Discovery of Female Laudakia papenfussi Zhao, 1998, with Insights into its Phylogenetic Relationships
Dahu ZOU, Fang YAN, Ke JIANG, Jing CHE, Robert W.MURPHY, 4, Theodore J.PAPENFUSS, Shuangquan DUANand Song HUANG
1School of Sciences, Tibet University, Lhasa 850000, China2College of Life and Environment Sciences, Huangshan University, Huangshan 245041, Anhui, China3State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming 650223, China4Centre for Biodiversity and Conservation Biology, Royal Ontario Museum, 100 Queen's Park, Toronto, Ontario M5S 2C6, Canada5Department of Integrative Biology, Museum of Vertebrate Zoology, University of California, Berkeley, CA 94720
Discovery of Female Laudakia papenfussi Zhao, 1998, with Insights into its Phylogenetic Relationships
Dahu ZOU1, 2, 3#, Fang YAN3#, Ke JIANG3, Jing CHE3, Robert W.MURPHY3, 4, Theodore J.PAPENFUSS5, Shuangquan DUAN1*and Song HUANG2*
1School of Sciences, Tibet University, Lhasa 850000, China
2College of Life and Environment Sciences, Huangshan University, Huangshan 245041, Anhui, China
3State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming 650223, China
4Centre for Biodiversity and Conservation Biology, Royal Ontario Museum, 100 Queen's Park, Toronto, Ontario M5S 2C6, Canada
5Department of Integrative Biology, Museum of Vertebrate Zoology, University of California, Berkeley, CA 94720
Abstract The sole voucher of Papenfuss' Rock Agama, Laudakia papenfussi (CIB 775001), an adult male, was collected from Zanda, Tibet, China on July 1, 1976 and described in 1998.No information on this species appeared since its description.In September 2014, we collected one female and six males at the type locality.Based on the original description and these newly collected specimens, we re-describe this species.Principal components analysis based on 33 morphological characteristics clearly diagnose L.papenfussi from closely related species.One way ANOVA test shows significant differences among four Tibetan rock agamas for 9 characters at 95% significant level, and 8 characters at 99% signifcant level.Molecular analyses recover three main clades in Laudakia.The four Tibetan rock agamas place into two geographical groups: Yarlung Zangbo group (L.sacra and L.wui) and the Himalayan group (L.tuberculata and L.papenfussi).
Keywords Papenfuss' Rock Agama, Tibetan Plateau, morphology, phylogeny
1.Introduction
The genus Laudakia Gray, 1845 (Agamidae Gray,1827) contains 20 species with a checkered taxonomic history dating back to more than 250 years, when the first member of this group, Lacerta stellio Linnaeus,1758 was described (Baig et al., 2012).Laudakia ranges from Greece and the Nile River delta in the west through the Middle East and Central Asia to Gobi Altai in the north-east and the Yarlung Zangbo in the east (Ananjeva and Tuniev, 1994).Generally, they are saxicolous andlive in either natural (rocks, screes, and clay slopes) or artifcial (stone walls, bridge parapets, ancient buildings)environments in arid zones.
Tibet hosts four species of rock agamas all of which are endemic to Qing Zang Gaoyuan (the valid name of the US Board on Geographic Names for the variant name Tibetan Plateau): L.sacra (Smith, 1935), L.wui Zhao,1998, L.tuberculata (Gray, 1827), and L.papenfussi Zhao, 1998.Ananjeva et al.(1990) elevated Agama himalayana sacra to a full species and assigned it to Laudakia as L.sacra.Zhao (1998a) described the new species L.wui.The sole voucher of L.papenfussi (adult male CIB 775001) was collected on 1 July 1976 from Zanda, Xizang (Tibet), China and was also described by Zhao (1998b).Since then, no information about this species appeared.Bahuguna (2008) documented elevational variation in morphology of L.tuberculata in the western Himalayas.
To better understand the diversity of Laudakia in Tibet, we conducted herpetofaunal investigations there from 2012 to 2014.Thirty-two specimens of four known species of rock agama were collected (Figure 1).Fortunately, one female and six male Papenfuss' Rock Agamas were sampled in September 2014 at its type locality (Figure 2).Because the species was described from a single specimen, nothing is known about variation within it.Here, we re-describe L.papenfussi based on its original description and our examination of the newly collected specimens.We employ principal component analyses (PCA) of 33 morphological characters to evaluate their differentiation.To quantify the variations among four species, we performed Analysis of Variance (ANOVA) and post-hoc test for each morphometric character.We also inferred their matrilineal relationships using mitochondrial DNA (mtDNA).

Figure 1 Sampling sites.Laudakia papenfussi: Zanda; L.tuberculata: Gyirong; L.sacra: Mainling, Qüxü; L.wui: Mêdog.

Figure 2 General views in life (A, B) and habitat (C, D) of a female Laudakia papenfussi.Photos by Diancheng YANG (Huangshan University).
2.Materials and Methods
2.1 Sampling A total of 31 rock agamas were collected.After euthanization and biopsies of tissues, all specimens were fixed in 10% formalin before final preservation in 70-80% EtOH.The Kunming Institute of ZoologyAnimal Care and Ethics Committee approved all work with the animals.All voucher specimens were deposited in Kunming Institute of Zoology, Chinese Academy of Sciences.
2.2 Morphometrics Terminology for the morphological description followed Ananjeva et al.(2011) and Baig (2012).Measurements and pholidosis, and their abbreviations were as follows: SVL (snout-vent length);TL (tail length, from vent to the tip of the tail); TRL (trunk length, distance from axilla to groin); LS (shoulder length); HL (distance from snout to posterior edge of tympanum); HW (maximum head width); HD (maximum head height); SL (snout length); ED (horizontal eye diameter); TD (tympanum diameter); IND (internarial distance); IOD (interorbital distance); UEW (upper eyelid width); EED (distance from anterior edge of ear opening to posterior corner of eye); EN (distance from anterior corner of eye to nostril); RW (rostral width);RH (rostral height); MW (mental width); MH (mental height); FL (total length of forelimb); LAL (lower-arm length); HAL (hand length, from proximal end of outer palmar tubercle to tip of the third finger); FEL (femur length); TIL (tibia length); FTL (length of foot and tarsus); NSF (number of subdigital lamellae on third finger); NST (number of subdigital lamellae on fourth toe); NSL (number of supralabials); NIL (number of infralabials); SBR (number of scales bordering the rostral scale); NSN (number of supranasals); SBM (number of scales bordering the mental scale); SBN (number of scales between the nasals); SBO (number of scales between the orbits); RV (rows of vertebral scales); NM (total number of scales around mid-body); NV (number of midventral scales from the posterior edge of the gular region to vent).All measurements were carried out by one person using dial calipers with a precision of 0.01 mm.PCAs of these morphological data were conducted.To eliminate potential effects of age, all measurements were standardized by dividing them by SVL.To compare the variations for all attributes among four species, we carried out one way ANOVA test.Data were firstly tested for normality (using Kolmogorov-Smirnov Test) and homogeneity (using Levene's test) to meet the assumptions of following ANOVA analysis.Post Hoc Multiple Comparisons were conducted by LSD or Dunnett T3 at 95% level of probability.All analysis above mentioned were conducted in SPSS v13.0 (SPSS, Inc.,Chicago, IL, USA).
2.3 DNA extraction, amplification and sequencing Total DNA was extracted using standard phenolchloroform protocols (Sambrook et al., 1989).Fragments of mtDNA gene encoding NADH dehydrogenase subunit 4 (ND4) and cytochrome c oxidase I (CO1) were amplified with primers ND4/Leu (for ND4; Arévalo et al., 1994) and Chmf4/Chmr4 (for CO1; Che et al., 2012).Polymerase chain reaction (PCR) mixtures (25 μL)contained 18.5μL ddH2O, 1μL (60 ng/μL) genomic DNA,2.5μL buffer, 1μL of each primer, and 1μL Taq DNA polymerase.PCR was performed on a DNA Engine Dyad Cycler as follows: 5 min of initial denaturation at 95 °C,35 cycles of denaturation for 1 min at 94 °C, annealing for 1 min at 45 °C for CO1 and 50 °C for ND4, and a fnal extension at 72 °C for 1 min.PCR products were purifed and then sequenced in both directions with BigDye Terminator Cycle Sequencing Kit and ABI PRISM 3730 (Applied Biosystems).All sequences were deposited in GenBank (Table 1).
2.4 Phylogenetic analyses Sequences of CO1 and ND4 from a closely related outgroup genus Phrynocephalus were downloaded from GenBank (Table 1).We also retrieved CO1 and ND4 sequences of other species of Laudakia from GenBank (Table 1).DNA sequences were aligned with CLUSTAL X v1.81 (Thompson et al.,1997) and optimized with BioEdit ver.7.1.9 (Hall, 1999).Before reconstructing the genealogy, we selected the bestft model of sequence evolution using JModelTest v2.1.1 (Darriba et al., 2012) with Akaike Information Criterion (AIC).Bayesian analysis was performed using MrBayes v3.2.1 (Ronquist and Huelsenbeck, 2003) with default priors and the appropriate model.We ran ten million generations of MCMC simulations with four separate chains (three heated, one cold) with a sample frequency of 1000 generations, and the frst 25% of the trees were discarded as burn-in.For more confdence, ML analyses with nonparametric bootstrap nodal support were performed using an internet implementation of RAxML v7.2.3 (Stamatakis et al., 2008).
3.Results and Discussion
3.1 Redescription of Laudakia papenfussi Zhao, 1998 Below, we redescribe our newly collected specimens and provide photographs of living individuals (Figures 3-4), as well as their habitat and microhabitat.Further,we present information on its distribution, ecology and conservation status.
Diagnosis Head and body depressed; dorsal and lateral surface scattered with yellow spots of equal size of vertebral scales; vertebral scales oblique towards midline and anterior; enlarged mucronate scales scatteredon the sides of body; dorsolateral fold present; gular pouch absent; chest and belly black mottled by orange;transverse mid-body scales more than 190; distinct tail segment of four whorls above becoming three below.
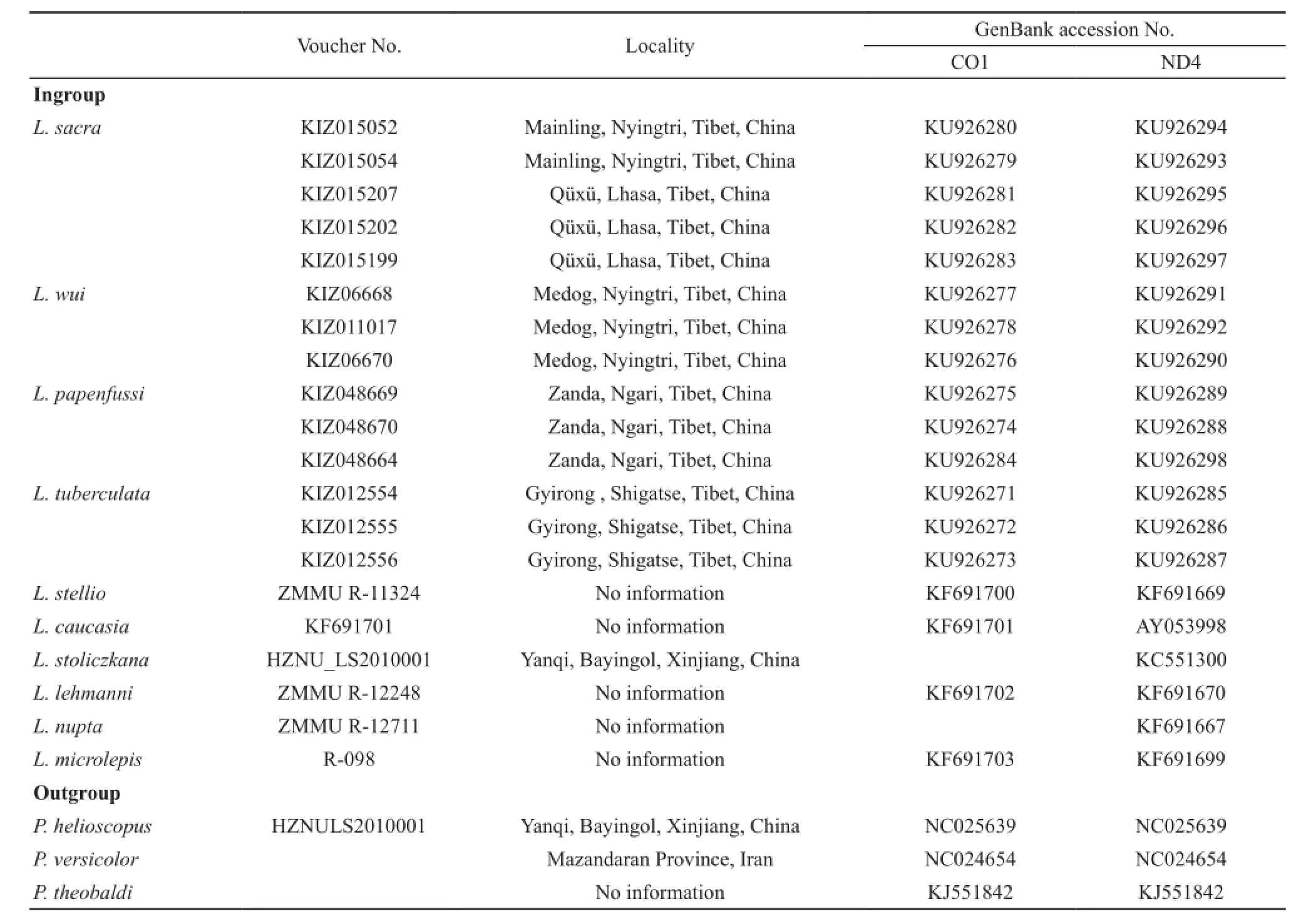
Table 1 Details of the individuals analyzed in molecular analysis, with localities and GenBank accession numbers.KIZ=Kunming Institute Zoology.
Redescription Habitus thin and depressed, head depressed and slightly triangular, HL 28.82±1.70 mm,HW 26.28±3.17 mm, HD 13.76±1.78 mm; SL 4.08±0.32 mm; nasal and nostril elliptic, nostril situated in the center of nasal, directed outwards and posteriorly, on canthus rostralis; 1 or 2 supranasals (KIZ017854 with 2 right supranasals); eye small with a rounded pupil and brown iris; eyelids covered with scales; a row of large and keeled scales arranged in a curve below the lower eyelid, separated from supralabials by 4 rows of small, smooth, slightly bulged scales; tympanum slightly rounded, and naked, more than half of eye diameter (except for juveniles), columella auris visible; tympanum surrounded by irregular conic scales, patches of these scales also on lateral side of neck; scales between eye and tympanum large and keeled on upper part, small and keeled on anterior lower part, granular on posterior lower part; rostral scale broad and low, upper margin straight;SBR 9-11; scales on dorsal head heterogeneous, large on snout, slightly large, bulging and smooth on frontoparietal region, small on supraocular region, distinctly keeled on occupit; parietal eye an indistinct small white spot.Superciliary ridge poorly developed, its margin blunt and not everted upwards; NSL 10-11 and NIL 10-12, mental large and triangular, 5-7 scales bordering mental; 4 or 5 rows of long and narrow scales parallel to infralabials; gular pouch absent, gular fold present,rudimentary nuchal crest.Vertebrals slightly enlarged,keeled and obliquely towards the midline on sacrum,arranged in 10-12 longitudinal rows; dorsals small and keeled, scattered with large and conic scales; skin of lateral sides wrinkly, forming dorsolateral fold; ventral scales all smooth, or weakly keeled, slightly larger than vertebrals; NM 196-228; NV 114-128.Limbs stout; thelongest toe of hind limb reaches the tympanum; dorsal side of limbs covered with large and strongly keeled scales; scales of ventral side of upper arm small; fngers and toes well developed, claws compressed and sharp,NSF 27-31, NST 31-37; fnger formula 4>3>5>2>1, toe formula 4>5>3>2>1.Tail cylindrical, slightly depressed at the base, covered with large and strongly keeled scales;scales of ventral side largest; scales arranged in rings and segments, each segment consisting of 4 rings of enlarged mucronate scales on dorsal side, with 3 on ventral side.Both callous preanal and abdominal scales present in males, only preanal scales in females; 4-6 rows of callous scales, abdominal one forming an oval patch.
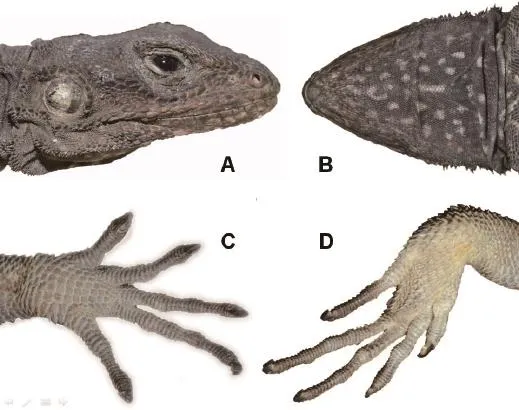
Figure 3 Head (A, B), fngers (C) and toes (D) of the female Laudakia papenfussi.Photos by Dahu ZOU.
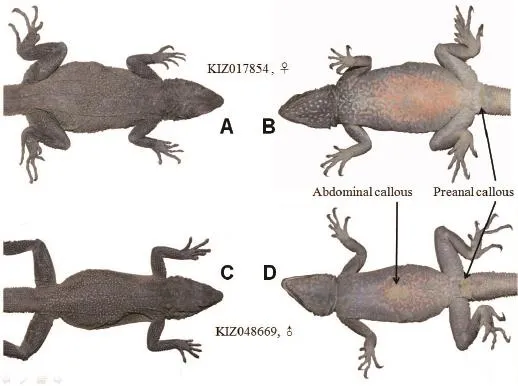
Figure 4 Dorsal and ventral views of female (A, B) and male (C, D) Laudakia papenfussi, showing the abdominal (only male) and preanal (both sexes) callous.
Coloration in life In adults, head is black or brownish black.Dorsum ranges from grayish-black to black with age, scattered with a series of small yellow spots composed of one keeled scale vertebrally.Laterally,yellow spots made of three to eight small keeled scales or one conical scale.Vertebral spots often form short random lines.Ventral surface speckled with orange patches.Coloration relates with age; young and juveniles may have a faded pattern with less black and more yellow.Gular region with white symmetric blotches on black ground.
The data of measurements and pholidosis of holotype (CIB775001) all fall in the ranges of newly collected specimens.Except for male abdominal callous, no significant difference in measurements, pholidosis and coloration was found between males and females.
Distribution and ecology This species is currently known only from type locality in the western Himalayas bordering India, where harsh climate dominates most of the year.It is most abundant around a small river between Mayang (3300 m a.s.l.) and Lugba (a.k.a.Luba, Diya;farm village (3050 m a.s.l.)).Lizards were observed mostly in roadside solid earthy cliffs basking or escaping and hiding in cracks.They bask in areas that offer long distances of sight.Consequently, they are far more diffcult to approach than L.sacra.Laudakia papenfussi appears to be omnivorous because one specimen was caught with a small yellow fower in its mouth and in the lab they readily preyed on worms.The only sympatric ectothermic vertebrate with L.papenfussi is Bufotes zamdaensis (Fei et al., 1999).This toad is also known only from Zanda and it was commonly encountered during our herpetofaunal surveys.
Comparisons Tail segment of Laudakia papenfussi consists 4 whorls dorsally or laterally, and 3 whorls on ventral side as L.sacra, L.wui, L.tuberculata and L.dayana do.In addition to the yellow spot on black color pattern, L.papenfussi can be also distinguished from L.sacra by gular pouch absent and having enlarged mucronate scales on both sides of body.By possessing more than 190 transverse mid-body scales, L.papenfussi can be distinguished from L.dayana.Laudakia papenfussi differs from L.wui by vertebrals arranged in oblique lines.Laudakia papenfussi differs from its most closely related species, i.e., L.tuberculata as follows, respectively: smaller yellow spots on back (~1 mm) vs.bigger spots (1-3 mm); brown iris vs.olive green; orange-yellow chest and belly vs.yellow.
3.2 Statistical analysis The measurements of 31 specimens (Laudakia.sacra, 17; L.wui, 5; L.tuberculata,3; L.papenfussi, 6) were shown in Table 2.The PCA determined that the first three components explained 41.22% of the total variance.Component 1 explained 21.17% of variation, with the following 12 characters (load factor > 0.5): HL, UEW, EN, LAL, TD, ED, RW,IND, RH, FTL, EED, and MW.Component 2 explained another 13.75% of variance, with a loading (factor >0.5) for the following 8 characters: SL, FEL, MH, NSL,SBR, HW, FL, TIL.Component 3 explained another 11.75% of variance, with a loading (factor > 0.5) for 5 characters: TIL, NSF, SBR, NM, NV.Loadings of each morphological character on the frst three principal components were showed in appendix Table S1.Plots with first three components showed in Figure 5.Intriguingly, PC2 and PC3 discriminate species better than PC1.Plot from PC2 and PC3 successfully identifed L.papenfussi, L.tuberculate and all individuals of L.sacra and L.wui but failed to distinguish L.sacra from L.wui.This is not surprising because qualitative traits (e.g.,color pattern) distinguish L.sacra and L.wui and these data were not included in our PCA dataset.According to results of normality test in appendix Table S3, we removed NIL, NSN and SBO from our dataset.The results of homogeneity of variables examined by Levene's test are shown in appendix Table S4.P-value of FTL and NV were less than 0.05, thus rejecting H0 (variances homogeneity among four species).One way ANOVA test show significant difference for 9 characters at 95% signifcant level, and 8 characters at 99% signifcant level (appendix Table S2).For these 17 characters, multiple comparisons were carried out.Significant differences (P-value < 0.05) were observed between species: Laudakia papenfussi and L.tuberculata (HD, SL, ED,IOD, UEW, EN, RH, SBN, NM, NV); L.papenfussi and L.wui (SL, ED, IOD, MH, FEL, NSL, SBR, SBM, SBN);Laudakia papenfussi and L.sacra (SL, ED, RH, MH,FEL, NSF, NST, NSL, SBR, NV); Laudakia tuberculata and L.wui (UEW, EN, MH, FEL, NSF, NST, NSL, SBR,NM, NV), L.tuberculata and L.sacra (HD, UEW, EED,EN, MH, NSF, NST, SBR, SBN, NM, NV), L.wui and L.sacra (EED, NSL, SBN, SBM) (appendix Table S5).Results of PCA, ANOVA and post-hoc test suggested that characters, such as MH, NSF, NSL and SBR, may be potential taxonomically important characters to rock agamas.
3.3 Molecular phylogenetic analysis PCRs of CO1 andND4 were successful for 14 individuals (L.sacra, 5; L.wui, 3; L.tuberculata, 3; L.papenfussi, 3).GTR+I+G and GTR+I were selected as the best ft models for CO1 and ND4 sequences, respectively.The genealogical tree constructed with the concatenated data united all species of Laudakia with strong support.The tree depicted matrilines A, B and C (Figure 6).Laudakia stellio from the Middle East (matriline C) rooted at the base of the tree.Matriline B was comprised of L.caucasia (Eichwald, 1831), L.lehmanni (Nikolsky, 1896), L.stoliczkana (Blanford, 1875) and L.microlepis (Blanford,1874).They were widely distributed from the Middle East, across Central Asia, to Gobi Altai.Finally, matriline A hosted the four Tibetan rock agamas plus L.nupta.Our
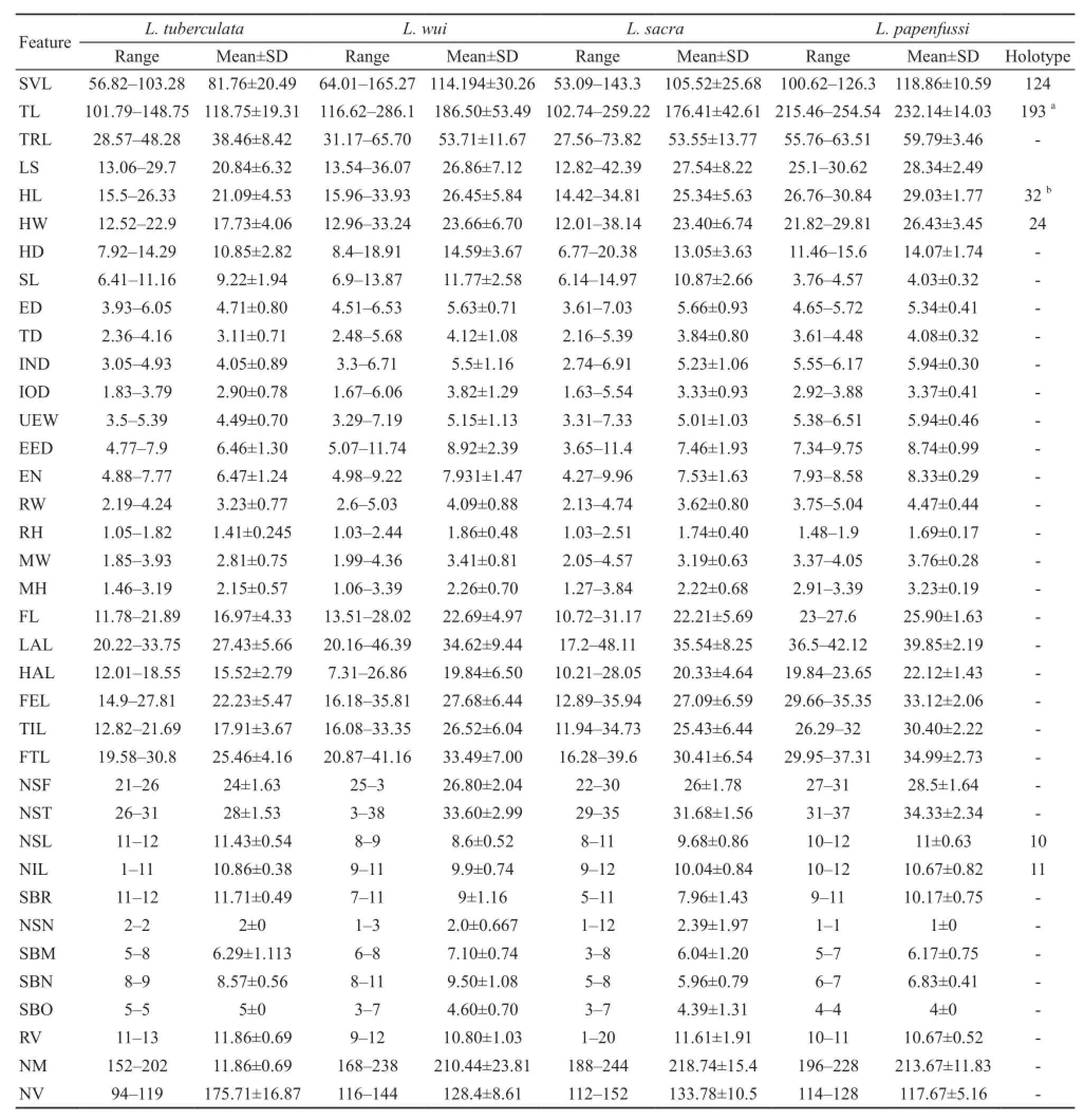
Table 2 Measurements (mm) and pholidosis of 31 specimens used in PCA and the measurements (mm) for the holotype of L.papenfussi (CIB775001, Zhao, 1998b).

Figure 5 Principal components analysis (PCA) toward 31 individuals of Laudakia corresponding to L.sacra, L.wui, and L.tuberculata, L.papenfussi with 33 morphometric variables.

Figure 6 Bayesian tree for species of Laudakia inferred using concatenated mitochondrial DNA CO1 and ND4 sequences.Number near nodes are the Bayesian posterior probabilities (BPP) and maximum likelihood bootstrap values.Photos beside the tree are L.wui, L.sacra, L.tuberculata, L.papenfussi and L.lehmanni from top to bottom.
genealogy corresponded to previous assessments (Baig et al., 2012; Pyron et al., 2013).Laudakia papenfussi and L.tuberculata, which occur on the south slope of the Himalayas, clustered together forming the Himalaya group (matriline A3).With a distribution along Yarlung Zangbo Jiang (Brahmaputra River), L.sacra and L.wuiformed matriline A1.Within this group, L.sacra occupies the upper and midstream reaches of the Yarlung Zangbo Jiang from Xigazê to Nyingchi while L.wui is located in downstream areas.Two snow covered mountains separate these two species, Namjagbarwa and Gyala Peri,that might be the geographical barrier for divergence of this two species (Zhao et al., 1999).However, the morphologically identifed L.sacra could not be clustered as a monophyletic clade based on mitochondrial DNA data.This suggested that population from Mainling,Nyingtri in Tibet might be a cryptical species, which need further investigation using nuclear data.
Conservation status The conservation status of L.papenfussi has not been assessed.Under IUCN assessment criteria (IUCN, 2012), there is only one area of occurrence for L.papenfussi (B1a), which encompasses 33 km2of which the species occupies about 15 km2.Moreover, this area is extremely vulnerable to disasters,such as debris fow, landslides and severe weather (B1b (i, ii, iii)).Interpreting these data using IUCN criteria, L.papenfussi should be considered Critically Endangered (CR Red List category, B1ab (i, ii, iii)).
Acknowledgements We thank Junxiao YANG (Kunming Institute of Zoology, CAS); Diancheng YANG, Xiaocun ZHOU (Huangshan University); Zeliang JU (Gansu Agricultural University); Shengling ZHOU, Yanjie ZHANG (Tibet University) for specimens collection.This work was supported by grants from the Ministry of Science and Technology of China (2014FY210200,2011FY120200), State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology,CAS (GREKF13-10), the Animal Branch of the Germplasm Bank of Wild Species of Chinese Academy of Sciences (the Large Research Infrastructure Funding),and the National Natural Science Foundation of China (NSFC 31060280).
References
Ananjeva N.B., Tuniev B.S.1994.Some aspects of historical biogeography of Asian rock agamids.Russ J Herpetol, 1: 42-52
Ananjeva N.B., Peters G., Macey J.R., Papenfuss T.J.1990.Steilio sacra (Smith 1935)—A distinct species of Asiatic rock agamid from Tibet.Asian Herpetol Res, 3: 104-115
Ananjeva N.N., Orlov N.L., Nguyen T.T., Ryabov S.A.2011.A new species of Acanthosaura (Agamidae, Sauria) from Northwest Vietnam.Russ J Herpetol, 18: 195-202
Arévalo, E., Davis, S.K., Sites Jr., J.W.1994.Mitochondrial DNA sequence divergence and phylogenetic relationships among eight chromosome races of the Sceloporus grammicus complex (Phrynosomatidae) in Central Mexico.Syst Biol, 43: 387-418
Bahuguna A.2008.Altitudinal Variations in Morphological Characters of Laudakia tuberculata Hardwicke and Gray, 1827 from Western Himalayas (Uttarakhand), India.Russ J Herpetol,15: 207-211
Baig K.J., Wagner P., Ananjeva N.B., Böhme W.2012.A morphology-based taxonomic revision of Laudakia Gray, 1845 (Squamata: Agamidae).Vert Zool, 62: 213-260
Che J., Chen H., Yang J., Jin J., Jiang K., Yuan Z., Murphy W.,Zhang Y.P.2012.Universal CO1 primers for DNA barcoding amphibians.Mol Ecol Res, 12: 247-258
Darriba D., Taboada G.L., Doallo R., Posada D.2012.JModelTest 2: More models, new heuristics and parallel computing.Nat Methods, 9: 772
Hall, T.A.1999.BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT.Nucleic Acids Symp Ser, 41: 95-98
IUCN.2012.IUCN Red List Categories and Criteria: Version 3.1.Second edition.Gland, Switzerland and Cambridge, UK: IUCN.iv + 32pp
Pyron R.A., Burbrink F.T., Wiens J.J.2013.A phylogeny and revised classifcation of Squamata, including 4161 species of lizards and snakes.BMC Evol.Biol., 13: 93
Ronquist F., Huelsenbeck J.P.2003.MrBayes 3: Bayesian phylogenetic inference under mixed models.Bioinformatics, 1: 1572-1574
Sambrook J., Fritsch E.F., Maniatis T.1989.Molecular Cloning: A Laboratory Manual, Sec Ed.Cold Springs Harbor, NY: Cold Spring Harbor Laboratory Press
Stamatakis A., Hoover P., Rougemont J.2008.A Rapid Bootstrap Algorithm for the RAxML Web-Servers.Syst Biol, 75(5): 758-771
Swofford D.L.2002.PAUP*.Phylogenetic Analysis using Parsimony (*and other methods).Version 4.0b10.Sinauer Associates, Sunderland, MA.
Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F.,Higgins D.G.1997.The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools.Nucleic Acids Res, 25: 4876-4882
Zhao E.M.1998a.A new species of the Genus Laudakia from Xizang (Tibet) Autonomous Region.Acta Zool Sin, 23: 440-444
Zhao E.M.1998b.Description of a new species of the genus Laudakia from Xizang (Tibet).Zool Res, 19: 401-404
Zhao E.M.1999.Laudakia.In Zhao E.M., Zhao K.T., Zhou K.Y.(Eds.), Fauna Sinica, Reptilia.Beijing: Science Press, 133-147 (In Chinese)
#These authors contributed equally to this work.
E-mail: dshq2007@aliyun.com (S.Q.DUAN); snakeman@hsu.edu.cn (S.HUANG):
Received: 30 June 2015 Accepted: 28 March 2016
DOI:10.16373/j.cnki.ahr.150047
*Corresponding author:Assoc.Prof.Shuangquan DUAN, School of Sciences, Tibet University, Lhasa, China, with his research focusing on plateau biodiversity; Prof.Song HUANG, Huangshan University,Huangshan, Anhui, China, with his research focusing on taxonomy and biogeography.
 Asian Herpetological Research2016年2期
Asian Herpetological Research2016年2期
- Asian Herpetological Research的其它文章
- Tracing the Origin of the Black-spotted Frog, Pelophylax nigromaculatus, in the Xinjiang Uyghur Autonomous Region
- Genetic Diversity and Population Structure for the Conservation of Giant Spiny Frog (Quasipaa spinosa) Using Microsatellite Loci and Mitochondrial DNA
- Coevolution of Male and Female Response Preferences to Sexual Signals in Music Frogs
- Ecological Niche Divergence between Trapelus ruderatus (Olivier,1807) and T.persicus (Blanford, 1881) (Sauria: Agamidae) in the Middle East
- The Effects of Chronic Hypoxia on Thermoregulation and Metabolism in Phrynocephalus vlangalii
- Comparison of Skull Morphology in Two Species of Genus Liua (Amphibia: Urodela: Hynobiidae), L.shihi and L.tsinpaensis
