Antibiotic resistance profile and RAPD analysis of Campylobacter jejuni isolated from vegetables farms and retail markets
Antibiotic resistance profile and RAPD analysis of Campylobacter jejuni isolated from vegetables farms and retail markets
Tel: +60 9 6993582
Fax: +60 9 6997881
E-mails: jyhtang@unisza.edu.my, jyhtang@gmail.com
Peer review under responsibility of Hainan Medical University.
Foundation Project: Supported by the International Foundation of Sciences, Sweden (Grant No. E/5237-1).
John Yew Huat Tang1*, Mohd Ikhsan Khalid1, Syazana Aimi2, Che Abdullah Abu-Bakar1, Son Radu21Department of Food Industry, Faculty of Bioresources and Food Industry, Universiti Sultan Zainal Abidin, Tembila Campus, 22200 Besut, Terengganu, Malaysia
2Food Safety Research Centre, Faculty of Food Science and Technology, Universiti Putra Malaysia, 43400 UPM Serdang,
Selangor, Malaysia
ARTICLE INFO
Article history:
Received 20 Jul 2015
Received in revised form 6 Aug,
2nd revised form 2 Sep 2015
Accepted 8 Oct 2015
Available online 10 Nov 2015
Keywords:
Campylobacter jejuni
Multiple antibiotic resistance index
Antibiotic resistance
RAPD analysis
ABSTRACT
Objective: To investigate antibiotic resistance profile and characterize Campylobacter jejuni (C. jejuni) isolates using random amplified polymorphic DNA (RAPD) analysis.
Methods: Ninety eight C. jejuni isolates from farms and retail outlets were screened against 10 antibiotics commonly used clinically and agriculturally by using disk diffusion method. RAPD analysis was done to characterize 98 C. jejuni isolates.
Results: Fifty-one percent of the isolates had multiple antibiotic resistance index 0.2 and below. This indicated that the isolates in the vegetables were not from the high risk environment or extensive farming practices. C. jejuni isolates found resistant towards penicillin G (93%), vancomycin (86%), ampicillin (35%), erythromycin (28%), gentamycin (4%), amikacin (3%), enrofloxacin (1%), norfloxacin (1%) and no resistance towards ciprofloxacin. RAPD clustering analysis showed that the contamination of C. jejuni in vegetables was likely due to cross contamination at retail markets.
Conclusions: C. jejuni contamination in vegetables at retail markets was due to cross contamination. Current finding proved that C. jejuni in small scale vegetables production was less expose towards antibiotic abuse.
1. Introduction
Campylobacter jejuni (C. jejuni) is a Gram-negative, spiralshape bacterium and requires microaerophilic growth condition [1,2]. C. jejuni nutrient utilisation has been fully elucidated, but its metabolic flexible processes allow survival in the environment which eventually causes infection in human[1,2]. C. jejuni is one of the most frequently implicated causative agent of Campylobacteriosis in human[1,2]. Major risk factors for causing Campylobacteriosis in humans are consumption of undercooked poultry, untreated or contaminated water and raw milk[2].
Campylobacter becomes more resistant toward antibiotics and some of it have formed multiple drug resistance[3,4]. Erythromycin and tetracycline are commonly administered in cases of Campylobacter infections, but high resistance among Campylobacter towards them has been reported [3,4]. Fluoroquinolones resistant C. jejuni was thought to be biologically stronger than susceptible strain and the usage of fluoroquinolones as prophylaxis in poultry has caused increase in resistance towards fluoroquinolones [3,4]. Chai et al. suggested C. jejuni resistance towards fluoroquinolone group of antibiotics is related to farming practices [4]. Krumperman reported the usefulness of multiple antibiotic resistance (MAR) indexing to identify bacteria isolates from high risk environment or fecal contamination[5].
The demand for ready-to-eat fresh produce has risen in recent years [6]. This might be due to the health awareness on the benefits of fresh produce intake. Several studies have reported the presence of Campylobacter spp. in fresh produce and the number of foodborne outbreaks associated with raw fruits and vegetables has also increased due to cross-contamination from fertilizer, soil and irrigation water [7–9]. Besides being reported presence in fresh produce that available at retail markets, Campylobacter spp. also detected to be present in vegetables at farms [9]. However, whether the presence of Campylobacter invegetables at retail market originated from farms has been rarely studied.
Therefore the goal of present study is to characterize C. jejuni isolates by antibiotic resistant profiles and random amplified polymorphic DNA-polymerase chain reaction (RAPD-PCR) to determine the genetic relatedness of C. jejuni isolates.
2. Materials and methods
2.1. C. jejuni isolates
A total of 98 C. jejuni isolates from various types of samples (vegetables and soils) from 5 small scale local vegetables farms and 12 retail markets in Terengganu, Malaysia from January 2013 to April 2014. It was comprised of 9 C. jejuni isolates from farms and 89 C. jejuni isolates from retail markets. All the isolates were confirmed by PCR using species specific primers targeting 23S rRNA [10]. The primers used were 23S rRNA F (5'-TATACCGGTAAGGAGTGCTGGAG-3') and 23S rRNA R (5'-ATCAATTAACCTTCGAGCACCG-3'). The PCR method was performed in 25 μL of reaction mixture as described in our previous study with a final concentration of 1×Green GoTaq Flexi buffer, 0.2 mmol/L concentration of the deoxynucleotide triphosphate mix, 0.2 mmol/L concentrations of each primer, 3 mmol/L MgCl2solution, 2 IU of GoTaq DNA polymerase, and 2 μL of DNA boiled lysate. All items used in the PCR were purchased from Promega (Madison, WI, USA) [8]. PCRs were performed on a Veriti 96-well Fast Thermal Cycler (Applied Biosystems, Foster City, CA, USA), with an initial denaturation step of 95°C for 5 min followed by 30 cycles of 95°C for 30 s, 55°C for 1 min, and 72°C for 30 s, and a final extension step at 72°C for 5 min. PCR products were electrophorised using 1.5% agarose gel at 70 V for 90 min. Bands were visualized with UV transilluminator (AlphaImager HP, Alpha Innotech, CA, USA) after staining with GelRed nucleic acid gel stain (Biotium, Hayward, CA, USA). A 100-bp DNA ladder (NL1405, Vivantis, Oceanside, CA, USA) was used as a DNA molecular ladder.
2.2. Antibiotic resistance test
Antibiotics resistance patterns were determined using disk diffusion method according to Clinical and Laboratory Standards Institute[11]. All isolates were grown in Bolton Broth with supplement (Oxoid, Hampshire, England) without lyse horse blood for 48 h at 42°C. Sterile cotton swabs were used to spread uniformly C. jejuni from broth into Mueller–Hinton agar plates (Merck, Germany). Ten antibiotic discs were selected to test on its susceptibility to C. jejuni.
All antibiotics discs were placed on the agar surface by using disc dispenser. The selected antibiotics were penicillin G (10 μg), tetracycline (30 μg), ciprofloxacin (5 μg), enrofloxacin (5 μg), erythromycin (15 μg), gentamicin (10 μg), norfloxacin (10 μg), amikacin (30 μg), vancomycin (5 μg), ampicillin (10 μg). Inoculated plates were incubated at 42°C for 48 h under microaerophilic condition generated by Anaerocult C system (Merck, Germany).
2.3. MAR index
MAR index of the isolates was determined as a/b, where‘a’represents the number of multiple antibiotics to which the particular isolates are resistant, and‘b’represents the number of multipleantibioticstowhichtheparticularisolatesareexposed[5].
2.4. Cluster analysis using RAPD-PCR
DNA was extracted using boiled cell method as described by Khalid et al. with minor modification[8]. A total of 1 mL Bolton broth from the turbid tubes was centrifuged at 12 000 r/min for 10 min in order to pellet the bacterial cells. The supernatant was discarded and the pellet was then resuspended with 300 μL of sterile distilled water and boiled for 10 min followed by freezing at−20°C for 10 min. It was then centrifuged at 12 000 r/min for 10 min to pellet the cell debris [8]. The supernatant was then kept for use in RAPD-PCR.
A 10-mer oligonucleotide primers of OPA 11 (5'-CAATCGCCGT-3') from Integrated DNA Technologies, Singapore were used to characterize the isolates. PCR amplification was done with following programme: initial denaturation of 95°C (5 min); 45 cycles of denaturation at 95°C (1 min), annealing at 36°C (1 min), and extension at 72°C (2 min);final extension at 72°C (5 min). All the PCR assays were performed with Veriti 96-well Thermal Cycler (Applied Biosystems, USA). PCR products were visualized by electrophoresis in a 1.5% agarose gel at 70 V for 90 min. Bands were visualized with UV transilluminator (AlphaImager HP, Alpha Innotech, CA, USA) after staining with GelRed™Nucleic Acid Gel Stain (Biotium, USA). A 100 bp-DNA ladder (NL1405; Vivantis, USA) was used as a DNA-molecular ladder. Cluster analysis was done using GelCompar II version 5.1 (Applied Maths, Belgium).
3. Results
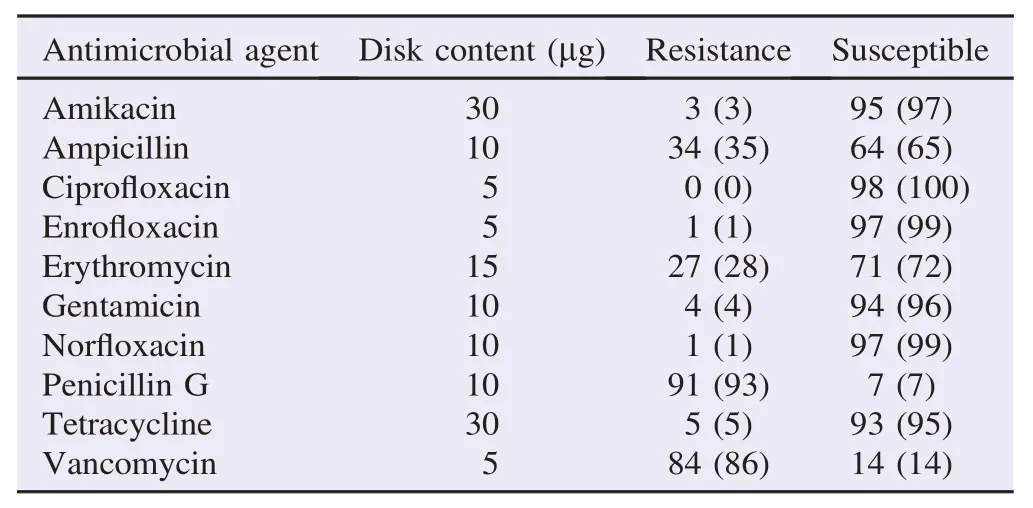
Table 1Number and percentages of antimicrobial-resistant C. jejuni isolated from various samples (N = 98). n (%).

Table 2Antibiotic resistance profile and multiple antibiotic resistance index of C. jejuni from vegetable and soil samples.
From Table 1, all 98 isolates of C. jejuni were tested against 10 types of antibiotics that frequently used in clinical andagricultural practices. C. jejuni isolates showed the highest resistance towards penicillin G (93%), followed by vancomycin (86%). All C. jejuni isolates were susceptible towards ciprofloxacin (100%) and showed highly susceptible to enrofloxacin, norfloxacin, amikacin, gentamicin and tetracycline with each recorded susceptibility level of 99%, 99%, 97%, 96% and 95%. Other antibiotics used in this study showed moderate percentage of C. jejuni resistance with ampicillin (35%) and erythromycin (28%).
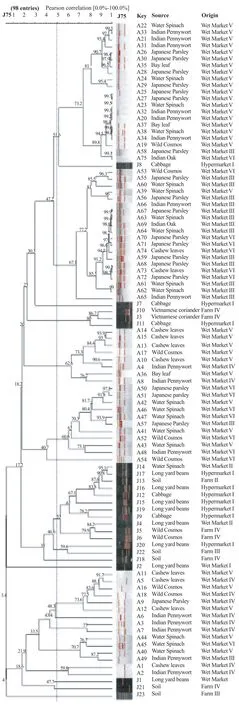
Figure 1. Dendrogram based on the hierarchic numerical analysis of the resistance profiles obtained for 98 C. jejuni isolates, employing the Pearson correlation coefficient and UPGMA for clustering.
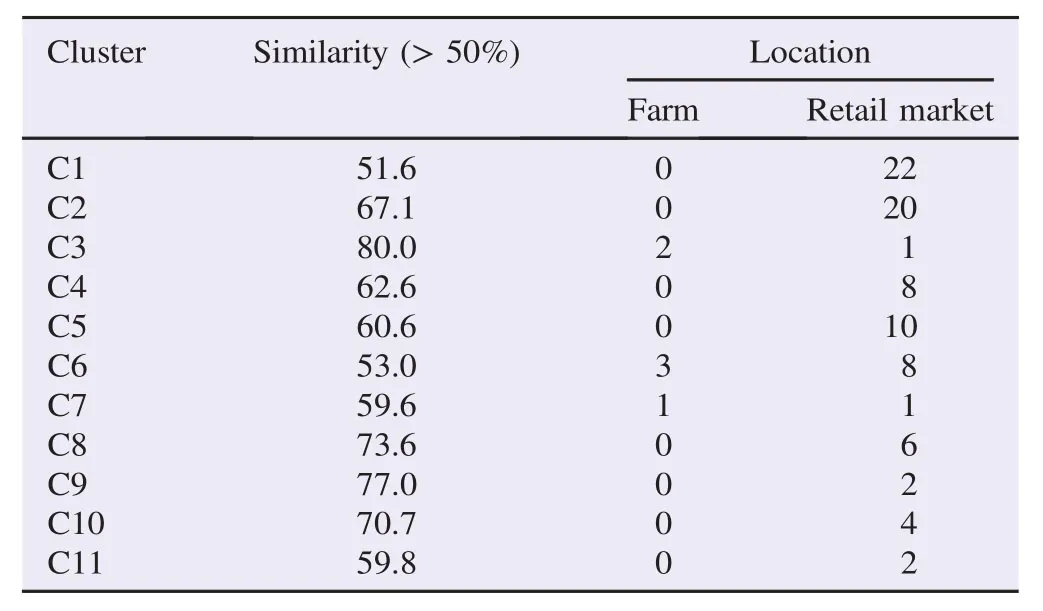
Table 3Characterization of C. jejuni clusters defined in the hierarchic analysis performed with location of sampling.
MAR index is shown in Table 2. Percentages of C. jejuni isolates recorded MAR index 0.1, 0.2, 0.3 and 0.4 were 3%, 48%, 31% and 18%, respectively.
The genetic diversity among C. jejuni strains isolated from various sources was investigated by RAPD-PCR using OPA 11 primer. All isolates generated bands with OPA 11 primer. The genetic diversity was observed among all strains. The dendrograms derived from the RAPD-PCR profiles generated with primers OPA 11 is shown in Figure 1. The dendrogram was constructed using GelCompar II version 5.1 by Applied Maths, Belgium with Pearson correlation and unweighted pair-group method with arithmetic means clustering to determine the genetic relatedness of the 98 isolates. According to Figure 1, the dendrogram branched into 11 major clusters at a similarity level of 50%. Ninety-one out of 98 isolates was grouped into this 11 major clusters and the remaining 7 isolates were not grouped due to similarity below 50%. Three out of 11 clusters demonstrated relatedness between isolates from farms and the retail markets is shown in Table 3.
4. Discussion
C. jejuni multidrug resistance especially towards quinolones and erythromycin had created concerns around the world [3,12]. Investigation of C. jejuni in vegetables was limited and most studies investigated antibiotic resistance of C. jejuni isolated from poultry and meat product [13–16]. Chai et al. had studied the biosafety of C. jejuni in‘ulam’and reported the same pattern which showed resistance of Campylobacter isolates towards erythromycin was higher compared to other antibiotics group [4]. In the present study, it has seen C. jejuni isolates from farms and retail outlets were susceptible towards fluoroquinolone, aminoglycosides and tetracycline groups. Our findings were contradicted with other studies that showed high resistance of C. jejuni towards fluoroquinolone due to the usage in clinical and animal farming [17,18]. Veterinary usageof third generation quinolones since 1980s to combat respiratory infection due to Escherichia coli has induced resistance among C. jejuni isolates [19]. Rodrigo et al. reported 86.6% Campylobacter spp. resistance towards ciprofloxacin [19]. Usage of untreated chicken manure as fertilizer has also been thought to be one of the contributing factor high resistance on quinolone among C. jejuni found in vegetables [4]. Low resistance towards these types of antibiotics in the present study might be due to small scale production of the vegetables.
Clinical and agriculture excessive use of antibiotics has been thought to cause increase resistance among C. jejuni isolates [4,17,18]. Fluoroquinolones-resistant Campylobacter strains have been demonstrated to be biologically stronger than fluoroquinolone-susceptible strains [20,21]. A study discovered inoculation of mixed population consists of fluoroquinolonesusceptible and fluoroquinolone-resistant Campylobacter isolates resulted in highly isolation rate of fluoroquinolone-resistant strain [21]. Antibiotic-free poultry farming discovered no Campylobacter in the chicken, but free-range farming showed no significant difference with regards to multiple antibiotics resistance among Campylobacter isolates compared to conventional farming [21,22].
Besides low fluoroquinolone resistance observed in this study, C. jejuni isolates also demonstrated similar low resistance towards aminoglycosides group. Penicillin G had the highest level of resistance (93%) among C. jejuni isolates. C. jejuni is inherently resistant to many β-lactam drugs making the use of drugs from β-lactamase group suboptimal, especially in serious infections [23].
It is shown that erythromycin-resistance among C. jejuni isolates were increasing, though earlier reports showed the minimal changes after years of testing for antimicrobial resistance in many parts of the world [24–27]. In Thailand, Campylobacter isolates from chicken, pig, dairy and human were found to be resistant to erythromycin and tetracycline at 38.3% and 66.2%, respectively [28].
However, this study suggested that there are differences in farming practices around Terengganu. Most of the vegetables farms are small scale to meet the local demands. This explains the low MAR index among C. jejuni isolates found in vegetables. Low MAR index would indicate that the isolates from vegetables were from low risks of animal waste contamination[5].
Dendogram from RAPD analysis which comprised of 98 C. jejuni isolates from farms and retail outlets were divided into 11 clusters (Table 3). Clusters 3, 6 and 7 comprised of isolates from both farm and retail outlet with various types of salad vegetable. Present study showed that C. jejuni isolates from retails and farm was less correlated to each other (Figure 1 and Table 3). Since majority of the cluster was unrelated, there was a little possibility that the strain from farm's sample had associated C. jejuni isolates at retail outlets. Only 3/11 (27%) of the clusters possibly have correlation. Most of C. jejuni isolates had demonstrated location specific or source specific either in retails or farm. This proved that C. jejuni isolated from raw salad vegetables (ulam) from the retail markets may have been exposed to cross-contamination due to poor handling practices among workers, quality of irrigation water and wash water[7,29,30].
This study showed that small scale vegetables production resulted in lower resistance profile among C. jejuni isolates, though there is an increase pattern of fluoroquinolone and macrolide resistance globally in Campylobacter[12]and growing threat in Southeast Asia region [4]. Cross-contamination is inevitable the major route of microorganism contamination in fresh produce. Ways to decontaminate or prevent growth of microorganisms in fresh produce at retail markets would be useful to reduce the risk of human infection from consumption of raw or minimally cooked fresh produce.
Conflict of interest statement
We declare that we have no conflict of interest.
Acknowledgments
This research was supported by the International Foundation of Sciences, Sweden (Grant No. E/5237-1).
References
[1] Epps SVR, Harvey RB, Hume ME, Phillips TD, Anderson RC, Nisbet DJ. Foodborne Campylobacter: infections, metabolism, pathogenesis and reservoirs. Int J Environ Res Public Health 2013; 10: 6292-304.
[2] Whiley H, van den Akker B, Giglio S, Betham R. The role of environmental reservoirs in human Campylobacteriosis. Int J Environ Res Public Health 2013; 10: 5886-907.
[3] Mansouri-najand L, Saleha AA, Wai SS. Prevalence of multidrug resistance Campylobacter jejuni and Campylobacter coli in chickens slaughtered in selected markets, Malaysia. Trop Biomed 2012; 29: 231-8.
[4] Chai LC, Fatimah AB, Ghazali FM, Lee HY, Tunung R, Shamsinar AT, et al. Biosafety of Campylobacter jejuni from raw vegetables consumed as Ulam with reference to their resistance to antibiotics. Int Food Res J 2008; 15: 125-34.
[5] Krumperman PH. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl Environ Microbiol 1983; 46: 165-70.
[6] Yoo JH, Choi NY, Bae YM, Lee JS, Lee SY. Development of a selective agar plate for the detection of Campylobacter spp. in fresh produce. Int J Food Microbiol 2014; 189: 67-74.
[7] Chai LC, Robin T, Ragavan UM, Gunsalam JW, Bakar FA, Ghazali FM, et al. Thermophilic Campylobacter spp. in salad vegetables in Malaysia. Int J Food Microbiol 2007; 117: 106-11.
[8] Khalid MI, Tang JY, Baharuddin NH, Rahman NS, Rahimi NF, Radu S. Prevalence, antibiogram, and cdt genes of toxigenic Campylobacter jejuni in salad styled vegetables (ulam) at farms and retail outlets in Terengganu. J Food Prot 2015; 78: 65-71.
[9] Chai LC, Ghazali FM, Bakar FA, Lee HY, Suhaimi LR, Talib SA, et al. Occurrence of thermophilic Campylobacter spp. on vegetables farms in Malaysia. J Microbiol Biotechnol 2009; 19: 1415-20. [10] Wang G, Clark CG, Taylor TM, Pucknell C, Barton C, Price L, et al. Colony multiplex PCR assay for identification and differentiation of Campylobacter jejuni, C. coli, C. lari, C. upsaliensis, and C. fetus subsp. fetus. J Clin Microbiol 2002; 40: 4744-7.
[11] Clinical and Laboratory Standards Institute. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria; approved guide. Line M45-A. Wayne: Clinical and Laboratory Standards Institute; 2006.
[12] Ge B, Wang F, Sjӧlund-Karlsson M, McDermott PF. Antimicrobial resistance in Campylobacter: susceptibility testing methods and resistance trends. J Microbiol Methods 2013; 95: 57-67.
[13] Keller J, Perreten V. Genetic diversity in fluoroquinolone and macrolide-resistant Campylobacter coli from pigs. Vet Microbiol 2006; 113: 103-8.
[14] Andersen SR, Saadbye P, Shukri NM, Rosenquist H, Nielsen NL, Boel J. Antimicrobial resistance among Campylobacter jejuni isolated from raw poultry meat at retail level in Denmark. Int J Food Microbiol 2006; 107: 250-5.
[15] Bae W, Kaya KN, Hancock DD, Call DR, Park YH, Besser TE, et al. Prevalence and antimicrobial resistance of thermophilicCampylobacter spp. from cattle farms in Washington State. Appl Environ Microbiol 2005; 71: 169-74.
[16] Ge B, White DG, McDermott PF, Girard W, Zhao S, Hubert S, et al. Antimicrobial-resistant Campylobacter species from retail raw meats. Appl Environ Microbiol 2003; 69: 3005-7.
[17] Wieczorek K, Denis E, Osek J. Comparative analysis of antimicrobial resistance and genetic diversity of Campylobacter from broilers slaughtered in Poland. Int J Food Microbiol 2015; 210: 24-32.
[18] Han K, Jang SS, Choo E, Heu S, Ryu S. Prevalence, genetic diversity and antibiotic resistance patterns of Campylobacter jejuni from retail raw chicken in Korea. Int J Food Microbiol 2007; 114: 50-9.
[19] Rodrigo S, Adesiyun A, Asgarali Z, Swanston W. Antimicrobial resistance of Campylobacter spp. isolated from broilers in small poultry processing operations in Trinidad. Food Control 2007; 18: 321-5.
[20] Luo N, Pereira S, Sahin O, Lin J, Huang S, Michel L, et al. Enhanced in vivo fitness of fluoroquinolone-resistant Campylobacter jejuni in the absence of antibiotic selection pressure. Proc Natl Adad Sci U S A 2005; 102: 541-6.
[21] Price LB, Johnson E, Vailes R, Silbergeld E. Fluoroquinoloneresistant Campylobacter isolates from conventional and antibioticfree chicken products. Environ Health Perspect 2005; 113: 557-60.
[22] Economou V, Zisides N, Gousia P, Petsios S, Sakkas H, Soultos N, et al. Prevalence and antimicrobial profile of Campylobacter isolates from free-range and conventional farming chicken meat during a 6-year survey. Food Control 2015; 56: 161-8.
[23] Siddique FM, Akram M, Noureen N, Noreen Z, Bokhari H. Antibiotic susceptibility profiling and virulence potential of Campylobacter jejuni isolates from different sources in Pakistan. Asian Pac J Trop Med 2015; 8: 197-202.
[24] Engberg J, Aarestrup FM, Taylor DE, Gerner-Smidt P, Nachamkin I. Quinolone and macrolide resistance in Campylobacter jejuni and C. coli: resistance mechanisms and trends in human isolates. Emerg Infect Dis 2001; 7(1): 24-34.
[25] Gibreel A, Taylor DE. Macrolide resistance in Campylobacter jejuni and Campylobacter coli. J Antimicrob Chemother 2006; 58: 243-55.
[26] Navarro F, Mir´o E, Mirelis B, Prats G. Campylobacter spp. antibiotic susceptibility. J Antimicrob Chemother 1993; 32: 906-7.
[27] Sj¨ogren E, Lindblom GB, Kaijser B. Norfloxacin resistance in Campylobacter jejuni and Campylobacter coli isolates from Swedish patients. J Antimicrob Chemother 1997; 40: 257-61.
[28] Padungtod P, Kaneene JB, Hanson R, Morita Y, Boonmar S. Antimicrobial resistance in Campylobacter isolated from food animals and humans in northern Thailand. FEMS Immunol Med Microbiol 2006; 47: 217-25.
[29] Garcia BC, Dimasupil MA, Vital PG, Widmer KW, Rivera WL. Fecal contamination in irrigation water and microbial quality of vegetable primary production in urban farms of Metro Manila, Philippines. J Environ Sci Health B 2015; 50: 734-43.
[30] Munther D, Luo Y, Wu J, Magpantay FM, Srinivasan P. A mathematical model for pathogen cross-contamination dynamics during produce wash. Food Microbiol 2015; 51: 101-7.
Original article http://dx.doi.org/10.1016/j.apjtb.2015.10.001
*Corresponding author:John Yew Huat Tang, Department of Food Industry, Faculty of Bioresources and Food Industry, Universiti Sultan Zainal Abidin, Tembila Campus, 22200 Besut, Terengganu, Malaysia.
 Asian Pacific Journal of Tropical Biomedicine2016年1期
Asian Pacific Journal of Tropical Biomedicine2016年1期
- Asian Pacific Journal of Tropical Biomedicine的其它文章
- The antibacterial activity of selected plants towards resistant bacteria isolated from clinical specimens
- Isolation of aerobic bacteria from ticks infested sheep in Iraq
- Rhinacanthus nasutus leaf improves metabolic abnormalities in high-fat diet-induced obese mice
- Influence of extraction solvents on antioxidant and antimicrobial activities of the pulp and seed of Anisophyllea laurina R. Br. ex Sabine fruits
- Antifungal activity of plant extracts with potential to control plant pathogens in pineapple
- Potent water extracts of Indonesian medicinal plants against PTP1B
