Association between TNF -308G/A polymorphism and susceptibility to pulmonary tuberculosis in the Lur population of Iran
Association between TNF -308G/A polymorphism and susceptibility to pulmonary tuberculosis in the Lur population of Iran
Tel: +98 66 33120172
Fax: +98 66 33120173
E-mail: avarzi@yahoo.co.uk
Peer review under responsibility of Hainan Medical University.
Foundation Project: Supported by the Lorestan University of Medical Sciences under Grant No 1328.
Farhad Shahsavar1, Ali Mohammad Varzi1*, Alireza Azargoon21Department of Immunology, Lorestan University of Medical Sciences, Khorramabad, Iran
2Department of Internal Medicine, Lorestan University of Medical Sciences, Khorramabad, Iran
ARTICLE INFO
Article history:
Received 15 Jan 2015
Received in revised form 29 Jan, 2nd revised form 21 Jul 2015
Accepted 20 Sep 2015
Available online 7 Nov 2015
Keywords:
TNFα polymorphism
Tuberculosis
Pulmonary tuberculosis
Lur population
Iran
1. Introduction
ABSTRACT
Objective: To investigate whether tumor necrosis factor-α(TNFα) -238G/A and -308G/ A polymorphisms are associated with susceptibility to pulmonary tuberculosis (TB) in the Lur ethnic population of Iran.
Methods: TNF polymorphisms genotyping was performed by polymerase chain reaction-restriction fragment length polymorphism method in 100 pulmonary TB patients and 100 healthy controls from the Lur population.
Results: The allelic and genotypic frequencies of TNFα-238G/A polymorphism were not significantly different between the pulmonary TB patients and the healthy controls. However, the TNFα-308G/A polymorphism showed a significantly higher frequency of genotype GG in TB subjects compared to healthy controls (94% in the patients vs. 62% in the controls, P = 0.0001, odds ratio = 0.104, confidence interval = 0.028–0.382). Moreover, in the TNFα-308G/A polymorphism, a significantly higher frequency of G allele was measured in the patient group compared with the control group (97% in the patient group vs. 81% in the control group, P = 0.0001, odds ratio = 0.132, confidence interval = 0.038–0.462).
Conclusions: Our findings suggest that TNFα-308G/A polymorphism may increase the susceptibility to pulmonary TB in the Lur population of Iran. Despite TNFα polymorphisms and susceptibility to pulmonary TB, we suggest that more studies with larger sample size are needed in the future. Increasing our understanding of susceptibility risk factors may help to improve current preventive measures and treatment for TB.
Tuberculosis (TB) is caused by the acid fast bacillus, Mycobacterium tuberculosis (MTB). TB is a common infectious disease. Worldwide, there are 9 million novel cases and 2 million mortalities annually [1]. It has been suggested that sensitivity to this disease is variable in the different populations and that contact with this microorganism does not always result in infection. Almost one third of the world population is infected by this bacterium of which 5%–10% are infected with the active form of TB. Additionally, the course and duration of disease vary in different individuals [2].
These differences may be due to host factors and genetic sensitivity of different individuals to this disease [3,4]. Different genetic factors are implicated in the susceptibility to and severity of TB, one of which is KIR3DS1 gene and it combines with HLA-B Bw4 and Ile80 ligand [5]. Moreover, human and mouse studies on MTB infection have demonstrated different loci in the susceptibility or resistance to TB such as toll like receptors[6–10]. It has been reported that tumor necrosis factor (TNF) is involved in the prevention of mycobacterial infection development and alsoinpreventingprogressionfromlatenttoactive TBform[11–14].
TNFα is a cytokine involved in the innate immunity. It is primarily formed against pathogens, and its genetic polymorphisms play an important role in the immune response efficacy against major infections. The principal source of TNFα production is the mononuclear phagocytes. The release of TNFαfrom these cells results in the recruitment of neutrophils and monocytes to the site of infection[15].
Theeffectof TNFαonthehumanbodyvariesfromactivationof inflammatory processes to activation of hepatocytes and tissue damagebasedonitslow,moderateandhighdoses.Italsocontributes to activation of macrophages and regulates interferonγ production [16]. Therefore, it initiates pro-inflammatory reactions implicated in the effect on disease and resistance to the Mycobacterium[17].
The human and animal TB investigations suggest that TNFα impresses the innate immune response to TB[18]. Hence, TNFα -238G/A and -308G/A polymorphisms have been analyzed widely as the nominated genes for susceptibility to TB in different populations [19–23]. Due to the differences between distribution of TNFα gene in different races and nations and also due to association of TNFα polymorphisms with pulmonary TB susceptibility, we were interested to investigate the prevalence of these polymorphisms in pulmonary TB patients and healthy controls of the Lur population dwelled in Lorestan Province. Therefore, the susceptibility to pulmonary TB infection was ascertained by the studying of TNFα-238G/ A and -308G/A polymorphisms in the TB group and the results were compared with the healthy control group.
2. Materials and methods
2.1. Patients and controls
This study was approved by the Ethics Committee of Lorestan University of Medical Sciences. All subjects gave informed written consent. We used case control study to implement this investigation. The patient group was comprised of 100 unrelated Lur individuals referred to the health center of Khorramabad city of Lorestan Province, with TB confirmed by sputum culture. All patients received TB standard treatment and none of them had drug resistance. The control group was comprised of 100 unrelated Iranian individuals of the same race and geographic region. The controls were asymptomatic, had normal radiologic results and their purified protein derivatives test was negative. The control group was matched on age and sex with the patient group. Additionally, all study subjects had parents of the same race. The blood samples were collected for analysis.
2.2. Genotyping
Patient and control DNA samples were extracted using QIAmp kit (Qiagen, Germany). The polymerase chain reactionrestriction fragment length polymorphism previously suggested by Fan et al. was performed to determine TNFα-238G/A and -308G/A polymorphisms in patients and controls using their genomic DNA [24]. The list of forward and reverse primer sequences (Qiagen, Germany), restriction enzymes (Biolabs, USA) and digestion patterns for different alleles were assorted in Table 1.

Table 1Primer sequences, restriction enzymes used and restriction digestion patterns for genotyping of TNF-α polymorphisms.
The amplification was carried out by using Mastercycler (BioRad, USA) in 20 μL reaction. Amplification conditions used were as follows: denaturation initiated at 94°C for 5 min and was followed by 5 cycles of denaturation at 94°C for 5 min, annealing at 60°C for 1 min, extension at 72°C for 1 min, and 25 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, extension at 72°C for 40 s with final extension at 72°C for 1 min. The PCR products were incubated with restriction enzymes Msp I and Nco I at 37°C for TNFα-238G/A and -308G/A, respectively for 24 h to digest the DNA. The electrophoresis of PCR products was accomplished on 3% agarose gel consisting of 0.5 mg/mL ethidium bromide. Finally, the products were visualized by using an ultraviolet light.
2.3. Statistical analysis
The genotypic and allelic frequencies of TNFα-238G/A and -308G/A polymorphisms were ascertained by direct counting in the TB population and healthy control population. All polymorphisms were consistent with values predicted by Hardy–Weinberg equilibrium in both patient and control groups. The differences in the genotypic and allelic frequencies of TNFα -238G/A and -308G/A polymorphisms were determined by the Chi-squared test and Fisher's exact test between TB population and healthy control population. Overall, P<0.05 was considered statistically significant after correction. The odds ratio (OR) was calculated by the cross-product ratio and exact confidence interval (CI) of 95% was obtained.
3. Results
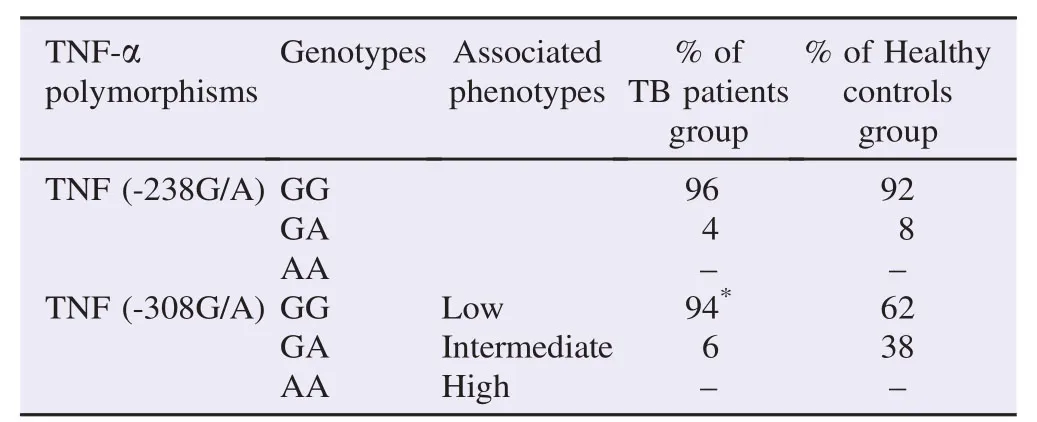
Table 2Distribution of TNF-α genotypes in TB patients group and healthy controls group.
The study subjects comprised of 100 healthy controls with the mean age, (30.21±2.55) years and 100 pulmonary TBpatients with the mean age, (39.65±3.87) years. Among the healthy controls, 50 individuals were males and 50 individuals were females, and among the pulmonary TB patients, 40 individuals were males and 60 individuals were females.
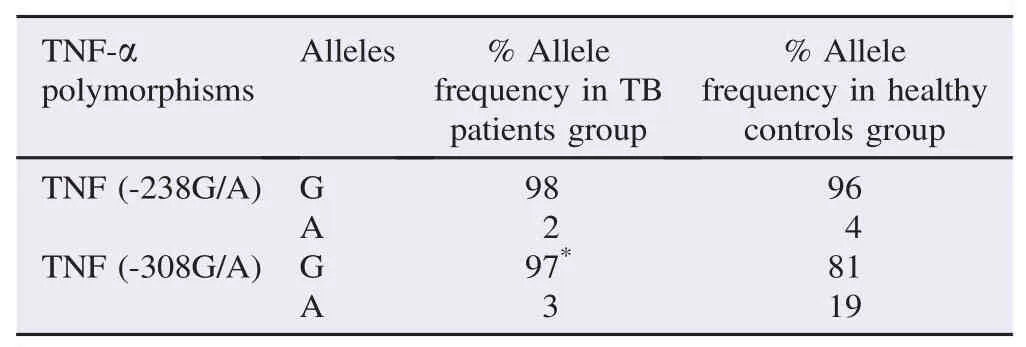
Table 3Distribution of TNF-α alleles in TB patients group and healthy controls group.
The genotypic and allelic frequencies of TNFα-238G/A and -308G/A polymorphisms were listed in Tables 2 and 3. The genotypic and allelic frequencies of TNFα-238G/A polymorphism did not have significant difference between the pulmonary TB patients and the healthy controls. Also, AA genotype of TNFα-238G/A polymorphism was not detected in the patient and control groups. Only, in TNFα-308G/A polymorphism, a significantly increased frequency of genotype GG was observed among patients compared with controls (94% in the patient group vs. 62% in the control group, P = 0.0001, OR = 0.104, CI = 0.028–0.382). Also AA genotype of TNFα -308G/A polymorphism was not observed in the patient and control group (Table 2).
In the TNFα-308G/A polymorphism, a significantly increased frequency of G allele was observed among the patient group compared with the control group (97% in the patient group vs. 81% in the control group, P = 0.0001, OR = 0.132, CI = 0.038–0.462) (Table 3).
4. Discussion
TNFα is one of the most significant cytokines involving in the primary host innate immunity against a pathogen. Moreover, it is shown that TNFα is characteristic in the prevention of mycobacterial establishment and preservation of TB in latent form [11–14]. Among this, TNFα-238G/A and -308G/A polymorphisms were widely determined as the nomination genes in the susceptibility to TB infection [19–23].
In this study, we showed the effect of TNFα-238G/A and -308G/A in the susceptibility to pulmonary TB in the Lur population of Iran. The genotypic and allelic frequencies of TNFα -308G/A polymorphism have significant difference between patients infected by pulmonary TB and the healthy control individuals. The TNFα-238G/A polymorphism had no significant differencebetweenpulmonary TBpatientsandthehealthycontrol group. In contrast, in the late studies carried out by Sharma et al. [12] and Kumar et al. [21] in the north of India, there were no association between these polymorphisms and pulmonary TB. This phenomenon is not surprising because of genetic nonidentity between Asians and Indians (racial differences).
Our results showed similar outcome as former studies in this field conducted by Fan et al.[24]and Qu et al.[25]on the Asian population which indicated the association of TNFα-308G/A with TB. One possible explanation for this is the genetic similarities between Asian populations. The study by Ben-Selma et al. [26] also indicated the association between TNFα -308G/A polymorphism and TB in the Tunisian population. Also, study by Amirzargar et al. [27] has demonstrated the association of TNFα-238G/A polymorphism with TB in other ethnic group of Iranian.
TNFα is an essential cytokine in the granuloma formation. The mice with deficiency in TNFα are impotent in the granuloma formation. This tragedy leads to MTB development and fulminant death in the infected animals. Furthermore, the former studies have demonstrated that TNFα blocking can lead to TB reactivation [28,29]. Hence, despite of paradoxical findings, the association of TNFα polymorphisms with TB seems undeniable. However, we can indicate other TNFα polymorphisms, the race of studied population and the size of studied samples as the reasons of these adverse results.
Finally, our findings show that TNFα-308G/A polymorphism may associate with pulmonary TB in the Lur population of Iran. Despite susceptibility to pulmonary TB, we suggest that more studies with larger sample size should be carried out to verify the role of TNFα polymorphisms, especially TNFα-308G/A polymorphism in the future.
Conflict of interest statement
We declare that we have no conflict of interest.
Acknowledgments
We would like to thank all the patients and healthy individuals for their participation. We are also indebted to Miss Delaram Varzi for her proofreading of the manuscript. This study was supported by the Lorestan University of Medical Sciences under Grant No. 1328.
References
[1] World Health Organization. Global tuberculosis report 2014. Geneva: World Health Organization; 2014. [Online] Available from: http://www.who.int/tb/publications/global_report/en/ [Accessed on 15th July, 2015]
[2] Azad AK, Sadee W, Schlesinger LS. Innate immune gene polymorphisms in tuberculosis. Infect Immun 2012; 80(10): 3343-59.
[3] Abel L, El-Baghdadi J, Bousfiha AA, Casanova JL, Schurr E. Human genetics of tuberculosis: a long and winding road. Philos Trans R Soc Lond B Biol Sci 2014; 369(1645): 20130428.
[4] Png E, Alisjahbana B, Sahiratmadja E, Marzuki S, Nelwan R, Balabanova Y, et al. A genome wide association study of pulmonary tuberculosis susceptibility in Indonesians. BMC Med Genet 2012; 13: 5.
[5] Shahsavar F, Mousavi T, Azargon A, Entezami K. Association of KIR3DS1+HLA-B Bw4Ile80 combination with susceptibility to tuberculosis in Lur population of Iran. Iran J Immunol 2012; 9(1): 39-47.
[6] Kleinnijenhuis J, Oosting M, Joosten LA, Netea MG, Van Crevel R. Innate immune recognition of Mycobacterium tuberculosis. Clin Dev Immunol 2011; 2011: 405310.
[7] Velez DR, Wejse C, Stryjewski ME, Abbate E, Hulme WF, Myers JL, et al. Variants in toll-like receptors 2 and 9 influence susceptibility to pulmonary tuberculosis in Caucasians, African-Americans, and West Africans. Hum Genet 2010; 127(1): 65-73.
[8] Shahsavar F, Azargoon A, Jafarzadeh M, Forutani S, Sabooteh T. [Toll-like receptor 2 Arg753Gln polymorphism is associated with susceptibility to pulmonary tuberculosis in the Lur population of Iran]. AFINIDAD 2014; 563: 53-7. Spanish.
[9] Vejbaesya S, Chierakul N, Luangtrakool P, Sermduangprateep C. NRAMP1 and TNF-alpha polymorphisms and susceptibility to tuberculosis in Thais. Respirology 2007; 12(2): 202-6.
[10] Zhang Y, Jiang T, Yang X, Xue Y, Wang C, Liu J, et al. Toll-like receptor -1, -2, and -6 polymorphisms and pulmonary tuberculosis susceptibility: a systematic review and meta-analysis. PLoS One 2013; 8(5): e63357.
[11] Shim TS. Diagnosis and treatment of latent tuberculosis infection in patients with inflammatory bowel diseases due to initiation of anti-tumor necrosis factor therapy. Intest Res 2014; 12(1): 12-9.
[12] Sharma S, Rathored J, Ghosh B, Sharma SK. Genetic polymorphisms in TNF genes and tuberculosis in North Indians. BMC Infect Dis 2010; 10: 165.
[13] Ahmad S. Pathogenesis, immunology, and diagnosis of latent Mycobacterium tuberculosis infection. Clin Dev Immunol 2011; http://dx.doi.org/10.1155/2011/814943.
[14] Bellofiore B, Matarese A, Balato N, Gaudiello F, Scarpa R, Atteno M, et al. Prevention of tuberculosis in patients taking tumor necrosis factor-alpha blockers. J Rheumatol Suppl 2009; 83: 76-7.
[15] Wiens GD, Glenney GW. Origin and evolution of TNF and TNF receptor superfamilies. Dev Comp Immunol 2011; 35(12): 1324-35.
[16] Varahram M, Farnia P, Nasiri MJ, Karahrudi MA, Dizagie MK, Velayati AA. Association of Mycobacterium tuberculosis lineages with IFN-gamma and TNF-alpha gene polymorphisms among pulmonary tuberculosis patient. Mediterr J Hematol Infect Dis 2014; 6(1): e2014015.
[17] Quesniaux VF, Jacobs M, Allie N, Grivennikov S, Nedospasov SA, Garcia I, et al. TNF in host resistance to tuberculosis infection. Curr Dir Autoimmun 2010; 11: 157-79.
[18] Saiga H, Shimada Y, Takeda K. Innate immune effectors in mycobacterial infection. Clin Dev Immunol 2011; 2011: 347594.
[19] Merza M, Farnia P, Anoosheh S, Varahram M, Kazampour M, Pajand O, et al. The NRAMPI, VDR and TNF-alpha gene polymorphisms in Iranian tuberculosis patients: the study on host susceptibility. Braz J Infect Dis 2009; 13(4): 252-6.
[20] Pacheco AG, Cardoso CC, Moraes MO. IFNG +874T/A, IL10 -1082G/A and TNF -308G/A polymorphisms in association with tuberculosis susceptibility: a meta-analysis study. Hum Genet 2008; 123: 477-84.
[21] Kumar V, Khosla R, Gupta V, Sarin BC, Sehajpal PK. Differential association of tumour necrosis factor-alpha single nucleotide polymorphism (-308) with tuberculosis and bronchial asthma. Natl Med J India 2008; 21(3): 120-2.
[22] Ates O, Musellim B, Ongen G, Topal-Sarikaya A. Interleukin-10 and tumor necrosis factor-alpha gene polymorphisms in tuberculosis. J Clin Immunol 2008; 28(3): 232-6.
[23] Wang Q, Zhan P, Qiu LX, Qian Q, Yu LK. TNF-308 gene polymorphism and tuberculosis susceptibility: a meta-analysis involving 18 studies. Mol Biol Rep 2012; 39(4): 3393-400.
[24] Fan HM, Wang Z, Feng FM, Zhang KL, Yuan JX, Sui H, et al. Association of TNF-alpha-238G/A and 308 G/A gene polymorphisms with pulmonary tuberculosis among patients with coal worker's pneumoconiosis. Biomed Environ Sci 2010; 23(2): 137-45.
[25] Qu Y, Tang Y, Cao D, Wu F, Liu J, Lu G, et al. Genetic polymorphisms in alveolar macrophage response-related genes, and risk of silicosis and pulmonary tuberculosis in Chinese iron miners. Int J Hyg Environ Health 2007; 210(6): 679-89.
[26] Ben-Selma W, Harizi H, Boukadida J. Association of TNF-alpha and IL-10 polymorphisms with tuberculosis in Tunisian populations. Microbes Infect 2011; 13(10): 837-43.
[27] Amirzargar AA, Rezaei N, Jabbari H, Danesh AA, Khosravi F, Hajabdolbaghi M, et al. Cytokine single nucleotide polymorphisms in Iranian patients with pulmonary tuberculosis. Eur Cytokine Netw 2006; 17(2): 84-9.
[28] Shim TS. Diagnosis and treatment of latent tuberculosis infection due to initiation of anti-TNF therapy. Tuberc Respir Dis Seoul 2014; 76(6): 261-8.
[29] Jo KW, Hong Y, Jung YJ, Yoo B, Lee CK, Kim YG, et al. Incidence of tuberculosis among anti-tumor necrosis factor users in patients with a previous history of tuberculosis. Respir Med 2013; 107(11): 1797-802.
Management and decision-making http://dx.doi.org/10.1016/j.apjtb.2015.09.019
*Corresponding author:Dr. Ali Mohammad Varzi, PhD, Department of Immunology, Lorestan University of Medical Sciences, Khorramabad, Iran.
 Asian Pacific Journal of Tropical Biomedicine2016年1期
Asian Pacific Journal of Tropical Biomedicine2016年1期
- Asian Pacific Journal of Tropical Biomedicine的其它文章
- The antibacterial activity of selected plants towards resistant bacteria isolated from clinical specimens
- Isolation of aerobic bacteria from ticks infested sheep in Iraq
- Rhinacanthus nasutus leaf improves metabolic abnormalities in high-fat diet-induced obese mice
- Influence of extraction solvents on antioxidant and antimicrobial activities of the pulp and seed of Anisophyllea laurina R. Br. ex Sabine fruits
- Antifungal activity of plant extracts with potential to control plant pathogens in pineapple
- Potent water extracts of Indonesian medicinal plants against PTP1B
