Proteomic profile of human monocytic cells infected with dengue virus
Viviana Martínez-Betancur,Marle´n Martínez-Gutierrez,2*
1Programa de Estudio y Control de Enfermedades Tropicales-PECET,Universidad de Antioquia,Medellín,Colombia
2Grupo de Investigaci´on en Ciencias Animales-GRICA,Facultad de Medicina Veterinaria y Zootecnia,Universidad Cooperativa de Colombia,Bucaramanga,Colombia
Proteomic profile of human monocytic cells infected with dengue virus
Viviana Martínez-Betancur1,Marle´n Martínez-Gutierrez1,2*
1Programa de Estudio y Control de Enfermedades Tropicales-PECET,Universidad de Antioquia,Medellín,Colombia
2Grupo de Investigaci´on en Ciencias Animales-GRICA,Facultad de Medicina Veterinaria y Zootecnia,Universidad Cooperativa de Colombia,Bucaramanga,Colombia
ARTICLE INFO
Article history:
Accepted 20 Oct 2015
Available online 8 Jan 2016
Dengue virus
Proteomic
Macrophages
Monocytes
U937 cells
Objective:To identify the changes in the proteome of U937 cells infected with dengue virus(DENV).
Methods:In this study,differentiated U937 cultures were infected with two DENV-2 strains,one of which was associated with dengue(DENV-2/NG)and the other one with severe dengue(DENV-2/16681),with the aim of determining the cellular proteomic profiles under different infection conditions.Cellular proteins were extracted and separated by two-dimensional electrophoresis,and those proteins with differential expression profiles were identified by mass spectrometry.The obtained results were correlated with cellular viability,the number of infectious viral particles,and the viral DNA/protein quantity.
Results:In comparison with non-infected cultures,in the cells infected with the DENV-2/NG strain,nine proteins were expressed differentially(five were upregulated and four were downregulated);in those cultures infected with the DENV-2/16681 strain,six proteins were differentially expressed(two were downregulated and four were upregulated).The downregulated proteins included fatty acid-binding protein,heterogeneous nuclear ribonucleoprotein 1,protein disulfide isomerase,enolase 1,heat shock 70 kDa protein 9,phosphotyrosyl phosphatase,and annexin IV.The upregulated proteins included heat shock 90 kDa protein AA1,tubulin beta,enolase 1,pyruvate kinase,transaldolase and phospholipase C-alpha.
Conclusions:Because the monocyte/macrophage lineage is critical for disease pathogenicity,additional studies on these proteins could provide a better understanding of the cellular response to DENV infection and could help identify new therapeutic targets against infection.
1.Introduction
Dengue is the most common disease in humans that is transmitted by arthropods.It is caused by the dengue virus(DENV)[1],which is transmitted by the bite of infected mosquitoes,principally those of the species Aedes aegypti andAedes albopictus[2].This virus,whose genome is made up of positive-strand RNA,belongs to the Flaviviridae family and the Flavivirus genus[3].Although,in general,the existence of four serotypes thatshare antigenic similarities butare genetically different has been reported in the urban viral cycle setting(DENV-1 to DENV-4),the existence of a fifth serotype has recently been reported(DENV-5)in a wild cycle setting[4].
Tel/Fax:+57 7 685 45 00
E-mail:marlen.martinezg@campusucc.edu.co
Foundation Project:Supported by the Administrative Department of Science,Technology,and Research-COLCIENCIAS(Projects 111549326092 and 111549326083).
Peer review under responsibility of Hainan Medical University.The journal implements double-blind peer review practiced by specially invited international editorial board members.
Dengue affects the populations of tropical countries,where more than 2500 billion people are at risk of being infected,and it is endemic in over 100 of these countries[5].Its prevalence has increased dramatically in the last few decades,and recent studies estimate that 96 million cases occur on a yearly basis,although this number may be underestimated in some regions due to poorclinical surveillance[1].From a clinical perspective,dengue is a self-limiting febrile illness characterized by headaches,muscle aches,and skin eruptions.The most severe form of the disease is known as severe dengue in which there is increased vascular permeability that results in plasma leakage;other symptoms include thrombocytopenia,hemorrhagic manifestations,and compromised organs,such as the liver,heart,and central nervous system[6].
2221-1691/Copyright©2016 Hainan Medical University.Production and hosting by Elsevier B.V.This is an open access article under the CC BY-NC-ND license(http:// creativecommons.org/licenses/by-nc-nd/4.0/).
Although the pathogenesis ofsevere dengue is not completely clear,some of the risk factors involved in its development have been described(some related to the host and some to the virus).Of the factors related to the virus,it has been reported that the viral genetic variations partly determine virulence and,therefore,epidemic potential[7],which allows certain strains with higher replicative potential to spread more easily in a primary infection.In addition,some serotypes and strains have been associated with the development of severe forms of the disease[8].In this regard,some DENV strains within serotype 2 have been reported to be associated with the development of mild or severe forms of the disease,as is the case for the New Guineastrain(DENV-2/NG)(originally isolated from a dengue patient)[9],and the DENV-2/16681 strain(isolated from a patient with severe dengue)[10].Based on their origin and etiology,the DENV-2/NG strain has been associated with the development of dengue[11],and the DENV-2/16681 strain has been associated with severe dengue[12].Regarding hostassociated factors,much has been discussed regarding the pathogenic role of the immune response to a secondary infection.In this regard,the principles of antibody-dependent enhancement and original antigenic sin are the most studied[13,14].Independent of the factors favoring the development of severe dengue,whether in a primary or secondary infection,there are some cells that play a very important role in the initial capture of the virus,its transport to specific sites within the host,and its replication and propagation.
Monocytes,macrophages,and dendritic cells are considered the primary targets of DENV infection in vivo[15,16],although the presence of the virus in different cell types such as hepatocytes,lymphocytes,endothelial,epithelial,neuronal,and muscle cells has also been reported[17].The monocyte/macrophage lineage,which is normally involved in the processes of the innate and adaptive immune responses,is successful in the elimination of mostpathogens,although pathogenssometimesuse this strategy to their advantage.In the case of dengue,macrophages are not only the main targets of DENV for its replication but also responsible for spreading the virus to different parts of the hosting organism after its transmission and for enhancing proinflammatory cytokine production,responsible in great measure for the development of severe dengue[15,18].Within this lineage,the U937 cell line has been widely used to study different aspects of DENV infection.
The U937 cell line,isolated from human histiocytic lymphoma,shows characteristics of immature monocytes,but its differentiation promoted with phorbol esters converts it to a cell line with the morphologic and functional characteristics of macrophages[19].This cell line can be infected with DENV by two different mechanisms:through cell surface receptors(heat shock 70 kDa protein and heat shock 90 kDa protein),thus allowing the study of conventional forms of viral entry into the host cell during primary or secondary infection,and through Fc receptors that bind viral complexes with antibodies during secondary infections[20,21].This cell line has been used for studies on viral replication[22],on the evaluation of antiviral agents[23-25],on the immune response[26],etc.
Despite the importance and broad use of the U937 cell line,no studies have been performed in which protein expression changes have been determined in this cell line when infected by DENV.For this reason,the present study identified the modifications in the U937 proteome after infection with two strains of DENV-2 clinically associated with dengue:the New Guinea strain DENV-2/NG(associated with dengue)and the DENV-2/ 16681 strain(associated with severe dengue)under primary infection conditions.
2.Materials and methods
2.1.Cell maintenance and viral stock production
Differentiated-U937 cells(human monocyte derived from lymphoma),and C6/36HT cells(derived from Aedes albopictus)were a kind gift from Dr.Jaime E.Castellanos at the El Bosque University(Bogot´a,Colombia)and Dr.Guadalupe Guzman at the Pedro Kouri Institute(Havana,Cuba).Both cell lines were grown in Dulbecco's modified Eagle medium(Gibco/Invitrogen,Grand Island,NY,USA)supplemented with 0.25μg/mL of amphotericin B,100μg/mL streptomycin,100 units/mL penicillin,and either 2%(U937 cells)or 10%(C6/36HT cells)fetal bovine serum(FBS,Gibco).Cell cultures were incubated with 5%CO2,at either 37°C(U937 cells)or 34°C(C6/36HT cells). For all assays,two serotype 2 reference strains were used:strain DENV-2/NG (dengue reference strain)and strain DENV-2/ 16881(severe dengue reference strain).Both strains were a kind gift from Dr.Jorge Osorio at the University of Wisconsin(Madison,WI,USA).The viral stocks were expanded in the C6/ 36HT cell line(five passages for each viral strain)and stored at-70°C until used.
2.2.MTT viability assay
A colorimetric MTT assay was used to measure the effect of the infection on U937cell viability.For this assay,2.5×104cells were seeded in 96-well plates and were infected[multiplicity of infection(MOI):1]at 24 h.The infected cells were incubated for an additional 48 h.Then,50μL of MTT(0.5 mg/mL)were added,and the cultures were incubated for 3 h at 37°C.Then,100μL of dimethyl sulfoxide(Fisher Scientific Inc.,Rockford,IL,USA)were added,and the absorbance was read at 450 nm using a Varioskan Flash reader(Thermo Scientific Inc.,Rockford,IL,USA).The data were processed by comparing the absorbance of the infected cultures with the absorbance of the uninfected control cultures.The results were expressed as the mean of at least three independent experiments,each with two replicates(n=6).
2.3.Cell infection
A total of 3×106U937 cells were seeded in 25 cm2cell culture flasks.After 24 h,the cultures were incubated with each of the reference stocks(DENV-2/NG or DENV-2/16681)for 2 h at 37°C at an MOI of 1.Afterwards,the virus was removed,and the cultures were cultured for an additional 48 h,after which the supernatant of each culture was collected and stored at-70°C for subsequent infectious viral particle quantitation(by the plaque method).The cell monolayers were used for viral genome quantitation(by real-time RT-PCR)and for protein extraction.There were five replicates of each experimental condition(DENV-2/NG-or DENV-2/16681-infected cultures),as well as five replicates for the uninfected control cultures.
2.4.Fluorescence microscopy
U937 cells were grown on glass coverslips at a density of 2.5×104cells per coverslip and infected with DENV-2/NG or DENV-2/16881 at a MOI of 1.At 48 h after inoculation,cell monolayers were washed with phosphate buffered saline(PBS)and fixed with 3.8%paraformaldehyde in PBS at 37°C for 30 min.The monolayers were permeabilized with 0.5%Triton X-100,blocked with 5%FBS and incubated with primary anti-DENV antibody(1:500)followed by incubation with anti-mouse secondary antibody conjugated with fluorescein isothiocyanate and Hoechst 33258 dye.Finally,the cells were washed with PBS and the coverslips were mounted.Images were obtained using an inverted microscope(IX-81 Olympus).
2.5.Viral quantitation by the plaque method
A total of 5×104U937 cells were seeded in 24-well plates. After 24 h,the cells were inoculated with 200μL serial dilutions(1×10-1to 1×10-5)of collected previously viral supernatants. After 2 h of incubation,the virus was removed and Dulbecco's modified Eagle medium was supplemented with 2%FBS,and 1.5%carboxymethyl cellulose(Sigma-Aldrich,St.Louis,MO,USA)was added.The plates were incubated for 8 days at 37°C and 5%CO2.Then,the monolayers were fixed with 4%paraformaldehyde(Sigma-Aldrich)and stained with a crystal violet solution(Sigma-Aldrich).The plaques were counted to determine the plaque-forming units(PFU/mL)of the viral inoculum. The results were expressed as the mean of five independent experiments,each with two replicates(n=20).
2.6.Viral quantitation by reverse transcription,followed by real-time PCR(RT-qPCR)
The viral genome was quantified in the collected previously cell monolayers using real-time RT-PCR according to the methodology previously described[27].For this,total RNA was extracted using the modified RNAzol method and a total of 0.5μg of RNA was reverse transcribed using 0.5μg/mL of random primers(Promega Corp.,Madison,WI,USA)and 200U of M-MLV reverse transcriptase(Promega).The cDNA was amplified by real-time PCR(qPCR)using SYBR Green and DENV-2-specific primers(mD1 and mTS2).These primers amplify a 119 bp segment of the C-prM region.Amplification was conducted in a SmartCycler(Chepeid,Sunnyvale,CA,USA),and the genomic copy amounts were calculated with an absolute quantitation,using a specific standard curve for DENV-2.The results were expressed as the mean of five independent experiments(n=5).
2.7.Protein extraction
The cells obtained from infected and uninfected cultures were dislodged using 0.25%trypsin,spun at 1500 r/min for 5 min,and washed twice with PBS.The cells were lysed using hypotonic PBS(13.6 mmol/L NaCl,0.27 mmol/L KCl,0.4 mmol/L Na2HPO4,and 0.15 mmol/L KH2PO4)and subjected to five freezing(in liquid nitrogen)and thawing(using ultrasound)cycles.The insoluble material was removed by centrifugation,and the supernatant was used to precipitate proteins using acetone.The precipitate was resuspended in hydration buffer(9 mol/L urea,4%CHAPS,1.6%3-10 ampholytes,1%4-7 ampholytes,and 40 mmol/L dithiothreitol),and the protein concentration was determined using a 2D Quant kit(GE Healthcare,Piscataway,NJ,USA).
2.8.Two-dimensional electrophoresis
A total of 200μg of protein obtained from each of the experimental conditions was loaded onto 7-cm DryStrip strips with a non-linear pH of 3-10(GE Healthcare)and allowed to hydrate passively for 12 h.The first dimension(isoelectric focusing)was conducted at 20°C and with a 50μA current per strip,using the Ettan IPGphor 3 system(GE Health-care)until 14 800 volt-hours was reached.The strips were equilibrated by incubating them in buffer I(6 mol/L urea,2%sodium dodecyl sulfonate,375 mmol/L Tris-HCl pH 8.8,20%glycerol,and 10 mg/mL of dithiothreitol)for 20 min,followed by incubation in buffer II(6 mol/L urea,2%sodium dodecyl sulfonate,375 mmol/L Tris-HCl pH 8.8,20%glycerol,25 mg/mL of iodoacetamide)for 20 min.After equilibration,the strips were placed on 12.5%acrylamide/bis-acrylamide gels,and the second dimension was conducted using the Mini-PROTEAN system(Biorad,Hercules,CA,USA)at 100 V for 90 min.The gels were stained using the Oriole fluorescent kit(Biorad,Hercules,CA,USA)for 90 min.A total of 5 gel replicates for each experimental condition was obtained(n=5).
2.9.Image analysis and protein expression comparison
Gel images were captured using a ChemiDoc XRS scanner(Biorad),and the images were analyzed using ImageMaster 2D Platinum 7.0 software(GE Healthcare).The parameters used to detect spots were as follows:smooth,4;saliency,8;and minimal area,3.To determine the spots displaying differential expression,the relative volume of spots expressed in U937 cells infected with each of the reference stocks and in uninfected cultures were compared using the image analysis software.
2.10.Protein digestion and MALDI-TOF/TOF mass spectrometry
The differentially expressed protein spots were excised from the gel,washed 3 times with 50%acetonitrile(ACN)and 50 mmol/L ammonium bicarbonate(NH4HCO3)for 10 min,and dehydrated with 100%ACN for 10 min.Then,20μg/mL trypsin in 25 mmol/L NH4HCO3was added,and the proteins were digested overnight at 36°C.The next day,ACN was added for 10 min,and the extracted peptides were dried in a vacuum centrifuge.After centrifugation,the peptides were resuspended in a 50%methanol/0.1%trifluoroacetic solution,and each sample was dropped on a MALDI plate containing a matrix solution(50%alpha-cyano-4-hydroxycinnamic acid in ACN,0.1%trifluoroacetic,10 mmol/L ammonium citrate).The samples were analyzed with an ABI 4800 MALDI-TOF/TOF mass spectrometer(MS),with a mass range of 700-4000 Da.Peptideswith a signal-to-noise ratio above 20 in MS mode were selected for tandem mass spectrometry(MS/MS).MS/MS was conducted using air as a collision gas and 2 kV collision energy.A maximum of 45 MS/MS spectra were allowed for each spot.
2.11.Protein identification
Protein identification based on the obtained MALDI-TOF/ TOF spectra was conducted using GPS Explorer™ protein analysis 3.6 software(Applied Biosystems,Foster City,CA,USA).This software conducts a combined search of the MS and MS/MS data against the non-redundant database of the National Center of Biotechnology(NCBI nr),using the MASCOT®search engine(Matrix Science,Boston,MA,USA).Trypsin was defined as the digesting enzyme,whereas methionine oxidation and lysine acetylation were defined as peptide modifications. Mass tolerances of 80 mg/kg for precursor ions and 0.5 Da for the corresponding fragments were allowed.The bioinformatics analysis was conducted by sequence homology of proteins belonging to Homo sapiens.The biological functions of the identified proteins were assigned according to gene ontology,and the identified proteins were classified according to their cell function.
2.12.Statistical analysis
Student's t-test was used to compare the cell viability between infected and uninfected cultures.Similarly,this test was used to compare the number of infectious viral particles and the numberofgenomiccopiesbetween DENV-2/NG-versus DENV-2/16681-infected U937 cells.In both cases,P<0.05 indicated a statistically significant difference.For the proteomic analysis,an ANOVA test was used to compare the level of intensity(volume intensity)for each spot in each condition(infection with DENV-2/NG,infection with DENV-2/16681 and non-infection).Finally,comparisons with P<0.05 were further assessed with Tukey's post-hoc test.
3.Results
3.1.Effect of DENV-2 infection on U937 cell viability
An MTT assay was conducted to determine the effect of virus infection on cell viability.No significant differences were observed at 48 h post-infection when DENV-2/NG-or DENV-2/ 16681-infected U937 cells were compared to the uninfected control culture(P<0.05)(Figure 1).DENV-2 infection did not affect U937 cell viability.Moreover,neither of two viral strains produced morphological changes in the cells U937(Figure 2). Based on these results,subsequent assays were conducted with a 48-h incubation period to guarantee that any proteome changes were due to viral infection and not due to infection-induced cell death.
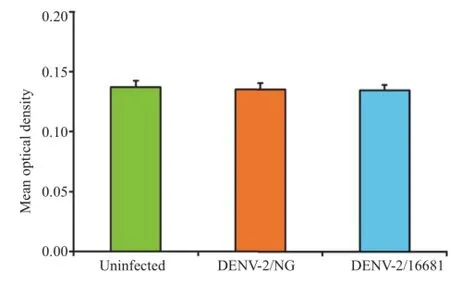
Figure 1.Effect of DENV serotype 2 on cell viability.The values represent the means of 3 independent experiments,each with 2 replicates for each condition±SEM(n=6).
3.2.Comparison of the viral genome replication rates of the DENV-2/16681 and DENV-2/NG strains
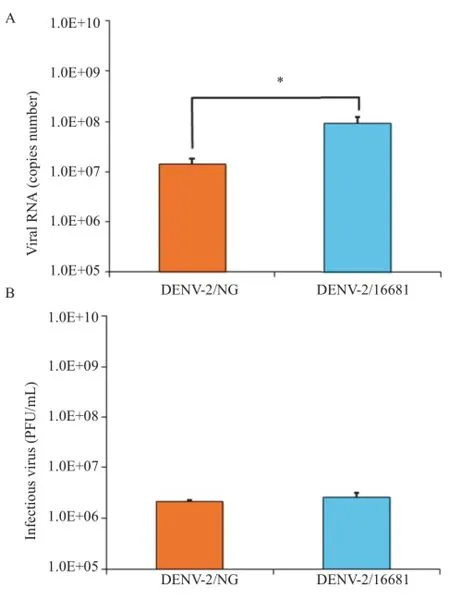
Figure 3.Comparison of the viral genome replication rates of the DENV-2/16681 and DENV-2/NG strains.Viral production was measured 48 h after cultures were infected with an MOI of 1.A:Viral genome quantitation measured by RT-qPCR in monolayers;B:Infectious viral particle quantitation was measured using a supernatant plaque assay.Significant differences were only detected when the genome copy numbers were compared.The asterisk indicates P<0.05(student's t-test).The values represent the means of 3 independent experiments,each with 2 replicates for each condition±SEM(n=20).
Figure 4.Comparison of the number of infected cells between cultures infected with DENV-2/16681 and cultures infected with DENV-2/NG.A:Uninfected cell cultures;B:Cell cultures infected with DENV-2/NG;C: Cell cultures infected with DENV-2/16681.
To compare the infection in the cultures infected with either DENV-2/NG or DENV-2/16681,RT-qPCR was conducted to quantify the viral genome(in monolayers).Viral titering by the plaque method was used to quantify the infectious viral particles released into the medium(in the supernatant).Significant differences were observed in the number of genomic copies obtained from DENV-2/NG-infected cells(1.43×107genomic copies/mL)compared with the genomic copies from DENV-2/ 16681-infected cells(9.01×107genomic copies/mL)(P<0.05).Infection was greater in the latter strain(Figure 3A). On the other hand,no significant differences were observed in the number of infectious viral particles in the supernatant of DENV-2/NG-infected cells(2.19×106PFU/mL)compared to DENV-2/16681-infected cells (2.54×106PFU/mL)(Figure 3B).These results were consistent with the number of infected cells that was higher in cultures infected with DENV-2/ 16681 than those infected with DENV-2/NG(Figure 4).
3.3.DENV-2 infection modifies protein expression in a strain-dependent manner
To obtain the proteomes,the proteins extracted from infected and uninfected U937 cell monolayers were separated by twodimensional electrophoresis.On average,189 spots were identified in uninfected U937 cells,156 spots in DENV-2/NG-infected cells,and 200 in DENV-2/16681-infected cells.The different proteins detected had isoelectric points(pI)between four and eight and molecular weights between 11 and 84 kDa. The differential expression analysis was conducted using the relative volumes of the expressed spots of the infected versus uninfected cultures.Only proteins with a differential expressionthat was reproducible in at least four of the five analyzed gels were included.The relative expression change was considered significant if the ANOVA and Tukey's post-hoc tests had P-values<0.05.
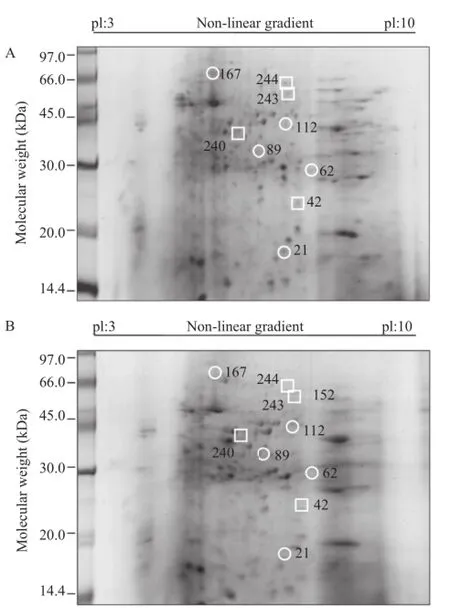
Figure 5.Differentially expressed proteins in uninfected cultures(A)compared with DENV-2/NG-infected cultures(B).A total of 200μg of protein was separated by two-dimensional electrophoresis for each sample. The differentially expressed spots are indicated in the figures.The squares indicate proteins with increased expression in infected cultures,and the circles indicate proteins with decreased expression in infected cultures.A representative gel of the 5 gels that were analyzed is shown for each biological condition.
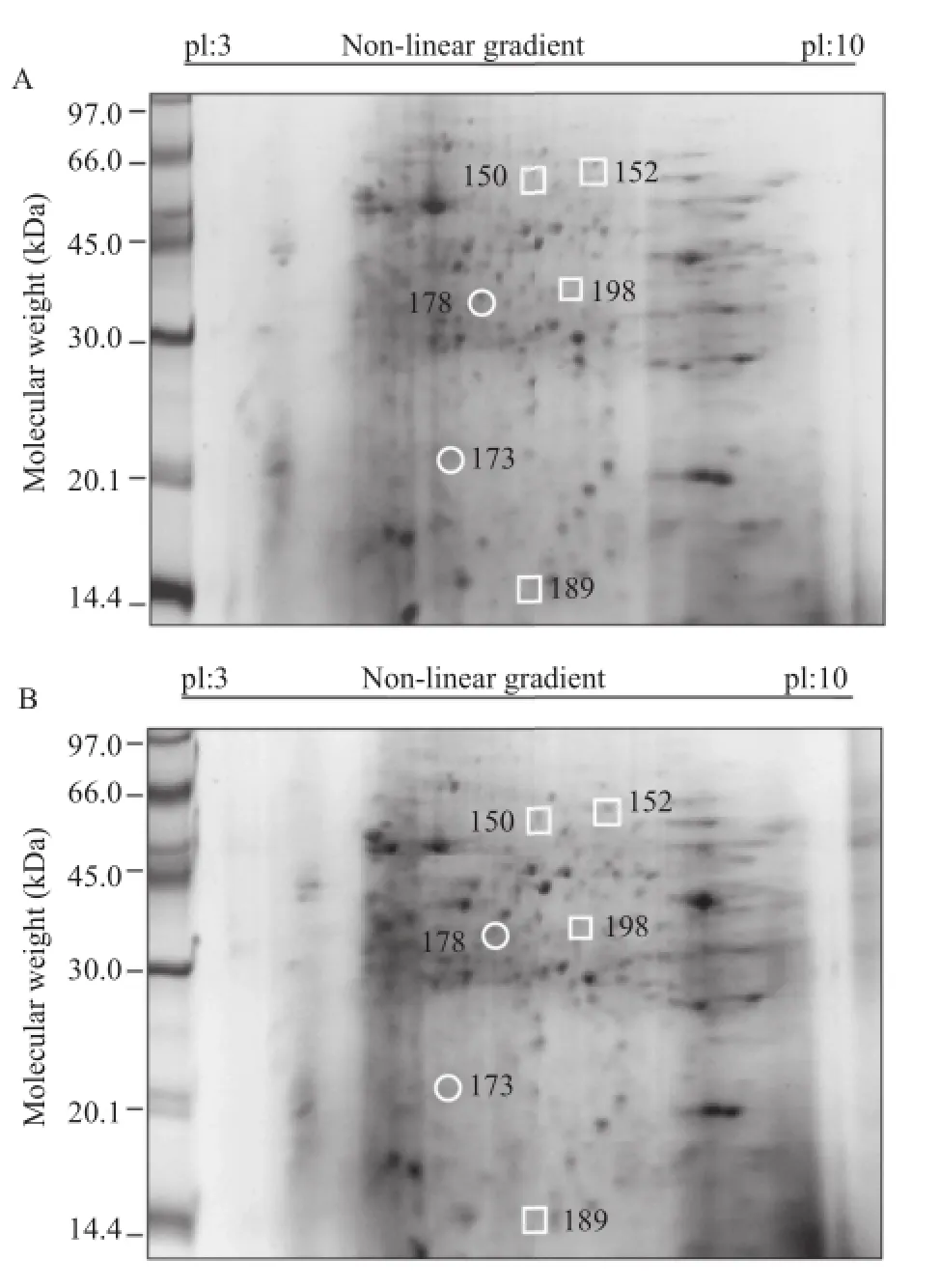
Figure 6.Differentially expressed proteins in uninfected cultures(A)compared with DENV-2/16681 infected cultures(B).A total of 200μg of protein was separated by two-dimensional electrophoresis for each sample. The differentially expressed spots are indicated in the figures.The squares indicate proteins with increased expression in infected cultures,and the circles indicate proteins with decreased expression in infected cultures.A representative gel of the 5 gels that were analyzed is shown for each biological condition.
The protein analysis indicated nine differentially expressed proteins in DENV-2/NG-infected U937 cells.The expression of five of these proteins(Spots 21,62,89,112 and 167)decreased in the infected cultures and four proteins(Spots 42,240,243 and 244)displayed increased expression compared with uninfected cultures(Figure 5A and B).The main percentage of differential expression was represented by proteins presenting decreased expression in the infected cultures(55.5%).
On the other hand,the protein analysis indicated that six proteins were differentially expressed in DENV-2/16681-infected U937 cells.The expression of two of these proteins(Spots 173 and 178)decreased in the infected cultures,whereas the expression of the remaining four proteins(Spots 150,152,189,and 198)increased in the infected cultures compared to the uninfected ones(Figure 6A and B).The main percentage of differential expression was represented by proteins displayed increased expression in the infected cultures(66.6%).
3.4.Protein identification by MS
Proteins with differential expression were identified by MALDI-TOF/TOF MS.The biological functions of these proteins were assigned according to gene ontology,and the identified proteins were classified into several different categories.
The proteins with decreased expression in DENV-2/NG-infected cultures compared with the uninfected cultures were as follows:fatty acid-binding protein(21),heterogeneous nuclear ribonucleoprotein 1(hnRNP 1)(62),protein disulfide isomerase(89),enolase 1(112)and heat shock 70 kDa protein 9(167).The proteins that displayed increased expression in the infected cultures were heat shock 90 kDa protein AA1(42),tubulin beta(240),enolase 1(243)and pyruvate kinase M2(244)(Table 1).
In DENV-2/16681-infected cultures,the proteins that displayed decreased expression were as follows:phosphotyrosyl phosphatase(173)and annexin IV (178).The proteins that displayed increased expression were as follows:pyruvate kinase(150),pyruvate kinase M2(152),transaldolase(189)and phospholipase C-alpha(198)(Table 2).
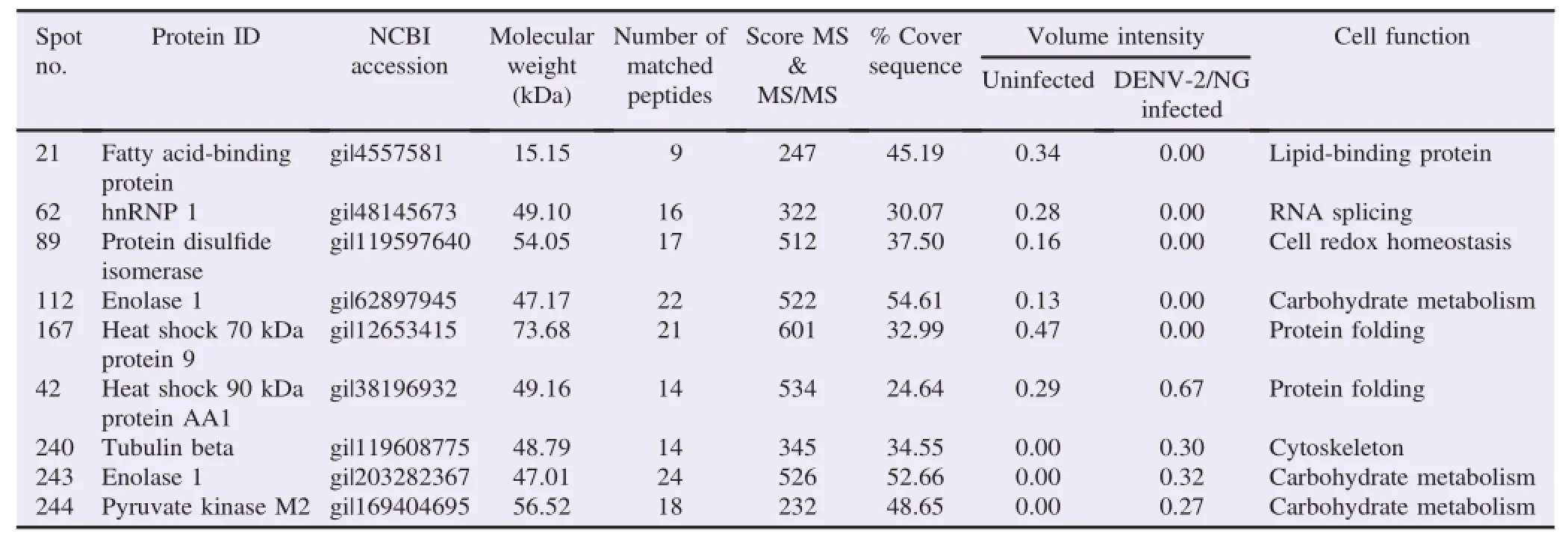
Table 1Comparison of proteins expressed in uninfected vs.DENV-2/NG-infected cultures.
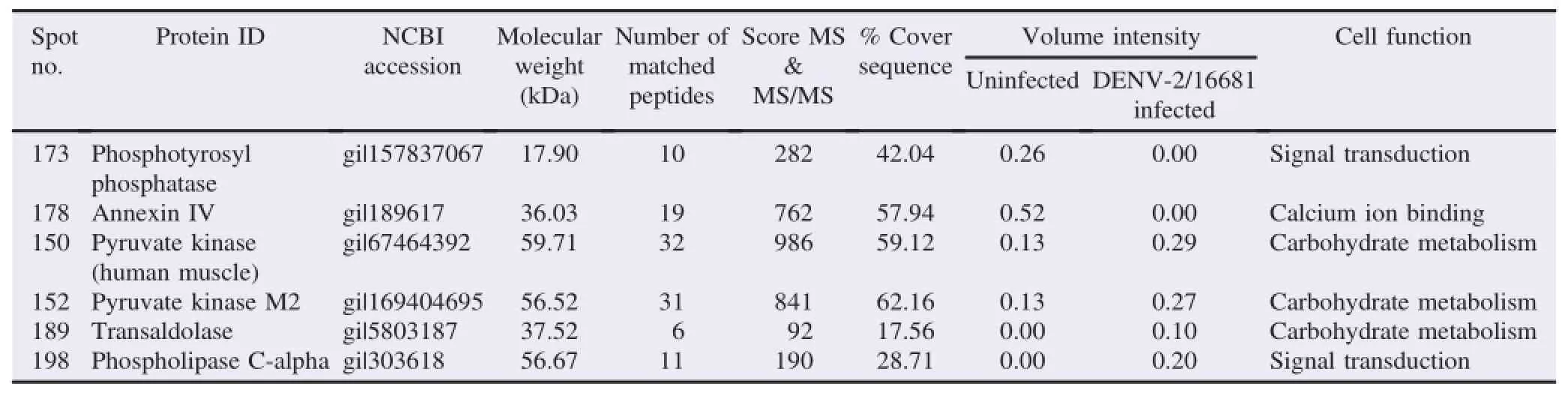
Table 2Comparison of proteins expressed in uninfected vs.DENV-2/16681-infected cultures.
4.Discussion
Cells of the monocyte/macrophage lineage are of great importance during dengue's development because they are one of the primary targets for DENV,responsible for spreading it from the site of infection toward different sections of the body[15].In addition,once infection has been established,these cells are the main responsible party for pro-inflammatory cytokine production,thus facilitating the development of severe forms of the disease[15,18].Despite the important role that these cells have on the disease's pathogenesis(in both primary and secondary infections),to date,there is only one study that shows how some proteins from the undifferentiated monocyte cell line THP-1 are affected under primary infection conditions[28].Therefore,this study evaluated the proteome changes in the differentiated monocyte cell line U937 after infection with two different viral strains,keeping in mind that the viral genetic variationshavebeenassociated withboththeepidemic potential of the infection and disease severity.
Because it has been previously reported that different strains from the same serotype can promote significant changes in both morphology and viability in cell cultures[27],an initial comparison was performed on the viability of the non-infected cell cultures with those infected for 48 h,revealing that viability was not significantly impaired in the infected cultures(Figure 1),similar to the behaviors found for other cell lines of epithelial(VERO)or hepatic origin(HepG2)[29,30].Significant changes in morphology were also not observed(Figure 2).This result offers assurance that the changes in the cellular proteome are caused by DENV infection and not cell death.
When performing the comparative analysis of the production of viral genome copies and infectious particles from each viral strain after infecting U937 cells,significant differences were detected only in viral RNA production.The cells infected with the DENV-2/16681 strain had a higher genomic copy number than those infected with the DENV-2/NG strain(Figure 3).This increased replication rate obtained with the strain associated with severe dengue had been previously reported in the VERO cell line[30]and could be related to the development of this clinical form of the disease,as some clinical studies have demonstrated higher levels of viral RNA in the plasma of patients with severe dengue in comparison with dengue patients[31].However,a higher number of cells were immunolabeled against viral proteins in cultures infected with DENV-2/16681(Figure 4),which could be correlated with the high viral protein levels detected in patients with severe dengue[32].
Previous studies have shown that the proteomes of epithelial cells(VERO)are significantly different when these cells are infected different viral strains[30];a similar behavior was detected in this study when U937 cells were infected with either the DENV-2/NG(Figure 5 and Table 1)or DENV-2/ 16681 strains(Figure 6 and Table 2).The most relevant findings will be discussed next.
The fatty acid-binding protein was downregulated in U937 cells infected with the DENV-2/NG strain.Keeping in mind that lipids and fatty acids induce the formation of replication complexes in DENV infection,their downregulation would be unfavorable for viral replication in this cell type.This finding could be related with observations from other studies showing that the percentage of infected monocytes in vitro are much lower compared with the percentages obtained from epithelial,hepatic,and endothelial cell types[33].However,keeping in mind that this same protein is upregulated in VERO cells infected with DENV-2/16681[30],it could be surmised that the regulation of its expression could be affected by the infecting viral strain,which,in turn,could be correlated with differences in viral genome quantity,which is lower in cells infected with the DENV-2/NG strain compared with the DENV-2/16681 strain(Figure 3).Regardless,in the present study,there was a significant increment in the quantity of this protein in the cultures of monocytes infected with the DENV-2/ 1668 strain,which makes it impossible to prove this hypothesis.
Another protein that was downregulated in the cultures infected with the DENV-2/NG strain was disulfide isomerase,a protein that regulates platelet function and helps to maintain their structural integrity[34].For this reason,its decreased expression could be related to the development of thrombocytopenia,which is characteristic of dengue[35]. However,decreased levels of this protein have also recently been detected in a pathologic condition known as macrothrombocytopenia[36],a finding that could be associated with what occurs in dengue infections,as it is known that platelet function is affected during disease development[37]. These results are also correlated with previous findings in VERO cells[30]in which downregulation of this protein was observed after infection with the DENV-2/16681 strain,which is related to platelet adhesion inhibition,thus allowing severe hemorrhage.
One of the isoforms of the enolase protein was upregulated in cultures infected with the DENV-2/NG strain,while another of the isoforms was downregulated in the same cultures.These findings agree with recent studies that showed increased levels of basic isoforms of enolase in hepatocytes,although its acidisoforms had a decreasing tendency[38].This multi-functional protein is not only involved in cytoplasmic processes such as glycolysis but is also expressed on the surfaces of diverse hematopoietic,epithelial and endothelial cells and serves as a receptor and plasminogen activator[39-41],thus fulfilling an important task in the intravascular coagulation and fibrinolytic system [41].It has been proposed that enolase upregulation during DENV infection could be associated with the compromised vascular and coagulation systems present during severe dengue cases.Theα-enolase protein could activate plasminogen,modify the coagulation-fibrinolysis balance and favor hyperfibrinolysis.In fact,several studies have reported increases in fibrin degradation products during DENV infection[42].Enolase downregulation could be correlated with a decrease in the intracellular viral load due to an increase in the tumor necrosis factor-αlevel,a recently proposed mechanism in THP-1 cells[28].
hnRNP 1 is another of the downregulated proteins detected in cultures infected with DENV-2/NG.This finding contrasts with the results obtained from other groups that have shown that several isoforms of this protein are upregulated in endothelial cells infected with the DENV-2/16681 strain or in undifferentiated monocytic cells[28,43].However,these differences could be explained considering that,as we previously demonstrated,the regulation ofthe expression ofcertain proteins is dependent on the cell type analyzed and the infecting viral strain[30].However,decreased expression of this protein in monocytes could prevent the migration of infected cells,which would limit,eliminate or delay the development of severe forms of the disease,as it has been reported that this protein,when interacting with intermediate filaments,plays an important role in cellular migration[44].
Heat shock protein 70 kDa(Hsp-70)was upregulated in VERO culturesinfected with DENV-2/16681[30],and interestingly,downregulated in U937 cells.Keeping in mind thatcells ofepithelialorigin replicate the virus more efficiently than monocytes as well as the role that this protein plays by interacting with the DENV E glycoprotein,facilitating viralreplication and the production ofviral particles[33,45],it could be postulated that its downregulation prevents the correct folding of viral proteins,making viral replication more difficult in these cells.In contrast,heat shock protein 90 kDa(Hsp-90)was upregulated;this protein is also required for viral replication,as has been previously reported[46].Thus,it could be proposed that Hsp-70 downregulation would be compensated with Hsp-90 upregulation.While Hsp-90 plays a very important role in DENV internalization[20],suggesting that the capture and internalization of the virus in macrophages could be made more efficient by Hsp-90 upregulation,and viral replication would be less efficient due to Hsp-70 downregulation.
The only protein identified in this study whose expression was increased independent of the infecting viral strain(DENV-2/NG or DENV-2/16681)was pyruvate kinase.Keeping in mind that this enzyme is involved in the process of glycolysis for energy production in the cell,it has been proposed that modifications in its expression are associated with a higher energy requirement for DENV replication[30,38].
Infection with DENV-2/NG also increased the expression of β-tubulin,a cytoskeletal protein.Keeping in mind that it has been postulated that tubulin(the main component of microtubules)facilitates the trafficking of the DENV E protein[47],its upregulation could facilitate viral assembly in these cells. However,its upregulation could also be correlated with a higher risk of developing severe forms of the disease,as increased tubulin expression has been detected in patient serum,an observation that has led to postulations on the suitability of tubulin as a biomarker for the development of severe forms of dengue disease[48].
The phosphotyrosyl phosphatase protein was downregulated,which could be related to an increase in nitric oxide(NO)production,as it has been demonstrated that this protein regulates NO production in macrophages[49].In turn,the increase in NO could be considered a cellular defense mechanism,as a correlation between NO production and the development of non-severe forms of dengue disease has been reported[50].
Anexin 4 was also downregulated in cultures infected with the DENV-2/16681 strain.It was recently shown that anexin A4 regulates interleukin 8(IL-8)production in macrophage cultures. When anexin is over-expressed in these cultures,IL-8 production is significantly decreased[51].For this reason,keeping in mind that the results obtained in this study demonstrate a decrease in anexin A4 expression in cultures infected with the DENV-2/16681 strain,it would be postulated that this downregulation could lead to increased IL-8 expression.In turn,the increase in IL-8 has been associated with the development of severe forms of dengue disease,as higher levels of IL-8 have been detected in patients with severe dengue[52].
Transaldolase protein expression was increased in cultures infected with the DENV-2/16681 strain.This enzyme participates in the phosphate pentose pathway,which allows the production of ribose 5-phosphate,an important precursor in nucleotide synthesis required for RNA replication[53].Thus,it is postulated that the increase in the expression of this enzyme may be associated with a higher replication rate of the DENV-2/ 16681 strain,which is correlated with the increase in the genome copy number of this strain(Figure 3A).
Finally,DENV-2/16681 infection also caused an increase in the expression of the phospholipase C protein,an enzyme that can cleave the viral non-structural protein 1(NS1)on the surfaces of cells infected with DENV[54].NS1 can be detected intracellularly,associated with the plasma membrane or in secreted form [55],the last of which is associated with the development of severe dengue,as the secreted NS1 protein can bind to prothrombin(a coagulation factor)and inhibit its activation,thus avoiding the developmentofa normal coagulation process and favoring hemorrhage[56].Therefore,it is suggested thatthe upregulation ofphospholipase C expression could induce an increase in the NS1 levels in patient serum and thereby contribute to the hemorrhagic symptoms characteristic of severe dengue.
This proteomic study identified changes in the expression levels of proteins in differentiated human monocytes in response to infection with two DENV strains.These proteins are involved in many biological processes,which may benefit viral replication or,alternatively,be related to disease development.Because monocytes are critical for the disease pathogenesis,additional studies on these proteins could lead to a better understanding of the cellular responses to DENV infection and to the identification of new therapeutic targets against this disease.
Conflict of interest statement
We declare that we have no conflict of interest.
Acknowledgments
The authors would like to thank Carolina Hern´andez,Carolina Quintero,and Lina Orrego for their technical assistance. This study was funded by the Administrative Department of Science,Technology,and Research-COLCIENCIAS(Projects 111549326092 and 111549326083).
References
[1]Guzman MG,Harris E.Dengue Lancet 2015;385(9966):453-65.
[2]Carrington LB,Simmons CP.Human to mosquito transmission of dengue viruses.Front Immunol 2014;5:290.
[3]Daep CA,Muñoz-Jord´an JL,Eugenin EA.Flaviviruses,an expanding threat in public health:focus on dengue,West Nile,and Japanese encephalitis virus.J Neurovirol 2014;20(6):539-60.
[4]Mustafa MS,Rasotgi V,Jain S,Gupta V.Discovery of fifth serotype of dengue virus(DENV-5):a new public health dilemma in dengue control.Med J Armed Forces India 2015;71(1):67-70.
[5]Messina JP,Brady OJ,Scott TW,Zou C,Pigott DM,Duda KA,et al.Global spread of dengue virus types:mapping the 70 year history.Trends Microbiol 2014;22(3):138-46.
[6]Lin CY,Huang CH,Chen YH.Classification of dengue:the clinical use of World Health Organization 2009 guideline. J Formos Med Assoc 2013;112(2):61-3.
[7]OhAinle M,Balmaseda A,Macalalad AR,Tellez Y,Zody MC,Saborío S,et al.Dynamics of dengue disease severity determined by the interplay between viral genetics and serotype-specific immunity.Sci Transl Med 2011;3(114):114ra128.
[8]Fried JR,Gibbons RV,Kalayanarooj S, Thomas SJ,Srikiatkhachorn A,Yoon IK,et al.Serotype-specific differences in the risk of dengue hemorrhagic fever:an analysis of data collected in Bangkok,Thailand from 1994 to 2006.PLoS Negl Trop Dis 2010;4(3):e617.
[9]Sabin AB,Schlesinger RW.Production of immunity to dengue with virus modified by propagation in mice.Science 1945;101(2634):640-2.
[10]Halstead SB,Simasthien P.Observations related to the pathogenesis of dengue hemorrhagic fever.II.Antigenic and biologic properties of dengue viruses and their association with disease response in the host.Yale J Biol Med 1970;42(5):276-92.
[11]Mladinich KM,Piaskowski SM,Rudersdorf R,Eernisse CM,Weisgrau KL,Martins MA,et al.Dengue virus-specific CD4+and CD8+T lymphocytes target NS1,NS3 and NS5 in infected Indian rhesus macaques.Immunogenetics 2012;64(2):111-21.
[12]Wu-Hsieh BA,Yen YT,Chen HC.Dengue hemorrhage in a mouse model.Ann N Y Acad Sci 2009;1171(Suppl 1):E42-7.
[13]Guzman MG,Alvarez M,Halstead SB.Secondary infection as a risk factor for dengue hemorrhagic fever/dengue shock syndrome: an historical perspective and role of antibody-dependent enhancement of infection.Arch Virol 2013;158(7):1445-59.
[14]Weiskopf D,Sette A.T-cell immunity to infection with dengue virus in humans.Front Immunol 2014;5:93.
[15]Chuang YC,Chen HR,Yeh TM.Pathogenic roles of macrophage migration inhibitory factor during dengue virus infection.Mediat Inflamm 2015;http://dx.doi.org/10.1155/2015/547094.
[16]Schmid MA,Diamond MS,Harris E.Dendritic cells in dengue virus infection:targets of virus replication and mediators of immunity.Front Immunol 2014;5:647.
[17]Cruz-Oliveira C,Freire JM,Conceição TM,Higa LM,Castanho MA,Da Poian AT.Receptors and routes of dengue virus entry into the host cells.FEMS Microbiol Rev 2015;39(2):155-70.
[18]Sun P,Kochel TJ.The battle between infection and host immune responses of dengue virus and its implication in dengue disease pathogenesis.Sci World J 2013;http://dx.doi.org/10.1155/2013/843469.
[19]St¨ockbauer P,Malaskov´a V,Soucek J,Chudomel V.Differentiation of human myeloid leukemia cell lines induced by tumorpromoting phorbol ester(TPA).I.Changes of the morphology, cytochemistry and the surface differentiation antigens analyzed with monoclonal antibodies.Neoplasma 1983;30(3):257-72.
[20]Reyes-del Valle J,Ch´avez-Salinas S,Medina F,Del Angel RM. Heat shock protein 90 and heat shock protein 70 are components of dengue virus receptor complex in human cells.J Virol 2005;79(8): 4557-67.
[21]Kyle JL,Beatty PR,Harris E.Dengue virus infects macrophages and dendritic cells in a mouse model of infection.J Infect Dis 2007;195(12):1808-17.
[22]Raheel U,Jamal M,Zaidi NU.A molecular approach designed to limit the replication of mature DENV2 in host cells.Viral Immunol 2015;28(7):378-84.
[23]Puerta-Guardo H,Medina F,DelaCruz Hern´andezSI,Rosales VH,Ludert JE,del Angel RM.The 1α,25-dihydroxyvitamin D3 reduces dengue virus infection in human myelomonocyte(U937)and hepatic(Huh-7)cell lines and cytokine production in the infected monocytes.Antiviral Res 2012;94(1): 57-61.
[24]Hern´andez-Castro C,Diaz-Castillo F,Martínez-Gutierrez M. Ethanol extracts of Cassia grandis and Tabernaemontana cymosa inhibit the in vitro replication of dengue virus serotype 2.Asian Pac J Trop Dis 2015;5(2):98-106.
[25]Farias KJ,Machado PR,de Almeida Junior RF,de Aquino AA,da Fonseca BA.Chloroquine interferes with dengue-2 virus replication in U937 cells.Microbiol Immunol 2014;58(6):318-26.
[26]Puerta-Guardo H,Raya-Sandino A,Gonz´alez-MariscalL,Rosales VH,Ayala-D´avila J,Ch´avez-Mungía B,et al.The cytokine response of U937-derived macrophages infected through antibody-dependent enhancement of dengue virus disrupts cell apical-junction complexes and increases vascular permeability. J Virol 2013;87(13):7486-501.
[27]Quintero-GilDC,OspinaM,Osorio-BenitezJE,Martinez-Gutierrez M.Differential replication of dengue virus serotypes 2 and 3 in coinfections of C6/36 cells and Aedes aegypti mosquitoes. J Infect Dev Ctries 2014;8(7):876-84.
[28]Mishra KP,Shweta Diwaker D,Ganju L.Dengue virus infection induces upregulation of hn RNP-H and PDIA3 for its multiplication in the host cell.Virus Res 2012;163(2):573-9.
[29]Conceição TM,El-Bacha T,Villas-Bˆoas CS,Coello G,Ramírez J,Montero-Lomeli M,et al.Gene expression analysis during dengue virus infection in HepG2 cells reveals virus control of innate immune response.J Infect 2010;60(1):65-75.
[30]Martínez-Betancur V,Marín-Villa M,Martínez-Gutierrez M. Infection of epithelial cells with dengue virus promotes the expression of proteins favoring the replication of certain viral strains.J Med Virol 2014;86(8):1448-58.
[31]Pal T,Dutta SK,Mandal S,Saha B,Tripathi A.Differential clinical symptoms among acute phase Indian patients revealed significant association with dengue viral load and serum IFN-gamma level. J Clin Virol 2014;61(3):365-70.
[32]ParanavitaneSA,GomesL,KamaladasaA,AdikariTN,Wickramasinghe N,Jeewandara C,et al.Dengue NS1 antigen as a marker of severe clinical disease.BMC Infect Dis 2014;14:570.
[33]Diamond MS,Edgil D,Roberts TG,Lu B,Harris E.Infection of human cells by dengue virus is modulated by different cell types and viral strains.J Virol 2000;74(17):7814-23.
[34]Zhou J,Wu Y,Wang L,Rauova L,Hayes VM,Poncz M,et al.The disulfide isomerase ERp57 is required for fibrin deposition in vivo. J Thromb Haemost 2014;12(11):1890-7.
[35]de Azeredo EL,Monteiro RQ,de-Oliveira Pinto LM.Thrombocytopenia in dengue:interrelationship between virus and the imbalance between coagulation and fibrinolysis and inflammatory mediators.Mediat Inflamm 2015;http://dx.doi.org/10.1155/2015/ 313842.
[36]Karmakar S,Saha S,Banerjee D,Chakrabarti A.Differential proteomics study of platelets in asymptomatic constitutional macrothrombocytopenia:altered levels of cytoskeletal proteins.Eur J Haematol 2015;94(1):43-50.
[37]Michels M,Alisjahbana B,De Groot PG,Indrati AR,Fijnheer R,Puspita M,et al.Platelet function alterations in dengue areassociated with plasma leakage.Thromb Haemost 2014;112(2): 352-62.
[38]Higa LM,Curi BM,Aguiar RS,Cardoso CC,De Lorenzi AG,Sena SL,et al.Modulation ofα-enolase post-translational modifications by dengue virus:increased secretion of the basic isoforms in infected hepatic cells.PLoS One 2014;9(8):e88314.
[39]Arza B,F´elez J,Lopez-Alemany R,Miles LA,Muñoz-C´anoves P. Identification of an epitope of alpha-enolase(a candidate plasminogen receptor)by phage display.Thromb Haemost 1997;78(3):1097-103.
[40]Qin J,Chai G,Brewer JM,Lovelace LL,Lebioda L.Structures of asymmetric complexes of human neuron specific enolase with resolved substrate and product and an analogous complex with two inhibitors indicate subunit interaction and inhibitor cooperativity. J Inorg Biochem 2012;111:187-94.
[41]Pancholi V.Multifunctionalα-enolase:its role in diseases.Cell Mol Life Sci 2001;58(7):902-20.
[42]Marchi R,Nagaswami C,Weisel JW.Fibrin formation and lysis studies in dengue virus infection.Blood Coagul Fibrinolysis 2009;20(7):575-82.
[43]Kanlaya R,PattanakitsakulSN,SinchaikulS,Chen ST,Thongboonkerd V.Alterations in actin cytoskeletal assembly and junctional protein complexes in human endothelial cells induced by dengue virus infection and mimicry of leukocyte transendothelial migration.J Proteome Res 2009;8(5):2551-62.
[44]Inoue A,Watanabe T,Tominaga K,Tsugawa K,Nishio K,Takahashi KP,et al.Association of hnRNP S1 proteins with vimentin intermediatefilamentsinmigratingcells.JCellSci2005;118:2303-11.
[45]Limjindaporn T,Wongwiwat W,Noisakran S,Srisawat C,Netsawang J,Puttikhunt C,et al.Interaction of dengue virus envelope protein with endoplasmic reticulum-resident chaperones facilitates dengue virus production.Biochem Biophys Res Commun 2009;379(2):196-200.
[46]Geller R,Taguwa S,Frydman J.Broad action of Hsp90 as a host chaperone required for viral replication.Biochim Biophys Acta 2012;1823(3):698-706.
[47]Foo KY,Chee HY.Interaction between Flavivirus and cytoskeleton during virus replication.Biomed Res Int 2015;http://dx.doi.org/ 10.1155/2015/427814.
[48]Thayan R,Huat TL,See LL,Khairullah NS,Yusof R,Devi S. Differential expression of aldolase,alpha tubulin and thioredoxin peroxidase in peripheral blood mononuclear cells from dengue fever and dengue hemorrhagic fever patients.Southeast Asian J Trop Med Public Health 2009;40(1):56-65.
[49]Blanchette J,Abu-Dayyeh I,Hassani K,Whitcombe L,Olivier M. Regulation of macrophage nitric oxide production by the protein tyrosine phosphatase Src homology 2 domain phosphotyrosine phosphatase 1(SHP-1).Immunology 2009;127(1):123-33.
[50]Valero N,Espina LM,Añez G,Torres E,Mosquera JA.Short report:increased level of serum nitric oxide in patients with dengue.Am J Trop Med Hyg 2002;66(6):762-4.
[51]Iwasa T,Takahashi R,Nagata K,Kobayashi Y.Suppression of MIP-2 or IL-8 production by annexins A1 and A4 during coculturing of macrophages with late apoptotic human peripheral blood neutrophils.Biochim Biophys Acta 2012;1822(2):204-11.
[52]Pandey N,Jain A,Garg RK,Kumar R,Agrawal OP,Lakshmana Rao PV.Serum levels of IL-8,IFNγ,IL-10,and TGFβand their gene expression levels in severe and non-severe cases of dengue virus infection.Arch Virol 2015;160(6):1463-75.
[53]Wamelink MM,Struys EA,Salomons GS,Fowler D,Jakobs C,Clayton PT.Transaldolase deficiency in a two-year-old boy with cirrhosis.Mol Genet Metab 2008;94(2):255-8.
[54]Jacobs MG,Robinson PJ,Bletchly C,Mackenzie JM,Young PR. Dengue virus nonstructural protein 1 is expressed in a glycosylphosphatidylinositol-linked form that is capable of signal transduction.FASEB J 2000;14(11):1603-10.
[55]Amorim JH,Alves RP,Boscardin SB,Ferreira LC.The dengue virus non-structural 1 protein:risks and benefits.Virus Res 2014;181:53-60.
[56]Lin SW,Chuang YC,Lin YS,Lei HY,Liu HS,Yeh TM.Dengue virus nonstructural protein NS1 binds to prothrombin/thrombin and inhibits prothrombin activation.J Infect 2012;64(3):325-34.
2 Sep 2015
inrevisedform28Sep2015
Original article http://dx.doi.org/10.1016/j.apjtb.2016.01.004
Marlen Martinez-Gutierrez,PhD,Calle 30A#33-51,Universidad Cooperativa de Colombia,Bucaramanga,Colombia.
 Asian Pacific Journal of Tropical Biomedicine2016年11期
Asian Pacific Journal of Tropical Biomedicine2016年11期
- Asian Pacific Journal of Tropical Biomedicine的其它文章
- Human and animal sarcocystosis in Malaysia∶A review
- Therapeutic applications of collagenase(metalloproteases)∶A review
- Cytotoxic activity and phytochemical standardization of Lunasia amara Blanco wood extract
- Antiacanthamoebic properties of natural and marketed honey in Pakistan
- GC-MS analysis of bioactive compounds present in different extracts of an endemic plant Broussonetia luzonica(Blanco)(Moraceae)leaves
- Anti-nitric oxide production,anti-proliferation and antioxidant effects of the aqueous extract from Tithonia diversifolia
