Screening of Preprocessing Method of Biolog for Soil Microbial Community Functional Diversity
Wen DANG, Chunhua GAO, Qiang ZHANG, Jianhua LI, Chaodong LU, Dongsheng JIN,Jinjing LU
1. School of Biological Engineering, Shanxi University, Taiyuan 030006, China;
2. Institute of Agricultural Environment & Resources, Shanxi Academy of Agricultural Sciences, Taiyuan 030006, China
Soil microorganism is the important component of soil ecosystem, which plays an irreplaceable role on animal and plant debris decomposition, nutrient transformation and circulation, soil structure and fertility keeping[1-3].Within a particular geographic area,the composition,structure and function of different microbial community showed a variety of differences in the same environment.Therefore, soil microbial diversity reflects the basic condition of the soil to some extent. Soil microbial diversity mainly includes species, genetic,community structure and functional diversity[4]. Currently, there are many kinds of methods used to explore microbial diversity. According to different objects of diversity research, it is roughly divided into plate dilution culture method, electron microscopy method and staining method that mainly on the microbial diversity;PLFA(phospholipid fatty acid analysis) that mainly on the community structural diversity; DG-GE (denaturing gradient gel electrophoresis), RFLP (restriction fragment length polymorphism),RAPD(random amplified polymorphic DNA marker)that mainly on the genetic diversity; Biolog microplate method and chloroform fumigation extraction method that mainly on the metabolic function diversity and activity and so on[5-7].All of these methods have advantages and disadvantages, but biolog method is favored by many researchers for itssimple operation, high sensitivity,strong resolution, no need to analysis pure microbial, rich data, directly reflect the overall activity of microbial population and clear the characteristic of microbial functional diversity[8]. Biolog microplate assay method was developed by the United States BIOLOG company in 1989, and it was initially applied to pure microbial identification. In 1991, Garland and Mills firstly applied this method to study the soil microbial communities, and indicated that the Biolog carbon source utilization method was an advanced method used to study the soil microbial community structure and diversity in different environment[9]. Due to different kinds of microorganisms and different microbial populations had different carbon sources utilization ability, it results in different carbon source utilization patterns that could characterize the differences of microbial communities[8]. Now Biolog technique was widely used in the analysis of microbial diversity. Jia xia’s research indicated that the culture time had a significant effect on the analysis results of microbial diversity index of Biolog Eco microplate, six kind of carbon sources utilization and PCA[10].There are many preprocessing methods used in the process of Biolog microplate research,but there were no comparative study about the preprocessing method. In this study, the results of three kinds of common preprocessing methods were compared,in order to select an optimal preprocessing method to analyze the soil in same area, and provide reference for future scientific experiments.
Materials and Methods
Soil and experimental treatments
Tested soil was collected from mining reclamation test base of Institute of Agricultural Environment and Resources, Academy of Agricultural Sciences,Luojianggou Village,Changzhi City, Shanxi Province. The corn planting area was treated by different bio-organic fertilizer, and the planting time was May 14thand the sampling time was July 8th in 2013. 0-20 cm of soil sample was collected after the topsoil (1-2 cm) and weeds were removed,then put into aseptic bags and took back to the laboratory.Stored at 4℃. The stones, roots and other debris of the rest soil sample were removed after air-dried, and sieved by 2 mm of sieve.Then put into sealed bags in order to determine the physical and chemical properties. In this test, two treatments were selected to screen preprocessing method. The basic physical and chemical properties of the tested soil was as follows: 0.037%of total nitrogen,0.051%of total phosphorus,1.78%of total potassium,7.71 mg/kg of available phosphorus, 142 mg/kg of rapidly available potassium,32.9 mg/kg of available nitrogen, 3.93 g/kg of organic matter,pH 8.29.
Test principle
Biolog EcoPlate has 96 micropores(8×12),and each 32 micropores was considered as a duplicate. Repeated 3 times. Added water to the control micropore.The other 31 micropores contained a different kind of organic carbon source and the same content of tetrazole purple dye. Under a certain temperature condition, the microorganism solution was inoculated into the micropores and cultured, then the electron generated by microbial respiration transferred and result in tetrazole purple dye turned purple. The shade of color reflected the ability of microorganism using corresponding carbon. OD value of Eco plant was measured at 590 nm wavelength by Biolog microplate reader system.Completed the data acquisition and storage. 31 kinds of single carbon in Eco plate could be divided into six categories: sugars and their derivatives(ten kinds), carboxylic acids (seven kinds), amino acids (six kinds), polymers (four kinds), polyamines (two kinds) and aromatic compounds (two kinds)[11-13].
Biolog Eco analysis
The main steps of the three kinds of different preprocessing methods selected in this study were same. Bacterium solution diluted by 1000 times and added to 96 micropores of Biolog Eco microplates by 8 channel pipetting device. Each micropore added 150 μl.Then the Biolog Eco microplate cultured at suitable temperature, and the data were collected at 24, 48, 72, 96,120, 144 and 168 h respectively by ELx808 microplate reader system. Analyzed the data which the scan wavelength was 590 nm.
Preprocessing methods
Method A:Added fresh soil(equal to 10 g of dired soil)to 90 ml of streilied saline (0.85% Nacl solution) in erlenmeyer flask.Shook for 1 min, then ice bathed for 1 min. Repeated 3 times.Standing for 2 min. Added 5 ml of supernatant to 45 ml of sterile saline,and shook for a moment.Repeated the last step. Then obtained 1 ∶1 000 dilution solution.After that,the solution was inoculated to Biolog Eco microplate and cultured at 30 ℃[14].
Method B:Added fresh soil(equal to 10 g of dired soil) to 90ml of streilied saline (0.85% Nacl solution)in erlenmeyer flask. The rotating speed of shaking table was 250 r/min and shook for 30 min. Standing for 10 min. Then diluted to 1 000 times in turn. After that, the solution was inoculated to Biolog Eco microplate and cultured at 28 ℃[15-16].
Method C: 10 g of fresh soil was added to 100 ml of 0.05 mol/L sterile phosphate buffer (pH 7.0).The rotating speed of the shaking table was 180 r/min and shook for 20 min.Then diluted to 1 000 times.After that,the solution was inoculated to Biolog Eco microplate and cultured at 25 ℃[17].
Added 10 g of fresh soil to the control group in each method.
Data analysis
The average well color development (AWCD value)of Biolog Eco microplate indicates the carbon source metabolism strength of microbial communities,which is an important index of soil microbial activity and function diversity[16].
In this formula,C refers to the absorbance value of each well of 31 well,R refers to the absorbance value of control well.
Average absorbance value of six kinds of carbon sources. AWCD = Σ(C-R)/n,n refers to the total number of carbon sources[18].
Diversity indexes. AWCD value reflects the overall activity of soil microorganism. The diversity index reflects the detailed microbial community species composition and the individuals distribution, and the different aspects of microbial functional diversity[16]. Common indexes include McIntosh index U,Simpson index D, Shannon richness H, Shannon evenness index E and Carbon source utilization richness index S et al[19-21].
(1) McIntosh index is used to measure the homogeneity degree of the community.
(2) Simpson index D=1-ΣPi2reflects the most common species of the community, and was often used to assess the dominance degree of microbial community.
(3) Shannon richness reflects the species richness. H=-ΣPilnP. In this formula,Pirefers to the ratio of the relative absorbance value of i well and the total relative absorbance value of the whole wells.
(4)Shannonevenness E=H/Hmax=H/lnS, H refers to the Shannon index,S refers to the number of well that the color changed.
(5) Carbon source utilization richness index S equal to the total number of the used carbon sources.
All data were proceeded variance analysis and principal component analysis[22-23]by Excel,Sigmaplot,SPSS17.0 software, repeated three times and calculated the average value.
Results and Analysis
Effect of different preprocessing method on AWCD value
The results showed that the change of AWCD value that refers to the use ability of soil microorganism on single carbon source present the similar trend with microbial metabolism,which increased with time extended[24]and changed gently in later stage and finally stabilized (Fig.1). Because after inoculation culturing, the soil microorganism via 24 h of lag phase,gradually adapt to the Biolog microplate environment,and went into the logarithmic growth phase until 96 h, then slowly grew and gradually stabilized. But the rise speed of different preprocessing method was different.The results obviously indicated that method B’s overall variation trend of AWCD value of 2 treatments was superior to A and C’s,which showed that metabolism intensity of microorganism carbon source was maximum under this preprocessing(Fig.1). Variance analysis results indicated that AWCD value of three kinds of preprocessing methods had no significant differences at first and then method B had significant differences with A and C method at 120 h.It indicated that the method B changed fast, even on the final stable stage,method B’s value was significantly higher than other two methods (Table 1). So method B was the better preprocessing method.

Table 1 The change of AWCD value of soil microbial communities under different treatments with the change of culture time
Carbon source utilization ratio of microorganism under different preprocessing method
The results of 1# showed that the carbon source utilization ratio of saccharides and their derivatives was B>A>C,the carboxylic acids was B>A>C,the amino acids was A >B >C, the polymers was B>A>C,the polyamines was B>C>A, the aromatic compounds was A>B>C (Fig.2).Method A’s carbon source utilization ratio of amino acids and aromatic compounds was more than other 2 method’s, but the polyamines was less. Method C’s carbon source utilization ratio was all less. The results of 2# showed the same t trends that method B’s microbial utilization of six carbon sources was higher by variance analysis, and the result was relatively stable.
Changes of soil microbial diversity index under different preprocessing method
According to the change dynamics of AWCD value under different preprocessing method, the resultsshowed that AWCD value appeared inflection point after cultured for 96 h,so the data of 96h were selected to do diversity index analysis[25]. The larger the McIntosh index, the better the community uniformity; the greater the Simpson index,the better the community diversity; the higher the Shannon richness index, the more homogeneous the community and the more uniform individual distribution; the greater the carbon source utilization richness index S, the more available carbon source and the better diversity(Table 2). So the Simpson index,Shannon richness index, Shannon evenness index and Carbon source utilization richness index of 1# sample were B >C >A, except McIntosh index was A>B>C. The variance of each diversity index of method B was smaller.The variance analysis results showed that the Shannon evenness had no significant difference, while the other indexes had significant difference.The results of 2# sample was similar to 1#sample, only the Shannon evenness index of method B was lower than the other two methods, but the difference was not significant,while the other indexes were higher than the other two methods. It indicated that different preprocessing methods had significant impact on the results of soil microbial diversity index. In conclusion, method B’s diversity indexes were higher than method A and C’s, so it was an ideal preprocessing methods.
Principal component analysis method
Principal component analysis(PCA) can better describe the carbon source utilization of different soil microbial communities and their metabolic characteristics, it is one of the widely used multivariate analysis[24]. It expresses the complicated data by two-dimensional or three-dimensional icon, because similar distribution has similar position,the difference of communities can be intuitively seen. Load factor refers to greater absolute value has larger impact on the carbon source. Generally 0.8 of load factor or more had greater impact on soil microorganisms.Referred to Dong liguo’s research,the change rate of the data in 96 h was the maximum[2],so the absorbance value of 96 h were selected to conduct principal component analysis. Extracted 8 principal components from each treatments in microplates.
The contribution rate of the first principal component of 1 # was 23.69% , which the weight was the biggest. The contribution rate of the second principal component was 21.35%, and the other principal component was lesser. So the former two principal components were chose to analyze, PC1 was horizontal axis, and PC2 was vertical axis. The score of two principal components of different treatments plotted as coordinate, then the principal component analysis diagram of soil microbial carbon source utilization treated by different preprocessing methods was obtained. The results showed that different preprocessing had obvious distribution difference in PC axis(Fig.3).In PC1 axis,A and B were distributed in the positive direction. In PC2 axis, B was all distributed in the positive direction of the first and second principal component,A was distributed in the negative direction. The results of 2# sample had the same conclusion, which indicated that method B’s carbon source utilization ability was strong and method B was better than other two methods.The results of 1#sample indicated that the weight of carboxylic acids and amino acids in PC1 axis was larger,which was the major carbon source used for discrimination. Carbon sources related to PC2 was mainly amino acids and polyamines. PC1of 2# sample mainly distinguished by amino acids and polymers, and carboxylic acids was the carbon source relate to PC2,and the relationship was relatively large.
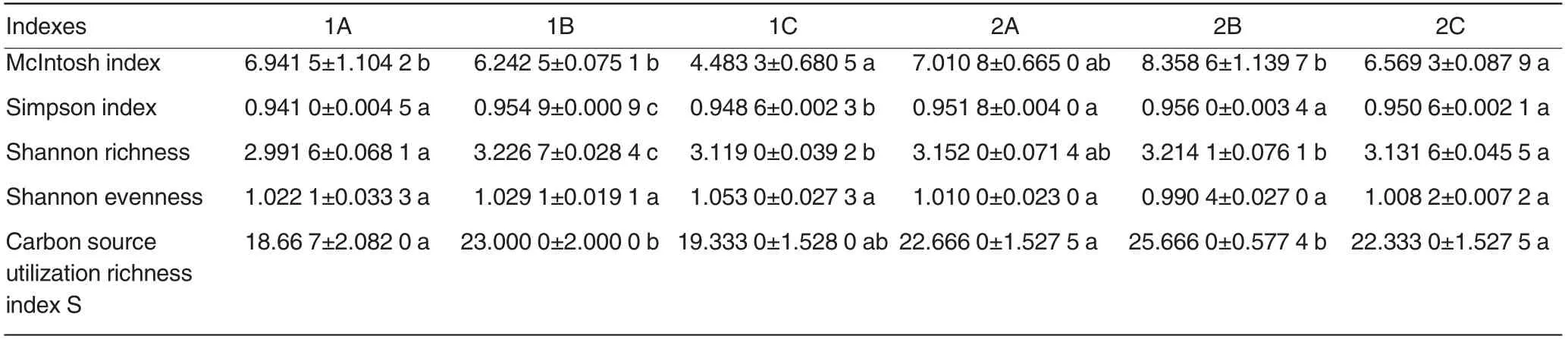
Table 2 The microbial diversity index of different preprocession at 96 h
Conclusions
Compared with the change of AWCD value, six kinds of carbon source utilization ability, five kinds of diversity indexes and principal component analysis, method B’s data were the best, method A and C were relatively poor, and A was bigger than C.Three methods were commonly used in all kinds of literatures,and were verified the correct methods. Compared with the three methods, although the overall trend of the conclusions obtained by these three methods were identical, there were differences be-tween the different methods. In addition, the reagent of method B was simple and easy to obtain, and the operation was easy.Seen from the data,that most of method B’s data were larger than A and C’s, the difference of 70% of diversity indexes reached significance level at 96 h,and the stability of the data was higher.Therefore,method B could be used as the effective method for similar soil analysis with the help of Biolog microplate automatic analysis system.
Discussion
Currently, there are many methods used to explore the microbial diversity. Because of the relatively simple operation of Biolog method, which can directly reflect the overall activity of microbial populations,it is commonly used in the analysis of functional diversity of microbial communities.However,the preprocessing methods were not the same in the process of experiments, and there was no research about the preprocessing method in the literatures. In order to screen the optimal preprocessing method, this study did the research of the effects of different methods on testing results. The results showed that all aspects of method B was superior to A and C.But this preprocessing method also had many problems, such as the culture time was short, the microplate has a certain selectivity that only the microorganisms took advantage of the carbon source could grow. However, it only represented a portion of the microbial community,while the various microorganisms cultured in microplates, the results of color was not simple addition of color caused by each various species, and these synergistic effects or antagonistic effects depended on cell density. There were many factors affected coloration, in addition to the preprocessing,inoculation density and incubation time were also the key factors, and the composition, quantity and activity of microbial populations,the number of interfering substances and other factors in nutrient solution would affect coloration of microplate,which would caused unstable testing results[26].
Therefore, it was necessary to combine with other methods and explore more rational data analysis methods,in order to deeply study the structure and function of microbial community and their relationships, so as to obtain more comprehensive results.
[1]DONG LG (董立国), JIANG Q (蒋齐),CAI JJ(蔡进军).Soil microbial functional diversity analysis of different restoration years of alfalfa field based on Biolog_ECO technology (基于Biolog_ECO 技术不同退耕年限苜蓿地土壤微生物功能多样性分析)[J].Arid Zone Research(干旱区研究),2011,28(4):630-637.
[2]LU SB (鲁顺保),ZHANG YJ (张艳杰),CHEN CR (陈成榕). Analysis of three different types of forest soil bacterial community functional differences based on BIOLOG fingerprint technology(基于BIOLOG 指纹解析三种不同森林类型土壤细菌群落功能差异)[J]. Acta Pedologica Sinica (土壤学报), 2013, 50(3):618-623.
[3]GARLAND J L.Analysis and interpretation of community-level physiological profiles in Microbial Ecology [J]. FEMS Microbiology Ecology, 1997 (24): 289-300.
[4]LIN XG(林先贵),HU JL(胡君利).Scientific connotation and ecosystem services of soil microbial diversity (土壤微生物多样性的科学内涵及其生态服务功能)[J]. Acta Pedologica Sinica (土壤学报),2008,45(5):892-900.
[5]JIAO XD (焦晓丹), WU FZ (吴凤芝).Progress of soil microbial diversity research methods (土壤微生物多样性研究方法的进展)[J]. Chinese Journal of Soil Science (土壤通报), 2004, 35(6):789-792.
[6]ZHANG JE (章家恩),CAI YF (蔡燕飞),GAO AX (高爱霞). Experimental research overview of soil microbial diversity(土壤微生物多样性实验研究方法概述)[J]. Soils (土壤), 2004, 36 (4):346-350.
[7]ZHANG RF(张瑞福),CUI ZL(崔中利),LI SP (李顺鹏).Research progress of soil microbial community structure (土壤微生物群落结构研究进展)[J].Soils(土壤),2004,36(5):346-350.
[8]WANG Q (王强),DAI JL (戴九兰),WU DQ (吴大千).Data analysis of microbial ecological study based on BIOLOG method (微生物生态研究中基于BIOLOG 方法的数据分析)[J].Ecology(生态学),2010,30(3):0817-0823.
[9]GARLAND J L, MILLS A L. Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization[J]. Applied and Environmental Microbiology,1991,57(8):2351-2359.
[10]JIA X (贾夏), DONG SM (董岁明),ZHOU CJ(周春娟).Effect of incubation time of Ecology Eco microplate on the analysis results in the research of microbial ecology(微生物生态研究中Biolog Eco 微平板培养时间对分析结果的影响)[J]. Journal of Basic Science and Engineering(应用基础与工程科学学报),2013,2(1):10-19.
[11]CHEN SN (陈胜男). Effect of azotobacter on soil enzyme activity and bacterial community functional diversity of corn rhizosphere(接种自生固氮菌对玉米根际土壤酶活性和细菌群落功能多样性的影响)[J]. Plant Nutrition and Fertilizer Science(植物营养与肥料学报),2012,18(2):444-450.
[12]KONG B (孔滨), YANG XJ (杨秀娟).The application principle and carbon constitution of Biolog ecological plants(Biolog 生态板的应用原理及碳源构成)[J]. Journal of Green Science and Technology(绿色科技),2011(7):231-234.
[13]CHIOI K H,Dobbs F C.Comparison of two kinds of Biolog microplates (GN and ECO)in their ability to distinguish among aquatic microbial communities[J]. Journal of Microbiological Methods,1999(36):203-213.
[14]HU K(胡可), WANG BL(王利宾). The application of BIOLOG microplate technique on the study of soil micro-ecology (BIOLOG 微平板技术在土壤微生态研究中的应用)[J]. Chinese Journal of Soil Science (土壤通报), 2007,38(4):819-821.
[15]LIU HM(刘红梅), LAI X(赖欣), SONG XL (宋晓龙),et al.Effect of transgenic cotton seeds (Bt-CpTI ) on functional diversity of soil microbial communities(转双基因Bt-CpTI 棉种省略际土壤微生物群落功能多样性的影响)[J]. Chinese Agricultural Science Bulletin(中国农学通报),2012,28(36):231-236.
[16]HUANG HY (黄红艳).The research of microbial diversity and improvement effects of secondary salinization soil(次生盐渍化土壤的微生物多样性及微生物改良效应研究)[D]. Shanghai:Shanghai Jiaotong University (上海:上海交通大学),2012:30-38.
[17]HUA JF(华剑锋), YANG YR(杨亦如),XU JH(徐建华),et al.Effect of the application of river sediment on soil microbial community functional diversity and the wheat growth(河流底泥施用对土壤微生物群落功能多样性和小麦生长的影响)[J]. Journal of Ecology and Rural Environment(生态与农村环境学报),2012,28(5):526-531.
[18]JACKSON L E, CALDERON F J,Steenwerth K L, et al. Responses of soil microbial processes and community structure to tillage events and implications for soil quality[J].Geoderma,2003(114):305-317.
[19]LUO XQ (罗希茜),HAO XH (郝晓晖),CHEN T (陈涛), et al. Effect of long term different fertilization on soil microbial community functional diversity in paddy (长期不同施肥对稻田土壤微生物群落功能多样性的影响)[J]. Acta Ecologica Sinica (生态学报),2009,29(2):740-748.
[20]SHI P(时鹏),GAO Q(高强),WANG SP(王淑平), et al. Effect of corn continuous cropping and fertilization on soil microbial community functional diversity (玉米连作及其施肥对土壤微生物群落功能多样性的影响)[J]. Acta Ecologica Sinica(生态学报),2010,30(22):6173-6182.
[21]YANG YH(杨永华),HUA XM(华晓梅).Effect of pesticide contamination on soil microbial community functional diversity (农药污染对土壤微生物群落功能多样性的影响)[J]. Journal of Microbiology(微生物学杂志), 2000(2): 23-25.
[22]BAI HQ (白慧强).Principal component analysis in SPSS application——Take herbaceous communities of Wenyu River riparian understory for example(主成分分析法在SPSS 中的应用——以文峪河河岸带林下草本群落为例)[J].Sci-Tech Information Development &Economy(科技情报开发与经济),2009,19(9):173-176.
[23]JI ZM(吉祝美),FANG L(方里),ZHANG J(张俊),et al.The application of principal component analysis in SPSS software and river water quality assessment(主成分分析法在SPSS 软件中的操作及在河流水质评价中的应用)[J].Environmental Technology(环保科技),2012,18(4):38-43.
[24]SHI XH(石贤辉).Soil microbial activity and community functional diversity of eucalyptus artificial forest (桉树人工林土壤微生物活性与群落功能多样性)[D].Guangxi:Guangxi University(广西:广西大学),2012:29-38.
[25]YAO Q (姚钦). Research of microbial diversity of black soil by means of crop rotation(黑土区轮作方式下土壤微生物多样性研究)[D]. Harbin: Northeast Forestry University(哈尔滨:东北林业大学),2012:18-19.
[26]ZHENG H(郑华),OUYANG ZY(欧阳志云),FANG ZG(方治国),et al.Application of BIOLOG on soil microbial community function diversity (BIOLOG 在土壤微生物群落功能多样性研究中的应用)[J].Acta Pedologica Sinica (土壤学报),2004,41(3):456-461.
 Agricultural Science & Technology2015年10期
Agricultural Science & Technology2015年10期
- Agricultural Science & Technology的其它文章
- Effects of Specific Gravity-based Seed Grading on Seed Germination,Seedling Emergence and Grain Yield of Hybrid Rice
- Effects of NaCl Stress on Seed Germination of Four Canavium album Raeuseh Cultivars
- Application Effects of Ultra-fine Powder Shaped Maize Seed Coating Agent in Spring Sowing areas in northeast China
- Breeding and Application of a Japonic Rice Cytoplasmic Male Sterility Line,E-Jing A
- Effect of Low Temperature and Sparse Light Conditions on Cold Tolerance of Different Rice Lines at Seedling Stage
- Molecular Marker Assisted Selection for Fusarium Wilt Resistance Breeding in Watermelon(Citrullus lanatus)
