Tracing the Origin of the Black-spotted Frog, Pelophylax nigromaculatus, in the Xinjiang Uyghur Autonomous Region
Supen WANG, Conghui LIU, Wei ZHU, Xu GAOand Yiming LI, *Key Laboratory of Animal Ecology and Conservation Biology, Institute of Zoology, Chinese Academy of Sciences, Beichen West Road, Chaoyang District, Beijing 000, ChinaUniversity of Chinese Academy of Sciences, 9 Yuquan Road, Shijingshan, Beijing 00049, China
Tracing the Origin of the Black-spotted Frog, Pelophylax nigromaculatus, in the Xinjiang Uyghur Autonomous Region
Supen WANG1#, Conghui LIU1, 2#, Wei ZHU1, Xu GAO1and Yiming LI1, 2*1Key Laboratory of Animal Ecology and Conservation Biology, Institute of Zoology, Chinese Academy of Sciences, 1 Beichen West Road, Chaoyang District, Beijing 100101, China
2University of Chinese Academy of Sciences, 19 Yuquan Road, Shijingshan, Beijing 100049, China
Abstract Identifying the origin of a biological invasion is critical for controlling the invaders.To explore the genetic diversity and identify the source region of introductions of Pelophylax nigromaculatus to the Xinjiang Uyghur Autonomous Region, we sequenced 695 bp of the mitochondrial Cyt b gene in 140 individuals of P.nigromaculatus and identifed 42 haplotypes in Heilongjiang, Beijing, Jiangsu, Shaanxi, Zhejiang provinces, Chongqing and the Xinjiang Uyghur Autonomous Region.We detected only four mitochondrial haplotypes in 20 specimens from Yining city in Ili Kazak Autonomous Prefecture.We traced the origin of Yining P.nigromaculatus to the Beijing and Chongqing area.Our results extend the known distribution range of this species in Asia.
Keywords Pelophylax nigromaculatus, invasion genetics, Xinjiang, invasion route
1.Introduction
Biological invasions are a major threat to global biodiversity and have evoked concerns among evolutionary ecologists and biological conservationists (Pyšek et al., 2008).Comparative studies that explore an invasive species in both its native and introduced ranges can reveal how an exotic species affects or is affected by a new environment (Meiners, 2007).Such studies not only establish the basic biological characteristics of an invader but also provide genetic information on the founding population of an invasive species, the invasion pathway of an exotic species throughout its introduced range, and its dispersal pattern (Campbell-Staton et al.,2012; Ficetola et al., 2008; Kolbe et al., 2004; Peacock et al., 2009).Therefore, these types of studies improve the prediction of the future distributions of invasive species.When studying biological invasions, population genetic analyses based on molecular markers can provide valuable information concerning population demographics,colonization history, and population structure (Ficetola et al., 2008; Moule et al., 2015; Rius et al., 2015; Sherwin et al., 2015).Moreover, this information can be used to detect the source populations for the original colonization and contemporary dispersal into the invaded range (Kolbe et al., 2004; Peacock et al., 2009).When developing management strategies, reconstructing the invasion history of invasive species can enhance our understanding of invasion risks by identifying the areas most susceptible to invasion and forecasting the future spread of such species based on past patterns of population expansion.Population genetics is a strong tool for enhancing the effectiveness and sustainability of management strategies (Liebl et al., 2015).
The black-spotted frog (Pelophylax nigromaculatus)is widely distributed in China, far-eastern Russia, the Korean peninsula and Japan.In China, this species originated in the north, east, central and southwest mainland and eastern islands (Zhang et al., 2008).Although there have been several (ecological, genetic and evolutionary) studies of the black-spotted frog in its native populations (Gao et al., 2015a; Gao et al., 2015b;Liu et al., 2010; Wang et al., 2014; Wang et al., 2009;Wang et al., 2008; Yang et al., 2003; Zhang et al., 2008;Zhang et al., 2004), no invasion genetics surveys have been performed in China.
Ili Kazakh Autonomous Prefecture is located in the west of Xinjiang Uyghur Autonomous Region (79º50'30"-84º56'50" E, 42º14'16"-44º50'30" N).Yining is located on the northern side of the Ili River in Ili Kazakh Autonomous Prefecture.The Ili River valley is wetter than most of Xinjiang and contains rich rice fields.Yining has a semi-arid climate without strong variations of seasonal precipitation.In winter, the average temperature is-8.8°C.However because of the infuence of the Dzungarian Altau and Boroboro Mountains to the northeast, the city is warmer than most of Xinjiang.Summers are hot with an average July temperature of 23.1°C.Species richness is lower in the Ili Kazakh Autonomous Prefecture than at similar latitudes elsewhere in China: 4 amphibian species (Bufo viridis, Rana chensinensis, R.asiatica and Pelophylax ridibundus) have been discovered in Ili Kazakh Autonomous Prefecture (Wang et al., 2006).
The aim of the present study was to investigate population genetic patterns underlying the expansion of P.nigromaculatus in Xinjiang Uyghur Autonomous Region as inferred from mtDNA markers.We a) assessed the genetic diversity and examined the genetic structure of P.nigromaculatus in its native range in China and in the invaded territory of Xinjiang Uyghur Autonomous Region; b) reconstructed the routes of species' expansion from its origin in East China to areas of recent colonization in West China; and c) discuss the application of these results to the planning of appropriate control measures.
2.Materials and Methods
Sampling of P.nigromaculatus In 2012, we collected black-spotted frogs from different locations in six native ranges: Qiqihaer, Beijing, Xuzhou, Xi'an, Wenzhou and Chongqing, because these locations encompass most of the distribution of this species in northeast,north, central, northwest, southeast and southwest China, and in 2014, we collected frogs from the region of introduction, Xinjiang Uyghur Autonomous Region (Figure 1).To determine whether a site has been invaded by P.nigromaculatus, we surveyed all accessible water bodies at each site for three consecutive nights with line transects.The third toe of the right hind-foot of each postmetamorphosed black-spotted frog was clipped, and the tissue samples were preserved separately in 95% ethanol and stored at-20ºC in the laboratory.
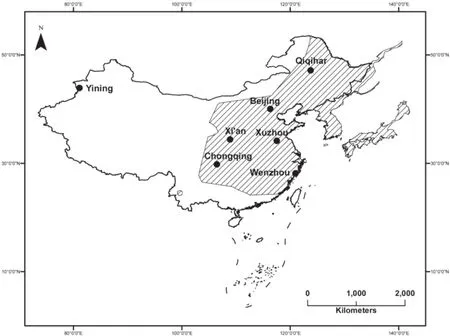
Figure 1 Sampled areas for P.nigromaculatus in China.Backward diagonal areas indicate distribution area in Asia.Closed circles denotes sampling site.
DNA extraction and amplification DNA was extracted according to the procedure described by Wang et al.(2014) and Zhu et al.(2014), with some modifications.Briefly, we placed approximately 3 mg (wet mass) ofeach sample in a 2 mL centrifuge tube with 100 µL of lysis buffer containing 0.5% Nonidet P-40, 0.1 M EDTA,0.01 M Tris-HCl (pH 8.0), 0.01 M NaCl, and 1 mg/mL proteinase K.The microtube was then shaken for 1 min using a vortex mixer at 16°C, followed by centrifugation for 1 min to concentrate all of the tissue at the bottom of the tube.Subsequently, the tube was incubated at 55°C for 24 h and at 100°C for 20 min.Finally, the microtube was centrifuged at 12,000 rpm for 3 min at 4°C, and the supernatant containing the DNA was diluted to one tenth of its original concentration for amplification via polymerase chain reaction (PCR).
We amplified a 695-bp segment of the mitochondrial cytochrome b (cyt b) gene from all specimens using the primers RanaLeuF5d (5'-AATMCC GWAAATCTCACCCCCT-3') and RanacytbB1 (5'-GCTGGTGTAAATTGTCTGGGTC-3') (Yang et al.,2003).The PCR products were then subjected to electrophoresis on 2% agarose gels.The resulting PCR products were directly sequenced using the same primers used for amplification (Beijing Genomics Institute,Beijing, China).
Data analysis The mitochondrial cyt b gene sequences were aligned and edited using MEGA 6 (Tamura et al.,2013).The sequences were aligned using Clustal X, and unique haplotypes were identified using DnaSP 5.10 (Rozas et al., 2003).The number of haplotypes (Hn),haplotype diversity (Hd) and nucleotide diversity (θπ)within each sampling population were estimated by ARLEQUIN ver3.5 (Excoffier and Lischer, 2010).We employed TCS 1.21 (Clement et al., 2000) to construct networks via statistical parsimony.
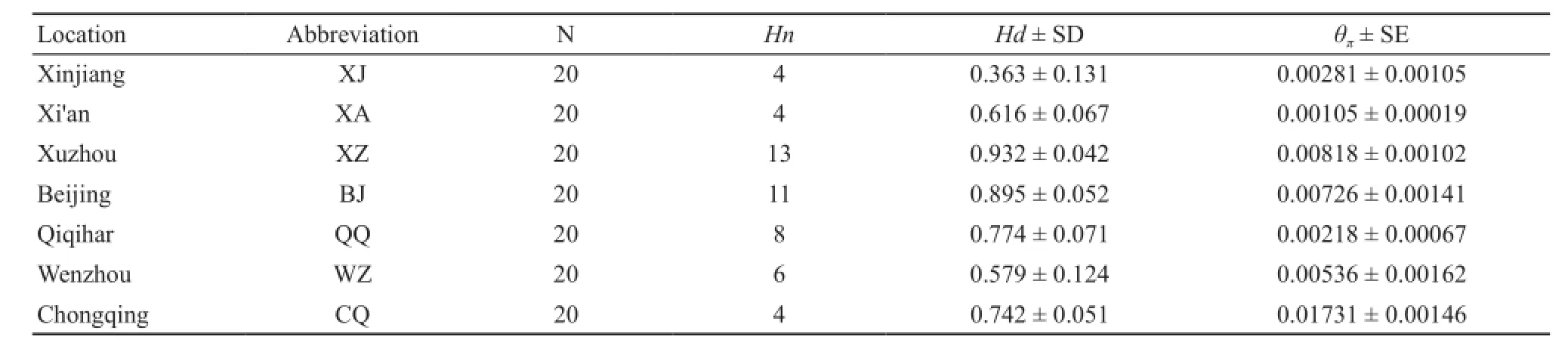
Table 1 Sampling information and mtDNA diversity of P.nigromaculatus.
3.Results
Hn, Hd and θπranged from 4 to 13, 0.363 to 0.932 and 0.00218 to 0.01731, respectively, among the sampled populations (Table 1).We also identified 42 haplotypes (H1-42), as defined by 116 polymorphic sites in the 695-bp cyt b fragment, among 140 P.nigromaculatus specimens from seven populations in China.Of these, 35 were unique haplotypes, and seven (H1, H2, H4, H5, H6,H7 and H13) were shared among local populations (Table 2).The statistical parsimony network showed that the H1 and H4 haplotypes occurred in the Chongqing population,while the H2 and H4 haplotypes occurred in the Beijing population, suggesting that the P.nigromaculatus found in Xinjiang most likely originated from Chongqing and Beijing (Figure 2).
4. Discussion
Our study is the frst to examine the invasion genetics of P.nigromaculatus in Ili Kazak Autonomous Prefecture in the Xinjiang Uyghur Autonomous Region.P.nigromaculatus is widely distributed in north, east,central and southwest China.However, P.nigromaculatus has only been noted within the area around Tcheng city of Xinjiang Uyghur Autonomous Region in the latest published records from northwest China (Jia et al., 1993).Therefore, the new record from Ili Kazak Autonomous Prefecture extends the known distribution range of this species in Asia by approximately 344 km into Tcheng city (Jia et al., 1993).
Historical records for the presence of P.nigromaculatus in Ili Kazak Autonomous Prefecture are not available,and therefore the pathways for invasion are unclear.The Shilei investigation report did not fnd P.nigromaculatus in the Xinjiang Uyghur Autonomous Region in 2006 (Shi et al., 2007), indicating that the black-spotted frog likely arrived subsequently and established its population.
Our mtDNA data demonstrate that the recently established populations of P.nigromaculatus have reduced genetic variability compared to most native China populations (Table 1).However, the haplotype diversity in the two native populations (XA and CQ) was identical to that in the XJ invasive population (four haplotypes in the native population vs.four haplotypes in the invasive population).Two of the four haplotypes were shared with native Chinese samples from CQ populations, and one ofthe four was shared with the BJ population.These results suggest that the Beijing and Chongqing areas in China are likely the source regions of P.nigromaculatus in Ili Kazak Autonomous Prefecture.The escape of alien animals from markets represents a significant source of introductions for alien animals (Hulme et al., 2008).Thus, the reason that these two areas are likely the source regions of P.nigromaculatus in Ili Kazak Autonomous Prefecture could be that they are both source areas for trade.

Figure 2 mtDNA haplotype parsimony network of P.nigromaculatus.Haplotype circle size is proportional to the number of individuals,numbers correspond to the haplotype numbers in Table 2.Solid represent Xinjiang population.Backward diagonal represent Qiqihar.Forward diagonal represent Xuzhou.Cross represent Xi'an.Diagonal cross represent Wenzhou.Horizontal represent Beijing.Vertical represent Chongqing.
Our mtDNA analysis of P.nigromaculatus revealed a clear genetic structure in native China (Figure 2).We detected few haplotypes that were shared among populations and significant genetic isolation in populations of China such as QQ, BJ, XA, XZ, WZ and CQ.These results indicate limited gene fow or migration among populations and are consistent with the results of our previous analysis of microsatellites (Wang et al.,2014).Furthermore, we observed high genetic diversity in the central populations but low diversity in the marginal populations.This discovery is consistent with studiesof the molecular phylogeography of P.nigromaculatus (Yang et al., 2003; Zhang et al., 2008).

Table 2 Population distribution of mtDNA haplotypes of P.nigromaculatus.
Monitoring the variations of the genetic diversity of these recently introduced populations is important for analyzing invasion genetics because the variability will increase over time as additional introduction events occur (Kolbe et al., 2004).Intraspecifc hybridization of individuals from genetically different populations (e.g.,from Beijing and Chongqing) may increase the fitness (e.g., higher fecundity) of P.nigromaculatus populations in Ili Kazak Autonomous Prefecture.Furthermore, mixed populations might be better adapted to new environments (Fitzpatrick and Shaffer, 2007), which could lead to a variety of serious problems associated with P.nigromaculatus in Ili Kazak Autonomous Prefecture.Identifying the origin of invasive P.nigromaculatus populations could facilitate the exploration of migratory pathways and prevent the potential influx of other haplotypes that would increase the genetic diversity of this species.
Invasive taxa threaten biodiversity by replacing native biota (Fitzpatrick et al., 2010; Rius and Darling, 2014).Reproductive interference (such as hybridization) is an important mechanism behind such replacement processes.Hybridization can have negative consequences (for example, the loss of locally adapted alleles, outbreeding depression and displacement by gene pool swamping)for the native population (Rius and Darling, 2014).Ili Kazak Autonomous Prefecture is home to another Pelophylax species (Pelophylax ridibunda).The results of the frst interactions between P.nigromaculatus and P.ridibunda will provide important insights on amphibian specialization.
Acknowledgements This research was funded by the Beijing Natural Science Foundation (5164036) and the National Natural Science Foundation of China (31370545 and 31200416).The collection and handling of frogs were conducted by the Animal Care and Use Committee of Institute of Zoology, Chinese Academy of Sciences (Project Nos.2012/17 and 2014/25).All staff received appropriate training before performing animal studies.
References
Campbell-Staton S.C., Goodman R.M., Backstrom N.,Edwards S.V., Losos J.B., Kolbe J.J.2012.Out of Florida: mtDNA reveals patterns of migration and Pleistocene range expansion of the Green Anole lizard (Anolis carolinensis).Ecol Evol, 2(9): 2274-2284
Clement M., Posada D., Crandall K.A.2000.TCS: A computer program to estimate gene genealogies.Mol Ecol, 9(10): 1657-1659
Excoffier L., Lischer H.E.L.2010.Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows.Mol Ecol Resour, 10(3): 564-567
Ficetola G.F., Bonin A., Miaud C.2008.Population geneticsreveals origin and number of founders in a biological invasion.Mol Ecol, 17(3): 773-782
Fitzpatrick B.M., Johnson J.R., Kump D.K., Smith J.J., Voss S.R.,Shaffer H.B.2010.Rapid spread of invasive genes into a threatened native species.P Natl Acad Sci USA, 107(8): 3606-3610
Fitzpatrick B.M., Shaffer H.B.2007.Hybrid vigor between native and introduced salamanders raises new challenges for conservation.P Natl Acad Sci USA, 104(40): 15793-15798
Gao X., Jin C., Camargo A., Li Y.2015a.Allocation trade-off under climate warming in experimental amphibian populations.PeerJ, 3: e1326
Gao X., Jin C.N., Llusia D., Li Y.M.2015b.Temperature-induced shifts in hibernation behavior in experimental amphibian populations.Sci Rep-Uk, 5
Hulme P.E., Bacher S., Kenis M., Klotz S., Kuhn I., Minchin D.,Nentwig W., Olenin S., Panov V., Pergl J.2008.Grasping at the routes of biological invasions: A framework for integrating pathways into policy.J Appl Ecol, 45(2): 403-414
Jia Z., Gao X., Yao J., Xu K., Zhang Z.1993.A New Record of Pelophylax nigromaculatus in Xinjiang, China.Arid Zone Research, 10(1): 2
Kolbe J.J., Glor R.E., Schettino L.R.G., Lara A.C., Larson A., Losos J.B.2004.Genetic variation increases during biological invasion by a Cuban lizard.Nature, 431(7005): 177-181
Liebl A.L., Schrey A.W., Andrew S.C., Sheldon E.L., Griffith S.C.2015.Invasion genetics: Lessons from a ubiquitous bird,the house sparrow Passer domesticus.Curr Zool, 61(3):465-476
Liu K., Wang F., Chen W., Tu L.H., Min M.S., Bi K., Fu J.Z.2010.Rampant historical mitochondrial genome introgression between two species of green pond frogs, Pelophylax nigromaculatus and P.plancyi.Bmc Evol Biol, 10
Meiners S.J.2007.Native and exotic plant species exhibit similar population dynamics during succession.Ecology, 88(5): 1098-1104
Moule H., Chaplin K., Rebecca D., Milller K.A., Thompson M.B., Chapple D.G.2015.A matter of time: Temporal variation in the introduction history and population genetic structuring of an invasive lizard.Curr Zool, 61(3): 456-464
Peacock M.M., Beard K.H., O'Neill E.M., Kirchoff V.S.,Peters M.B.2009.Strong founder effects and low genetic diversity in introduced populations of Coqui frogs.Mol Ecol,18(17): 3603-3615
Pyšek P., Richardson D.M., Pergl J., Jarošík V., Sixtová Z.,Weber E.2008.Geographical and taxonomic biases in invasion ecology.Trends Ecol Evol, 23(5): 237-244
Rius M., Darling J.A.2014.How important is intraspecifc genetic admixture to the success of colonising populations? Trends Ecol Evol, 29(4): 233-242
Rius M., Bourne S., Hornsby H.G., Chapman M.A.2015.Applications of next-generation sequencing to the study of biological invasions.Curr Zool, 61(3):488-504
Rozas J., Sánchez-DelBarrio J.C., Messeguer X., Rozas R.2003.DnaSP, DNA polymorphism analyses by the coalescent and other methods.Bioinformatics, 19(18): 2496-2497
Sherwin W.B., Frommer M., Sved J.A., Raphael K.A.,Oakeshott J.G., Shearman D.C.A., Gilchrist A.S.2015.Tracking invasion and invasiveness in Queensland fruit flies: From classical genetics to ‘omics'.Curr Zool, 61(3):477-487
Shi L., Yang J., Hou M., Zhao H., Dong B., Xiong J., Wang X., Wang X.2007.Herpetological surveys of Xinjiang Uygur autonomous region.Sichuan Journal of Zoology, 26(4): 812-818
Tamura K., Stecher G., Peterson D., Filipski A., Kumar S.2013.MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0.Mol Biol Evol, 30(12): 2725-2729
Wang S.P., Zhu W., Gao X., Li X.P., Yan S.F., Liu X., Yang J., Gao Z.X., Li Y.M.2014.Population size and time since island isolation determine genetic diversity loss in insular frog populations.Mol Ecol, 23(3): 637-648
Wang X., Ai S., Yuan L., Zhang Y.2006.The Progress in the Research of Xinjiang Amphibian Animals.Journal of Xinjiang Normal University (Natural Sciences Edition), 25(2): 50-53
Wang Y., Li Y., Wu Z., Murray B.R.2009.Insular shifts and trade-offs in life-history traits in pond frogs in the Zhoushan Archipelago, China.J Zool, 278(1): 65-73
Wang Y., Wu Z., Lu P., Zhang F., Li Y.2008.Breeding ecology and oviposition site selection of black-spotted pond frogs (Rana nigromaculata) in Ningbo, China.Frontiers of Biology in China,3(4): 530-535
Yang Y., Zhang D., Li Y., Ji Y.2003.Mitochondrial DNA diversity and preliminary biogeographic inference of the evolutionary history of the black-spotted pond frog Rana nigromaculata populations in China.Curr Zool, 50(2): 193-201
Zhang H., Yan J., Zhang G.Q., Zhou K.Y.2008.Phylogeography and demographic history of chinese black-spotted frog Populations (Pelophylax nigromaculata): Evidence for independent refugia expansion and secondary contact.Bmc Evol Biol, 8(1): 1
Zhang X., Zhou K., Chang Q.2004.Population genetic structure of Pelophylax nigromaculata in Chinese mainland based on mtDNA control region sequences.Acta genetica Sinica, 31(11): 1232-1240
Zhu W., Xu F., Bai C.M., Liu X., Wang S.P., Gao X., Yan S.F., Li X.P., Liu Z.T., Li Y.M.2014.A survey for Batrachochytrium salamandrivorans in Chinese amphibians.Curr Zool, 60(6): 729-735
#These authors contributed equally to this work.
E-mail: liym@ioz.ac.cn
Received: 11 December 2015 Accepted: 6 May 2016
DOI:10.16373/j.cnki.ahr.150071
*Corresponding author:Prof.Yiming LI, from Institute of Zoology,Chinese Academy of Sciences, Beijing, China, with his research focusing on conservation biology and ecology of amphibians.
 Asian Herpetological Research2016年2期
Asian Herpetological Research2016年2期
- Asian Herpetological Research的其它文章
- Genetic Diversity and Population Structure for the Conservation of Giant Spiny Frog (Quasipaa spinosa) Using Microsatellite Loci and Mitochondrial DNA
- Coevolution of Male and Female Response Preferences to Sexual Signals in Music Frogs
- Ecological Niche Divergence between Trapelus ruderatus (Olivier,1807) and T.persicus (Blanford, 1881) (Sauria: Agamidae) in the Middle East
- The Effects of Chronic Hypoxia on Thermoregulation and Metabolism in Phrynocephalus vlangalii
- Comparison of Skull Morphology in Two Species of Genus Liua (Amphibia: Urodela: Hynobiidae), L.shihi and L.tsinpaensis
- Discovery of Female Laudakia papenfussi Zhao, 1998, with Insights into its Phylogenetic Relationships
