretical and Applied Genetics, 2015, 128: 2367-2374.
[12] DUBCOVSKY J, LUKASZEWSKI A J, ECHAIDE M, ANTONELLI E F, PORTER D R. Molecular characterization of two Triticum speltoides interstitial translocations carrying leaf rust and greenbug resistance genes. Crop Science, 1998, 38: 1655-1660.
[13] NOORI S A S. Assessment for salinity tolerance through intergeneric hybridisation: Triticum durum × Aegilops speltoides. Euphytica, 2005, 146: 149-155.
[14] YUDINA R S, LEONOVA I N, SALINA E A, KHLESTKINA E K. Change in salt tolerance of bread wheat as a result of the introgression of the genetic material of Aegilops speltoides and Triticum timopheevii. Russian Journal of Genetics: Applied Research, 2016,6(3): 244-248.
[15] PSHENICHNIKOVA T A, SIMONOV A V, ERMAKOVA M F, CHISTYAKOVA A K, SHCHUKINA L V, MOROZOVA E V. The effects on grain endosperm structure of an introgression from Aegilops speltoides Tausch. into chromosome 5A of bread wheat. Euphytica, 2010, 175: 315-322.
[16] AWLACHEW Z T, SINGH R, KAUR S, BAINS N S, CHHUNEJA P. Transfer and mapping of the heat tolerance component traits of Aegilops speltoides in tetraploid wheat Triticum durum. Molecular Breeding, 2016, 36: 78.
[17] FRIEBE B, BADAEVA E D, KAMMER K, GILL B S. Standard karyotypes of Aegilops uniaristata, Ae. mutica, Ae. comosa subspecies comosa and heldreichii (Poaceae). P1ant Systematics Evolution, 1996, 202: 199-210.
[18] FERNÁNDEZ-CALVIN B, ORELLANA J. Metaphase-I bound-arm frequency and genome analysis in wheat-Aegilops hybrids. 2. Cytogenetical evidence for excluding Ae. sharonensis as the donor of the B genome of polyploid wheats. Theoretical and Applied Genetics, 1993, 85(5): 587-592.
[19] FRIEBE B, QI L L, NASUDA S, ZHANG P, TULEEN N A, GILL B S. Development of a complete set of Triticum aestivum-Aegilops speltoides chromosome addition lines. Theoretical and Applied Genetics, 2000, 101: 51-58.
[20] TEOH S B, MILLER T E, READER S M. Intraspecific variation in C-banded chromosomes of Aegilops comosa and Ae. Speltoides. Theoretical and Applied Genetics, 1983, 65: 343-348.
[21] FRIEBE B, SCHUBERT V, BLIITHNER W D, HAMMER K. C-banding pattern and polymorphism of Aegilops caudata and chromosomal constitutions of the amphiploid T. aestivum-Ae. caudata and six derived chromosome addition lines. Theoretical and Applied Genetics, 1992, 83: 589-596.
[22] FRIEBE B, JIANG J, TULEEN N, GILL B S. Standard karyotype of Triticum umbellulatum and the characterization of derived chromosome addition and translocation lines in common wheat. Theoretical and Applied Genetics, 1995, 90: 150-156.
[23] FRIEBE B, TULEEN N A, GILL B S. Standard karyotype of Triticum searsii and its relationship with other S-genome species and common wheat. Theoretical and Applied Genetics, 1995, 91: 248-254.
[24] BADAEVA E D, DEDKOVA O S, ZOSHCHUK S A, AMOSOVA A V, READER S M, BERNARD M, ZELENIN A V. Comparative analysis of the N-genome in diploid and polyploid Aegilops species. Chromosome Research, 2011, 19: 541-548.
[25] MIRZAGHADERI G, HOUBEN A, BADAEVA E D. Molecularcytogenetic analysis of Aegilops triuncialis and dentification of its chromosomes in the background of wheat. Molecular Cytogenetics, 2014, 7: 91.
[26] TANG Z X, YANG Z J, FU S L. Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. Journal of Applied Genetics, 2014, 55: 313-318.
[27] FERNFINDEZ-CALVFN B, ORELLANA J. Metaphase I-bound arms frequency and genome analysis in wheat-Aegilops hybrids. 3. Similar relationships between the B genome of wheat and S or SLgenomes of Ae. speltoides, Ae. longissima and Ae. sharonensis. Theoretical and Applied Genetics, 1994, 88: 1043-1049.
[28] 王玉海, 何方, 鲍印广, 明东风, 董磊, 韩庆典, 李莹莹, 王洪刚.高抗白粉病小麦-山羊草新种质TA002的创制和遗传研究. 中国农业科学, 2016, 49(3): 418-428.
WANG Y H, HE F, BAO Y G, MING D F, DONG L, HAN Q D, LI Y Y, WANG H G. Development and genetic analysis of a novel wheat-Aegilops germplasm TA002 resistant to powdery mildew. Scientia Agricultura Sinica, 2016, 49(3): 418-428. (in Chinese)
[29] WANG Y H, WANG H G. Characterization of three novel wheat-Thinopyrum intermedium addition lines with novel storage protein subunits and resistance to both powdery mildew and stripe rust. Journal of Genetics and Genomics, 2016, 43: 45-48.
[30] KRUPPA K, TÜRKÖSI E, MAYER M, TÓTH V, VIDA G, SZAKÁCS É, MOLNÁR-LÁNG M. McGISH identification and phenotypic description of leaf rust and yellow rust resistant partial amphiploids originating from a wheat × Thinopyrum synthetic hybrid cross. Journal of Applied Genetics, 2016, 57: 427-437.
[31] LI G R, WANG H J, LANG T, LI J B, LA S X, YANG E N, YANG Z J. New molecular markers and cytogenetic probes enable chromosome identification of wheat-Thinopyrum intermedium introgression lines for improving protein and gluten contents. Planta, 2016, 244: 865-876.
[32] 陈雷, 李萌, 王洋洋, 邱玲, 汤述尧, 唐宗祥, 符书兰. 小麦-黑麦1BL/1RS 易位系中的染色体结构变异. 麦类作物学报, 2015, 35(8): 1038-1043.
CHEN L, LI M, WANG Y Y, QIU L, TANG S Y, TANG Z X, FU S L. Structural variation of chromosomes in wheat-rye 1BL/1RS translocation lines. Journal of Triticeae Crops, 2015, 35(8): 1038-1043. (in Chinese)
[33] SCHNEIDER A, RAKSZEGI M, MOLNÁR-LÁNG M, SZAKÁCS É. Production and cytomolecular identification of new wheat-perennial rye (Secale cereanum) disomic addition lines with yellow rustresistance (6R) and increased arabinoxylan and protein content (1R, 4R, 6R). Theoretical and Applied Genetics, 2016, 129: 1045-1059.
[34] ZHUANG L F, LIU P, LIU Z Q, CHEN T T, WU N, SUN L, QI Z J. Multiple structural aberrations and physical mapping of rye chromosome 2R introgressed into wheat. Molecular Breeding, 2015, 35: 133.
[35] FU S L, REN Z L, CHEN X M, YAN B J, TAN F Q, FU T H, TANG Z X. New wheat-rye 5DS-4RS·4RL and 4RS-5DS·5DL translocation lines with powdery mildew resistance. Journal of Plant Research, 2014, 127: 743-753.
[36] DELGADO A, CARVALHO A, MARTÍN A C, MARTÍN A, LIMA-BRITO J. Use of the synthetic Oligo-pTa535 and Oligo-pAs1 probes for identification of Hordeum chilense-origin chromosomes in hexaploid Tritordeum. Genetic Resources and Crop Evolution, 2016, 63: 945-951.
[37] CUADRADO A, JOUVE N. Evolutionary trends of different repetitive DNA sequences during speciation in the genus secale. The Journal of Heredity, 2002, 93(5): 339-345.
[38] KATO A, JONATHAN C L, BIRCHLER J A. Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proceedings of the National Academy of Sciences of the USA, 2004, 101(37): 13554-13559.
[39] MOLNÁR I, KUBALÁKOVÁ M, ŠIMKOVÁ H, FARKAS A, CSEH A, MEGYERI M, VRÁNA J, MOLNÁR-LÁNG M, DOLEŽEL J. Flow cytometric chromosome sorting from diploid progenitors of bread wheat, T. urartu, Ae. speltoides and Ae. Tauschii. Theoretical and Applied Genetics, 2014, 127: 1091-1104.
[40] BADAEVA E D, AMOSOVA A V, MURAVENKO O V, SAMATADZE T E, CHIKIDA N N, ZELENIN A V, FRIEBE B, GILL B S. Genome differentiation in Aegilops. 3. Evolution of the D-genome cluster. Plant Systematics and Evolution, 2002, 231: 163-190.
[41] BADAEVA E D, AMOSOVA A V, SAMATADZE T E, ZOSHCHUK S A, SOSHTAK N G, CHIKIDA N N, ZELENIN A V, RAUPP W J, FRIEBE B, GILL B S. Genome differentiation in Aegilops. 4. Evolution of the U-genome cluster. Plant Systematics Evolution, 2004, 246: 45-76.
[42] LINC G, SEPSI A, MOLNÁR-LÁNG M. A FISH karyotype to study chromosome polymorphisms for the Elytrigia elongata E genome. Cytogenetic and Genome Research, 2012, 136: 138-144.
[43] 颜济, 杨俊良. 小麦族生物系统学(第一卷第二版), 小麦-山羊草复合群. 北京: 中国农业出版社, 2013: 43-53.
YAN J, YANG J L. Triticeae Systematics (volume 1 2ndedition), Triticum- Aegilops complx. Beijing: China Agriculture Press, 2013: 43-53. (in Chinese)
[44] 董玉琛, 郑殿升. 中国小麦遗传资源. 北京: 中国农业出版社, 2000: 152.
DONG Y C, ZHENG D S. The Wheat Genetic Resource in China. Beijing: China Agriculture Press, 2000: 152. (in Chinese)
(责任编辑 李莉)
Karyotypic Analysis of Aegilops speltoides Revealed by FISH
DONG Lei1,2, DONG Qing1, ZHANG WenLi1, HU XiaoLong1, WANG HongGang2, WANG YuHai1
(1Zaozhuang University, Zaozhuang 277160, Shandong;2Agronomy College, Shandong Agricultural University/State Key Laboratory of Crop Biology of Shandong Agricultural University, Tai’an 271018, Shandong)
【Objective】The objective of this study is to reveal the karyotypic polymorphism of Aegilops speltoides (Aegilops short for Ae. hereafter) and the karyotypic difference between common wheat and Ae. speltoides via the establishment of FISH karyotype of Ae. speltoides.【Method】Multicolor fluorescence in situ hybridization (mc-FISH) was employed to detect the distribution of Oligo-pSc119.2 and Oligo-pTa535 in chromosomes of Ae. speltoides; centromere-specific oligonucleotide CCS1 wasused to identify the location of centromeres on chromosomes; FISH karyotype comparison was conducted to show the karyotypic differences between Ae. speltoides and wheat.【Result】In wheat, oligo-pTa535 signals were observed mainly on chromosomes in A and D genomes, and only very sporadic signals were found in B genome. However, oligo-pTa535 signals were absent in Ae. speltoides of five accessions. Oligo-pSc119.2, compared to a little distribution in A and D genomes, shined plentiful fluorescence in the whole genome B in wheat, and especially in S genomes in Ae. speltoides of five accessions used in this study. Different pairs of chromosomes in wheat could be distinguished from each other according to the distribution of Oligo-pSc119.2 and Oligo-pTa535 on chromosomes of wheat. FISH patterns produced by Oligo-pSc119.2 in wheat showed similarity among wheat materials whether of different ploidy or of different varieties of same ploidy, and that in Ae. speltoides varied depending on accessions. Even homologous chromosomes in one cell in Ae. speltoides exhibited differences in FISH pattern. Oligo-pSc119.2 FISH patterns of five accessions each showed obvious differences from that of B genomes in wheat. Four of five Ae. speltoides accessions possess six pairs of metacentric chromosomes except for homologous pairs 4S of submetacentric chromosomes, which were involved in the karyotype formula 2n = 14 = 12m + 2sm, and the rest one, PI542238, however, houses seven pairs of metacentric chromosomes which resulted in the karyotype formula 2n = 14 = 14m. 【Conclusion】 Chromosomes of Ae. speltoides house rich repetitive DNA sequences highly homologous to pSc119.2 and lack that highly homologous to pTa53. The distribution of pSc119.2 on chromosomes of Ae. speltoides showed differences between accessions, between plants of one accession and even between homologous chromosomes in one plant. FISH patterns produced by Oligo-pSc119.2 on Ae. speltoides chromosomes exhibited significant difference from that on chromosomes of B genomes in wheat. FISH analysis, using Oligo-pSc119.2 and Oligo-pTa535 as probes, not only could differentiate the chromosomes in wheat from that in Ae. speltoides, but also could dishtinguish the chromosomes from each other whether in wheat or in Ae. speltoides.
Triticum aestivum; Aegilops speltoides; karyotypic analysis; FISH; oligonucleotides
2017-01-03;接受日期:2017-02-17
国家“十三五”重点研发计划(2016YFD0102004)、山东省博士基金(BS2011SW053)、作物生物学国家重点实验室开放课题(2015KF06)、枣庄学院国家基金预研究项目(2016YY14)
联系方式:董磊,E-mail:dong2012lei@163.com。通信作者王洪刚,E-mail:hgwang@edu.sdau.cn。通信作者王玉海,E-mail:yhwang92@163.com

