Identification of antigenic proteins from salivary glands of female Anopheles maculatus by proteomic analysis
Yunita Armiyanti,Renam Putra Arifianto,Elisa Nurma Riana,Kartika Senjarini,Widodo Widodo,Loeki Enggar Fitri,Teguh Wahju Sardjono
1Doctoral Program in Medical Science,Faculty of Medicine,Universitas Brawijaya,Malang,Indonesia
2Department of Biology,Faculty of Mathematics and Natural Sciences,University of Jember,Jember,Indonesia
3Department of Biology,Faculty of Mathematics and Natural Sciences,Universitas Brawijaya,Malang,Indonesia
4Department of Parasitology,Faculty of Medicine,Universitas Brawijaya,Malang,Indonesia
Identification of antigenic proteins from salivary glands of female Anopheles maculatus by proteomic analysis
Yunita Armiyanti1*,Renam Putra Arifianto2,Elisa Nurma Riana2,Kartika Senjarini2,Widodo Widodo3,Loeki Enggar Fitri4,Teguh Wahju Sardjono4
1Doctoral Program in Medical Science,Faculty of Medicine,Universitas Brawijaya,Malang,Indonesia
2Department of Biology,Faculty of Mathematics and Natural Sciences,University of Jember,Jember,Indonesia
3Department of Biology,Faculty of Mathematics and Natural Sciences,Universitas Brawijaya,Malang,Indonesia
4Department of Parasitology,Faculty of Medicine,Universitas Brawijaya,Malang,Indonesia
ARTICLE INFO
Article history:
revised form 18 Aug 2016
Accepted 22 Aug 2016
Available online 14 Sep 2016
Saliva
Immunogenic
Proteins
Anopheles
Objective:To identify antigenic proteins from the salivary glands of female Anopheles maculatus using a proteomic approach to find the biomarker candidate for serological tools.
Methods:The identification of antigenic proteins of Anopheles maculatus salivary gland used these techniques:one-dimensional gel electrophoresis(sodium dodecyl sulfate polyacrylamide gel electrophoresis),western blot,and liquid chromatography-mass spectrometry.
Results:The proteins that have molecular weight(MW)43 and 34 kDa were the antigenic protein.Computational bioinformatic analysis by Mascot Server revealed seven novel hypothetical proteins(MW:43 kDa)and two novel hypothetical proteins(MW: 34 kDa).Further analysis(BLASTP,antigenicity,epitope mapping,and specificity analysis)showed that two novel proteins were identified as apolipoprotein D and cathepsin D in Anopheles darlingi.
Conclusions:The identified proteins are potential to be developed as a biomarker of mosquito bite's exposure.
1.Introduction
Malaria is an important health problem of the world since approximately half of the world's population have a risk of infection.Malaria is an infectious disease caused by Plasmodium and spreads by female Anopheles mosquitoes.There are 30 from 400 species that can be major vectors,including Anophelesmaculatus(An.maculatus)[1].An.maculatus is one of major malaria vectors in Asia that widely spread in Afghanistan,Pakistan,South China,India,Indonesia,Malaysia,Thailand and the Philippines[2].
Tel:+62 816596475
E-mail:yunita.fk@unej.ac.id
The study protocol was performed according to the Helsinki declaration and approved by the Health Research Ethics Committee of the Faculty of Medicine,Universitas Brawijaya in statement of ethical clearance No.251/EC/KEPK-S3/03/ 2014 dated 26 March 2014.Informed written consent was obtained from Health Research Ethics Committee of the Faculty of Medicine,Universitas Brawijaya.
Foundation Project:Funded by a Doctorate Research Grant of the Directorate General of Higher Education Indonesia(Grant No.435/UN25.3.1/LT.6/2014).
Peer review under responsibility of Hainan Medical University.The journal implements double-blind peer review practiced by specially invited international editorial board members.
Anopheles mosquitoes prefer in tropic climate and humid temperature.So,Anopheles mosquitoes can spread easily in a tropical country,including Indonesia.As a consequence,there are many malaria-endemic areas found in the tropical countries.The society that stays in the malaria-endemic areas mostly used repellent,insecticides,and insecticide-treated nets to avoid Anopheles mosquitoesbites[3].Thehealthprogramtocontrolthevectorspreading isa crucialproject since the vaccine was still not available yet.The malaria control programs based on ento-mological methods,such as mosquito abundance,blood feeding rates,and mortality are difficult to apply on a large scale.Another method,such as human landing catch is commonly used for evaluating individual human exposure.However,this method is limited ethically and not representative for children or under-aged[4].Therefore,a program to control malaria vector needs a new approach to assessmalaria risk by evaluating the efficacy of vector control at both population and individual levels[5].
2221-1691/Copyright©2016 Hainan Medical University.Production and hosting by Elsevier B.V.This is an open access article under the CC BY-NC-ND license(http:// creativecommons.org/licenses/by-nc-nd/4.0/).
Malaria infection mechanism begins when Anopheles mosquito salivary proteins inject into the host during its bloodfeeding which is able to modulate the host immune response. The antibody protein,immunoglobulin G(IgG)anti-salivary protein,has been detected in malaria patients and the population of malaria-endemic areas[6].Apyrase,Anopheles gambiae(An.gambiae)salivary gland protein 6(gSG6),TRIO protein,Anopheles antiplatelet protein,glycine-and glutamate-rich salivary gland protein,and anophensin are salivary proteins that are antigenic[7].Therefore,the salivary gland protein that is able to elicit the host immune response has been proposed as a marker of exposure to mosquito bites,even as a biomarker of malaria risk[8].As a biomarker of malaria risk,the level of anti-gSG6 IgG antibody was also associated with malaria transmission according to strong positive association with merozoite surface protein-1 and glutamate-rich protein[6].
Recently,some studies showed gSG6 protein is a potential candidate for An.gambiae exposure marker[6,9].The gSG6 is a small protein first described in An.gambiae with mature peptide/protein weighed 10 kDa.The human antibody response to gSG6 protein indicated a reliable indicator of human exposure to the three main malaria vectors in tropical Africa,i.e.,An.gambiae,Anopheles arabiensis(An.arabiensis),and Anopheles funestus[9].The recombinant protein of gSG6 has been studied by gSG6-P1 peptide as a biomarker of low exposure to Anopheles bites.The IgG response against gSG6-P1 peptide was detectable in most individuals in malaria-endemic regions in a low-rated season of mosquitoes bites exposure(September-December)in Senegal[10].
In Africa,salivary gland proteins from An.gambiae have been studied as an immunological marker of Anopheles bites exposure.But,in Asia region the study about Anopheles mosquitoes(An.maculatus)is limited.The gSG6 salivary protein has been detected in Anopheles barbirostris species A2 salivary glands by mass spectrometry(MS)-based proteomic analysis,but it has not been proved to be antigenic protein[11]. The first step to determine the candidate protein for biomarker of exposure to Anopheles mosquito bites is identification of salivary gland proteins that possess antigenic properties.The identification can be performed by the proteomic approach that provides a direct measurement of protein expression level[12]. Therefore,the study aimed to identify the antigenic proteins of An.maculatus salivary gland as biomarker candidate for serological tools.
2.Materials and methods
2.1.Mosquitoes collection and salivary gland dissection The adult female An.maculatus were collected from Kalirejo village,Kokap District,Kulonprogo Regency in Yogyakarta by an aspirator.That mosquito species was a major vector of malaria and the dominant species in the research location.The mosquitoes were maintained under standard conditions at(27±2)°C with(70±10)%relative humidity and fed with 10%sucrose solution in the insectariums of the Entomology Laboratory of Parasitology Department,Faculty of Medicine,Gajah Mada University.The salivary glands of An.maculatus were dissected using fine entomological needles under a stereo microscope(4×)and pooled into a microcentrifuge tube containing phosphate buffer saline with protease inhibitor(phenylmethylsulfonyl fluoride).The salivary glands were stored at-80°C.
2.2.Salivary gland extraction and protein quantification
Dissected salivary glands(100 pairs)in phosphate buffer saline and phenylmethylsulfonyl fluoride were mixed(1:1)in lysis buffer(1.5 mmol/L MgCl2,10 mmol/L Tris-HCl,10 mmol/L NaCl,1%Nonidet P-40,and 2 mmol/L ethylenediaminetetraacetic acid,NaOH)and homogenized using a micropestle.The mixture was sonicated by a water sonicator for 30 min.Afterward,the suspension was centrifuged at 10000 r/min and 4°C,for 15 min.The extracted supernatant was collected and concentrated using a spin concentrator(cut-off of 10 kDa;Corning)and centrifugated(10000 r/min,4°C,for 30 s).The protein concentrations of salivary gland extracts(SGEs)were determined using a nanophotometer(Implen NanoPhotometer®P 360,Germany).
2.3.Human serum samples
The blood was collected from 15 healthy adult residents living in Kalirejo village,Kokap District,Kulonprogo Regency in Yogyakarta.The human subject protocol for this study was approved by the Ethical Committee of Medical Research,Faculty of Medicine,Universitas Brawijaya.The level of antibodies anti-salivary gland extract IgG of An.maculatus in the serum samples was measured by ELISA as described by Fontaine et al.[13].Five serum samples with a high level of IgG antibodies antisalivary gland extract were used as primary antibodies in western blot[13].
2.4.Sodium dodecyl sulfate polyacrylamide gel
electrophoresis(SDS-PAGE)
A total of 40μg of SGE sample was mixed in sample buffer(1:1)consisting of 0.5 mol/L Tris-HCl,10%SDS,glycerol,distilled water,1%bromophenol blue andβ-mercaptoethanol. Then,the mixture was heated at 95°C for 5 min.Then,20μL sample was loaded and separated onto 12%SDS-PAGE in VGES(WEALTEC,USA).Molecular weight(MW)protein marker(Nacalai)was loaded on the gel stained with Coomassie Brilliant Blue.
2.5.Western blotting
Gels were transferred to polyvinylidene difluoride membranes(MACHEREY-NAGEL,Germany)using semidry blotting(Bio-Rad)for 1 h at 100 mA.The membranes were saturated at room temperature for 1 h with 5%w/v non-fat dried milk in blocking buffer,which consisted of Tris-buffered saline and 0.05%Tween 20(TBS-T),then washed with TBS-T three times.Membranes were incubated with serum samples at dilution 1:20 in blocking buffer overnight at 4°C.Subsequently,blots were washed three times with TBS-T,then incubated with alkaline phosphatase goat anti-human IgG secondary antibody(KPL,USA)at dilution 1:2000 for 2 h at room temperature.The blot stained with nitro blue tetrazolium/bromo-4-chloro-3-indolyl phosphate and prestained broad range MW markers(9-200 kDa)(Nacalai,Japan)used to estimate the protein size.
2.6.In-gel protein digestion and MS analysis
The gels were digested using 10 mL trypsin digest solution(12.5 mg/mL trypsin,25 mmol/L ammonium bicarbonate)and incubated overnight at 37°C.The digested peptides were extracted by 10-20 mL acetonitrile containing 1%trifluoroacetic acid and incubated for 20 min.The extracts were dried by rotary evaporation and stored at-20°C until further analysis by MS[14].
Peptideswereanalyzedby electrospray ionizationMSusing the Agilent1260InfinityHPLCsystem(Agilent)coupledtoanAgilent 6540 mass spectrometer(Agilent).Tryptic peptides were loaded onto a C18 column 300 SB,5μm(Agilent)and separated with a linear gradient of water/acetonitrile/0.1%formic acid(v/v).Spectra wereanalyzedtoidentifyproteinsofinterestusingMascotsequence matching software(Matrix Science)with Ludwig NR database.
2.7.MS data analysis
The parameters were:(a)trypsin as the specific enzyme,(b)peptide mass tolerance:±0.2 Da,(c)fragment mass tolerance: ±0.2 Da,(d)variable modification oxidation of methionine carbamidomethyl and database used Ludwig NR and MSPnr100.The amino acid sequence was retrieved from Mascot Server(https:// sysbio-mascot.wehi.edu.au/mascot).For identifying the protein profile,the amino acid sequence samples were analyzed by comparing with the non-redundant protein sequence from Anopheles(taxid:7164)through BLASTP(http://blast.ncbi.nlm.nih.gov/ Blast.cgi).All identifications were manually validated based on MW,signal peptide(SignalP 4.1:http://www.cbs.dtu.dk/services/ SignalP/)and subcellular location.Data reliability was measured by the query coverage and percent identity of each sample.
2.8.Analysis of antigenicity and epitope mapping
The antigenicity of proteins was investigated using Kolaskar and Tongaonkar antigenicity(http://www.iedb.org)and a default threshold value was 1.0[15].The epitope mapping was performed by using Bepipred Linear Epitope Prediction with a default threshold of 0.35.BepiPred method is a computational method to predict linear B-cell epitopes that consist a linear sequence of amino acids that can be recognized by the antibodies[16].The similarity between all novel proteins and human's protein was also analyzed by using BLASTP(http://blast.ncbi.nlm.nih.gov/ Blast.cgi).The antigenic sites and epitopes were also visualized by 3D structure profiles using PyMol software.
3.Results
3.1.Antigenic proteins of An.maculatus salivary gland
The first step to identify the candidate proteins was antigenic proteins determination by western blot.The antigen from the salivary gland of An.maculatus(Figure 1)and sera from five individuals from malaria-endemic areas(Kalirejo village)were used.Based on SDS-PAGE result,the proteins from salivary glands of An.maculatus showed numerous bands ranging from 14 kDa to 200 kDa.The MW of seven major bands was 62,50,43,39,37,34 and 14 kDa(Figure 2).The results of western blotting showed that five bands(43,37,34,20 and 14 kDa)were cross-reacted with pooled serum of five individuals from malaria-endemic areas(Kalirejo village)(Figure 3A).Theseantigenic proteins were also recognized by serum as well as by pooled serum.According to the intensity of antigenic bands that showed up at the individual responses,proteins with MWs of 34 and 43 kDa showed up at all of serum samples(Figure 3B). Furthermore,the negative result appeared in the reaction between pooled sera of seven individuals living in non-malaria endemic areas with SGEs of An.maculatus(Figure 3C).The antigenic proteins with MWs of 34 and 43 kDa would be further identified with MS-based in gel digestion approach.

Figure 1.A pair of the salivary gland from adult female An.maculatus. DL:Distal region of lateral lobe;PL:Proximal region of lateral lobe;ML: Median lobe;DS:Ductus.
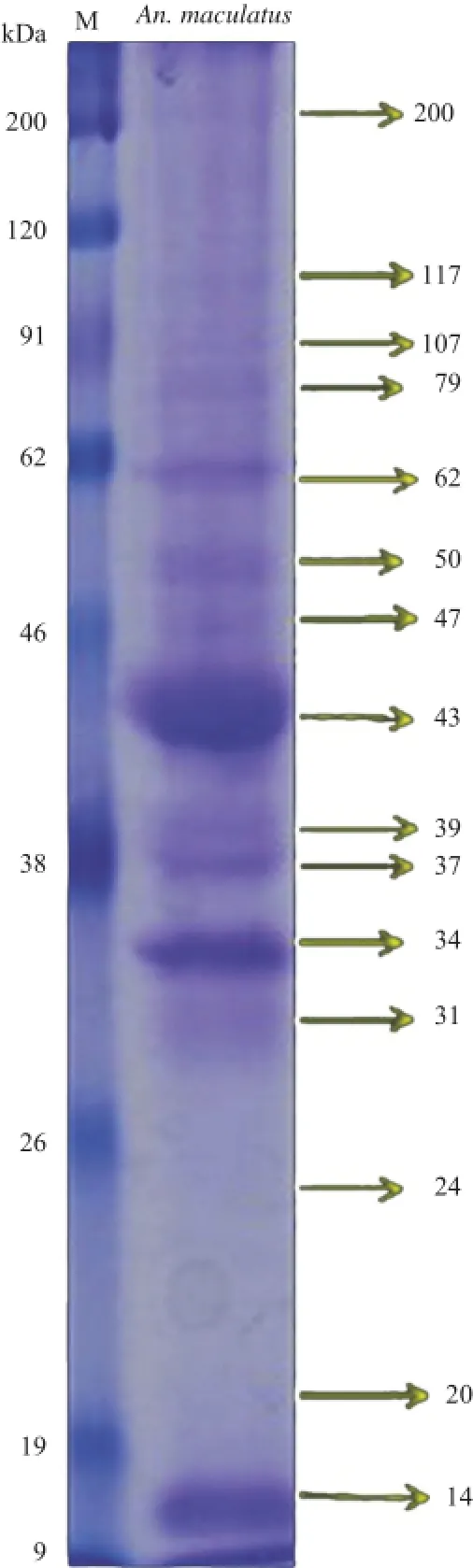
Figure 2.Salivary gland proteins of female An.maculatus mosquitoes were separated on 12%SDS-PAGE(right lane)and stained with Coomassie Blue.Lane M:Protein standard marker(kDa).

Figure 3.Western blotting of anti-salivary gland protein IgG antibodies in a pool of sera from 5 individuals living in malaria-endemic area as positive control(A),individual response from individuals living in malaria-endemic area(B)and a pool of sera from 7 individuals living in non-malaria endemic area as negative control(C).
3.2.MS analysis for antigenic proteins identification
MS data suggested that the study has identified two hypothetical proteins from 34 kDa band and seven hypothetical proteins from 43 kDa similar to proteins of An.gambiae,Anopheles sinensis(An.sinensis),and Anopheles darlingi(An. darlingi)(Tables 1 and 2).The data showed the amino acid residues SREELNSGGIGEMIRPYR from 34 kDa band were matched(64%)with FMRFamide variant 1(gi:656339519)and AGAP005518-PA(gi:118786597)proteins of An.gambiae.
Among seven novel hypothetical proteins from 43 kDa,three of them had the same function in carboxylic ester hydrolase activity,one of them had a function in aspartic-type endopeptidase activity,and the rest proteins had unknown function.One novel hypothetical protein that had been identified was similar to lipase of An.darlingi(gi:568251178)(64%similarity).Another novel hypothetical protein was identified similar to An.darlingi cathepsin D(aspartic proteases enzymes).This protein matched with the MS/MS spectrum that assigned the amino acid sequence AEDLVPTLVSRLASQQLMAILDPPR(Figure 4). All identified proteins were selected based on their similarity to human's surface cell protein using BLASTP tools(http://blast. ncbi.nlm.nih.gov/Blast.cgi).

Figure 4.MS/MS spectrum of the peak at m/z 911.79 corresponds to the peptide sequence,AEDLVPTLVSRLASQQLMAILDPPR which matches a novel protein(gi:568251273)that designated as cathepsin D in An.darlingi.

Table 1A list of novel proteins from 34 kDa antigenic band identified by LC-MS/MS using in-gel digestion approach.
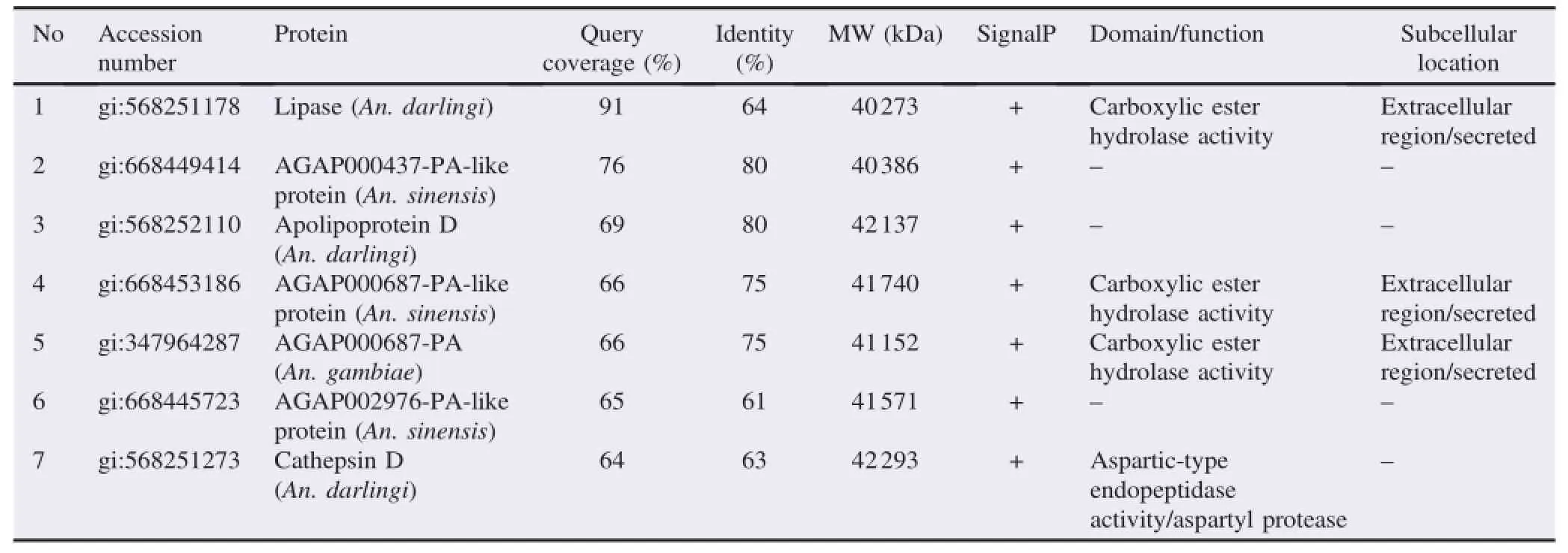
Table 2A list of novel proteins from 43 kDa antigenic band identified by LC-MS/MS using in-gel digestion approach.
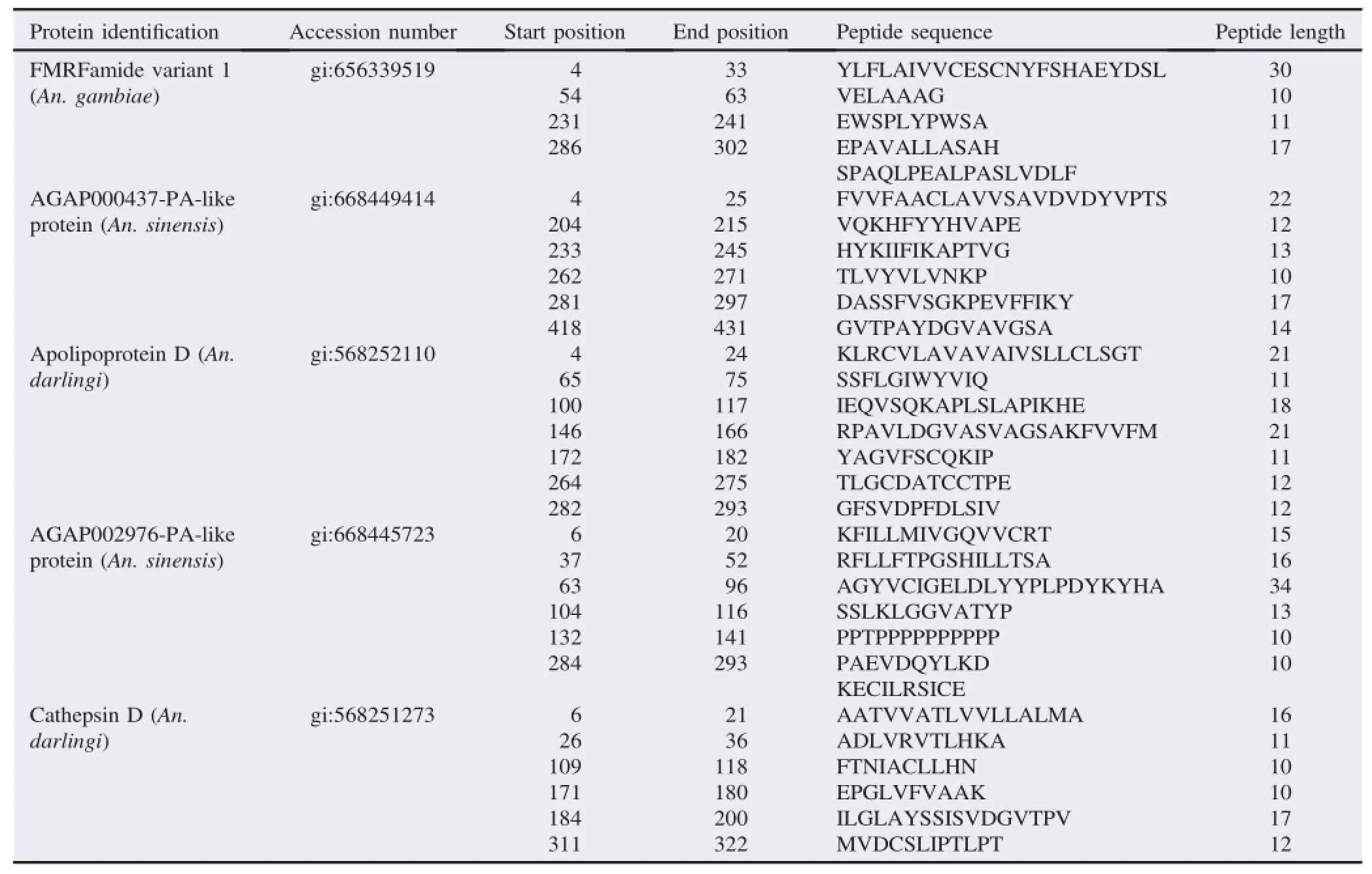
Table 3Antigenic region of selected proteins as biomarkers of An.maculatus bites.
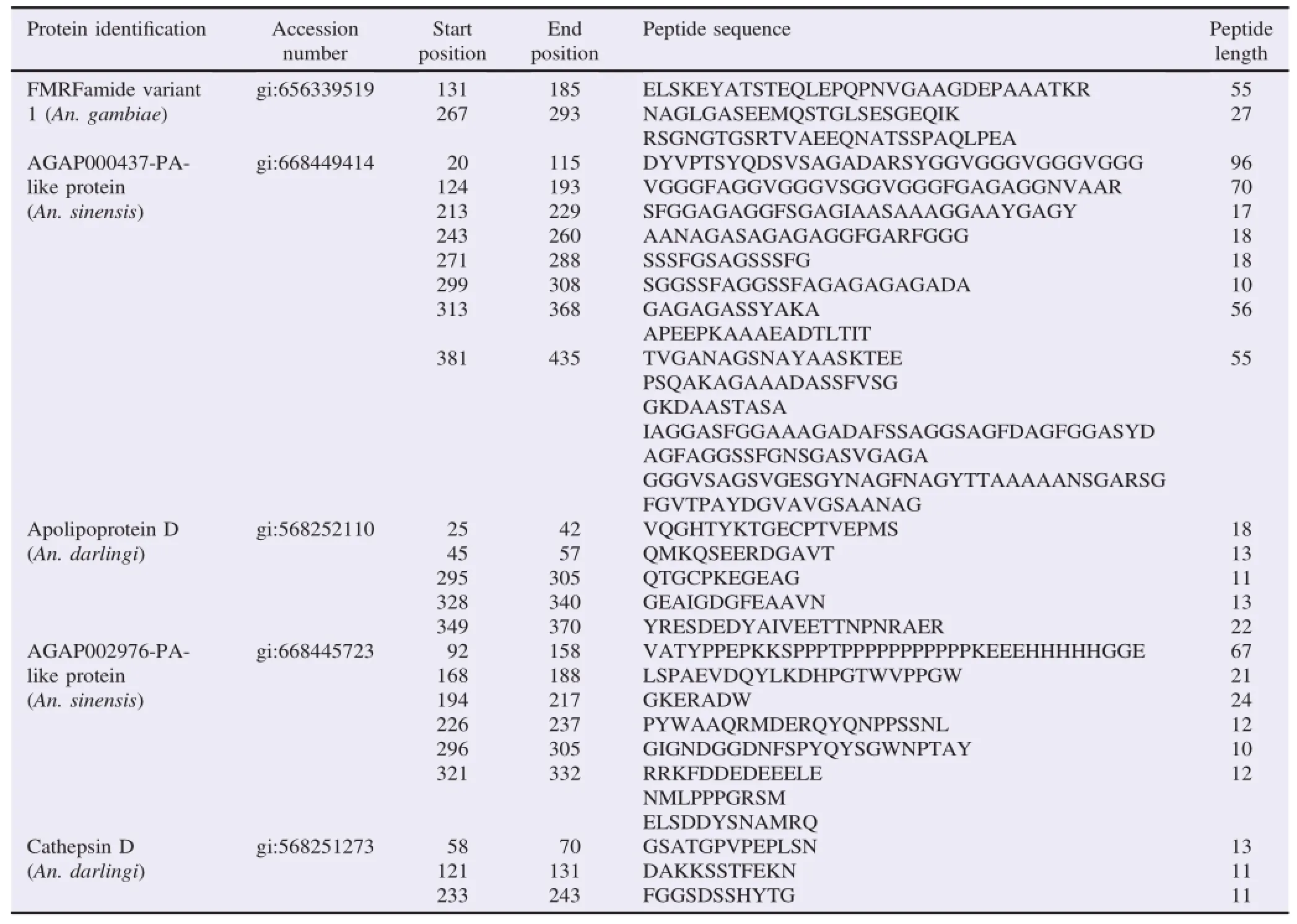
Table 4Epitope mapping of selected proteins as biomarkers of An.maculatus bites.
3.3.Antigenicity and epitope mapping analysis of identified proteins as potential candidate biomarker
The selected antigenic proteins of An.maculatus salivary gland that were not similar to human's surface cell proteins were further analyzed using bioinformatic tools to assess their antigenicity and epitope mapping.Epitope mapping using BepiPred method was performed to determine epitope of selected proteins as the binding portion of the antigen.The result of antigenicity analysis showed that selected proteins have a multi-antigenic region and polyepitope(Tables 3 and 4).However,apolipoprotein D and cathepsin D have higher antigenicity properties than other selected protein based on score value.The antigenic site and epitope region of cathepsin D protein as the best model were visualized using PyMol(Figure 5).

Figure 5.Predicted epitopes and antigenic region of cathepsin D(gi:568251273)as one of candidate proteins for biomarker tool of Anopheles mosquito bites exposure visualized by PyMol.
4.Discussion
The morphology of female An.maculatus salivary gland is similar to other Anopheles mosquitoes like Anopheles stephensi(An.stephensi)[17].Anopheles dirus A [18],and Anopheles sundaicus[19]that have three lobes and every lobe has a different region to maintenance function of saliva secretory.The proximal lateral lobe secretes amylase andα1-4 glucosidase that are essential for sugar feeding,while the distal lateral lobe and the medial lobe can secrete enzymes such as apyrase,anticoagulant,and vasodilator substances involved in blood-feeding[11].
The proteins of Anopheles salivary glands were detected as major bands with the MW,i.e.,62,43,39,36-37,33-34 kDa[18,19].These proteins could belong to conserved proteins at the genus,subgenusorspecies-specific level[7].Some proteins are ubiquitous salivary proteins that can be found in the salivary glands of many types of blood sucking insects and ticks,such as enzymes that are involved in sugar feeding(maltase),degradation of platelet aggregation(apyrase,5′-nucleotidases),and inflammation(adenosine deaminase)[20].Salivary proteins conserved in the genus, subgenus,and species-specific level can be used to develop immunological marker of individual mosquito's bites exposure.However,the proteins will be an optimal candidate as biomarker depends on the purpose of serological tool utilization.Proteins that were found exclusively in anophelines,such as gSG6 can be applied as biomarker tool for a wide area with several Anopheles species,for example An.gambiae,Anopheles funestus,and An.stephensi[9].Species-specific anopheline salivary proteins could be useful to determine the predominant mosquito populations[21].
The result of western blot showed that IgG antibodies recognized the proteins which have MW of 43,37,34,20 and 14 kDa.In addition,proteins which have MW of 43 and 34 kDa were antigenic and appeared on all of the individual responses. Those proteins would act as an antigen that stimulates B-cells to produce antibodies[16].This antigen-antibody reaction is the basic concept to develop a biomarker tool of human exposure to malaria vector.The salivary gland proteins which have a MW in the range of 35-40 kDa of An.maculatus,An. gambiae,An.arabiensis and An.stephensi showed antigenic activity towards the host.But,salivary gland proteins which have MW of 40,35 and 11 kDa from An.gambiae and An. arabiensis exhibited high reactivity with the pooled serum[7]. The existence of cross-reactivity between some Anopheles species should be a consideration to determine the optimal candidate protein as biomarker tool of mosquito bites exposure.
The two novel hypothetical proteins from this study(34 kDa)are similar with proteins in other Anopheles mosquito species,e.g.An.gambiae.FMRFamide variant 1(gi:656339519)and AGAP005518-PA (gi:118786597)proteins of An.gambiae contain FMRFamide domain.FMRFamide is a neuropeptide from FMRFamide-related peptide family sharing an RFamide peptide at their C-terminus and involving in myotropic activities[22].The proteins of 43 kDa might have a function on carboxylic ester hydrolase activity,aspartic-type endopeptidase,and lipase. The member of this lipase group included secretory phospholipase A2in Phlebotomus,triacylglycerol lipases in the mosquito[Culex quinquefasciatus(Cx.quinquefasciatus)and An.stephensi],carboxylesterase in Aedes aegypti,and phospholipase C in Cx.quinquefasciatus.Phospholipase C in Cx.quinquefasciatus has an activity to hydrolyze platelet aggregation factor,but its molecular nature remains unknown[20].Another novel hypothetical protein was identified similar to An.darlingi cathepsin D.Cathepsin D that has activity as aspartic-type endopeptidase is major catalytic classes of proteases,which are widely distributed not only in plants but also among vertebrates including insects,such as Musca domestica,Stomoxys calcitrans,and Aedes aegypti[23].Cathepsins were also found in Simulium nigrimanum and Culex tarsalis,but their function has not been characterized[20].
Apolipoprotein D and cathepsin D have higher antigenicity properties than other proteins.Moreover,both proteins had epitope length ranging from 10 to 96 residues.Structural studies of B-cell epitope showed that 10-22 amino acid residues are considered to be in contact with the atoms of antibody[24]. Apolipoprotein D has five peptides with length range 11-22 residues,and cathepsin D has three peptides with length range 11-13 residues as the antigenic site.Thus,apolipoprotein D and cathepsin D proteins have the potency to be developed as candidateproteinsforan immunologicalmarkerofAn. maculatus mosquito bites exposure based on their antigenicity properties and epitope mapping.This study is the first thatrevealed the candidate proteins of serological marker for human exposure to An.maculatus mosquito bites from antigenic proteins of the salivary gland.However,further analysis is necessary to determine the genome of An.maculatus salivary gland and their biological functions in blood-feeding and malaria transmission by the transcriptomic approach.
We found nine novel hypothetical proteins from 34 to 43 kDa protein bands as antigenic bands that are similar to the protein in An.gambiae,An.sinensis,and An.darlingi.The two expected proteins were apolipoprotein D and cathepsin D proteins that have the highest antigenicity that warrants for developing immunological marker of malaria vector bites exposure.
Conflict of interest statement
We declare that we have no conflict of interest.
Acknowledgments
The authors would like to acknowledge Didik Huswo Utomo for helping the proteomics analysis.This study was funded by a Doctorate Research Grant of the Directorate General of Higher Education Indonesia(Grant No.435/UN25.3.1/LT.6/2014).
References
[1]World Health Organization.World malaria report 2013.Geneva: World Health Organization;2013.[Online]Available from:http:// www.who.int/malaria/publications/world_malaria_report_2013/en/[Accessed on 25th March,2016]
[2]Elyazar IR,Sinka ME,Gething PW,Tarmidzi SN,Surya A,Kusriastuti R,et al.The distribution and bionomic of Anopheles malaria vector mosquitoes in Indonesia.Adv Parasitol 2013;83: 173-266.
[3]Raghavendra K,Barik TK,Reddy BP,Sharma P,Dash AP.Malaria vector control:from past to future.Parasitol Res 2011;108(4):757-79.
[4]Onyango SA,Kitron U,Mungai P,Muchiri EM,Kokwaro E,King CH,et al.Monitoring malaria vector control interventions: effectiveness of five different adult mosquito sampling methods. J Med Entomol 2013;50(5):1140-51.
[5]Drame PM,Poinsignon A,Besnard P,Le Mire J,Dos-Santos MA,Sow CS,et al.Human antibody response to Anopheles gambiae saliva:an immuno-epidemiological biomarker to evaluate the efficacy of insecticide-treated nets in malaria vector control.Am J Trop Med Hyg 2010;83(1):115-21.
[6]Stone W,Bousema T,Jones S,Gesase S,Hashim R,Gosling R,et al.IgG responses to Anopheles gambiae salivary antigen gSG6 detect variation in exposure to malaria vectors and disease risk. PLoS One 2012;7(6):e40170.
[7]Fontaine A,Fusai T,Briolant S,Buffet S,Villard C,Baudelet E,et al.Anopheles salivary gland proteomes for major malaria vectors.BMC Genomics 2012;13:614.
[8]Drame PM,Poinsignon A,Marie A,Noukpo H,Doucoure S,Cornelie S,et al.New salivary biomarkers of human exposure to malaria vector bites.In:Manguin S,editor.Anopheles mosquitoes-new insights into malaria vectors.Rijeka:In Tech;2013.
[9]Rizzo C,Ronca R,Fiorentino G,Mangano VD,Sirima SB,N`ebi`e I,et al.Wide cross-reactivity between Anopheles gambiae and Anopheles funestus SG6 salivary proteins support exploitation of gSG6 as a marker of human exposure to major malaria vectors in tropical Africa.Malar J 2011;10:206.
[10]Poinsignon A,Samb B,Doucoure S,Drame P,Sarr JB,Sow C,et al.First attempt to validate the gSG6-P1 salivary peptide as an immuno-epidemiological tool for evaluating human exposure to Anopheles funestus bites.Trop Med Int Health 2010;15(10): 1198-203.
[11]Jariyapan N,Roytrakul S,Paemanee A,Junkum A,Saeung A,Thongsahuan S,et al.Proteomic analysis of salivary glands of female Anopheles barbirostris species A2(Diptera:Culicidae)by two-dimensional gel electrophorasis and mass spectrometry.Parasitol Res 2012;111(3):1239-49.
[12]Sehrawat N,Gakhar SK.Mosquito proteomics:present and future prospective.Res Biotechnol 2014;5(4):25-33.
[13]Fontaine A,Pascual A,Orlandi-Pradines E,Diouf I,Remoue F,Pages F,et al.Relationship between exposure to vector bites and antibody response to mosquito salivary gland extracts.PLoS One 2011;6(12):e29107.
[14]Vijay S,Rawat M,Sharma A.Mass spectrometry based proteomic analysis of salivary glands of urban malaria vector Anopheles stephensi.Biomed Res Int 2014;2014:686319.
[15]Gomase VS,Changbhale SS,Chitlange NR,Sherkhane AS. Prediction of antigenic epitopes from Tityus serrulatus venom allergen 5:an aid to antitoxin vaccines.J Toxicol Res 2012;2(1): 20-4.
[16]Kavithak V,Saritha R,Vinod Chandra SS.Computational methods in linear B-cell epitope prediction.Int J Comput Appl 2013;63(12):28-32.
[17]Wells MB,Andrew DJ.Salivary gland cellular architecture in the Asian malaria vector mosquito Anopheles stephensi.Parasit Vectors 2015;8:617.
[18]Cotama S,Dekumyoy P,Samung Y,Lek-Uthai U.Salivary glands proteins expression of Anopheles dirus A fed on Plasmodium vivax-and Plasmodium falciparum-infected human blood. J Parasitol Res 2013;2013:535267.
[19]ArmiyantiY,NuryadyMM,ArifiantoRP,NurmarianaE,Senjarini K,Fitri LE,et al.Detection of immunogenic proteins from Anopheles sundaicus salivary glands.Rev Soc Bras Med Trop 2015;48(4):410-6.
[20]Ribeiro JMC,Mans BJ,Arca B.An insight into sialome of blood feeding Nematocera.Insect Biochem Mol Biol 2010;40(11):767-84.
[21]Ali ZM,Bakli M,Fontaine A,Bakkali N,Vu Hai V,Audebert S,et al.Assessment of Anopheles salivary antigen as individual exposure biomarkers to species-specific malaria vector bite.Malar J 2012;11:439.
[22]Altstein M,Nassel DR.Neuropeptide signaling in insects.Neuropeptide systems as targets for parasite and pest control.Berlin: Springer;2010.
[23]Seddigh S,Darabi M.Proteomics comparison of aspartic protease enzyme in insects.Turk J Biol 2016;40(1):69-83.
[24]Kringelum JV,Nielsena M,Padkjaerb SB,Lund O.Structural analysis of B-cell epitopes in antibody:protein complexes.Mol Immunol 2013;53(1-2):23-34.
4 Apr 2016
inrevisedform17Aug,2nd
Original article http://dx.doi.org/10.1016/j.apjtb.2016.08.012
Yunita Armiyanti,Doctoral Program in Medical Science,Faculty of Medicine,Universitas Brawijaya,Malang,Indonesia.
 Asian Pacific Journal of Tropical Biomedicine2016年11期
Asian Pacific Journal of Tropical Biomedicine2016年11期
- Asian Pacific Journal of Tropical Biomedicine的其它文章
- Human and animal sarcocystosis in Malaysia∶A review
- Therapeutic applications of collagenase(metalloproteases)∶A review
- Cytotoxic activity and phytochemical standardization of Lunasia amara Blanco wood extract
- Antiacanthamoebic properties of natural and marketed honey in Pakistan
- GC-MS analysis of bioactive compounds present in different extracts of an endemic plant Broussonetia luzonica(Blanco)(Moraceae)leaves
- Anti-nitric oxide production,anti-proliferation and antioxidant effects of the aqueous extract from Tithonia diversifolia
