Molecular Marker Assisted Selection for Fusarium Wilt Resistance Breeding in Watermelon(Citrullus lanatus)
Jiming WANG, Jianli SHANG, Yaling DONG, Na LI, Yongyang XU, Shuangwu MA
Zhengzhou Fruit Research Institute, Chinese Academy of Agriculture Sciences, Zhengzhou 450009, China
Fusarium oxysporum f. sp. N iveum is a devastating disease in watermelon production. In recent years, watermelon Fusarium wilt disease has occurred all over the country. The rate of diseased plant is generally about 10%-20% and it will be up to 80%-90% when the disease happens seriously, and even lead to no harvest[1].Since the 1980s,a number of watermelon varieties resistant to Fusarium wilt have been bred and applied in actual production. The resistant varieties play a very positive role in prevention of Fusarium wilt and are important materials in modern Fusarium wilt-resistant breeding in watermelon. Currently, artificial inoculation technique is mainly used to select resistant plants in watermelon Fusarium wilt-resistant breeding. However, the method is vulnerable to environment and inoculation conditions with some disadvantages including time waste,poor accuracy, low efficiency and difficulties in distinguishing between resistant homozygote and heterozygote.The development of marker“4451_fon” closely linked with resistance gene of Fusarium wilt race 1 provided a tool for molecular marker assisted selection(MAS)in watermelon Fusarium wilt-resistant breeding[2].In this study,marker “4451_fon”was used to analyze resistant genotype and screen resistant plants from part monophyletic group of the innovative Fusarium wilt-resistant germplasm in order to provide a basis for large-scale application of MAS in watermelon Fusarium wilt-resistant breeding.
Materials and Methods
Materials
The tested watermelon materials were monophyletic groups selected from the innovative germplasm with Fusarium wilt resistance includingdiploid monophyletic B61-B68 (sticky seed),tetraploid monophyletic D5-D13 and D16-D17,resistant parent"Sugarlee", susceptible parent "14CB11" and the 22 F1 hybrid generations of the two parents.
Methods
Thirty plants were chosen from each watermelon material,which were inoculated with Fusarium wilt and then the resistance was identified at seedling stage. In addition, resistant genotypes were detected at molecular level.
(1) Artificial inoculation identification of Fusarium wilt resistance at seedling stage. Race 1 of Fusarium oxysporum f.sp.niveurm was used as the pathogenic bacteria of Fusarium wilt to infect the watermelon materials.The inoculation and identification were referred to the method of inoculation with the mixture of soil and diseased wheat grain reported by Ji[3]. Sterilized soil and diseased wheat grains were mixed with a ratio of 0.5%, then which were put into nutritional bowls. The watermelon seeds to be identified were sowed after the bowls were watering and put at 28 ℃for culture. The disease level was investigated after four weeks. Percentage of diseased plants [(Total number of seedlings -Number of live seedlings)/total number of seedlings × 100%]was counted.The grading standard of resistance was referred to Zhang[4]. Highly resistance (HR): percentage of diseased plants was 0 -20% ; resistance (R):percentage of diseased plants was 21%-40%;moderate resistance(MR):percentage of diseased plants was 41%-60%; susceptible (S): percentage of diseased plants was 61%-80%;high-susceptible (HS): percentage of diseased plants was 81% -100% ,among which, above the moderate resistance (MR)defined as resistant and below the susceptible (S) defined as susceptible.
(2)DNA extraction.CTAB method[5]was adopted and slightly improved for DNA extraction of watermelon. 0.2-0.5 g young leaves were taken, grinded in liquid nitrogen and transferred into a 2 ml centrifuge tube.1 ml CTAB extracting solution was added to the centrifuge tube, and then which was put at 65 ℃water bath for 1 h. Mixed solution of chloroform: isopropanol(24∶1)of the same solution was added to the centrifuge tube and mixed thoroughly. The centrifuge tube was centrifuged at 10 000 rpm for 5 min. 75%ethyl alcohol was used to wash the DNA after the liquid supernatant was discarded.200 μl TE was added to the centrifuge tube to dissolve the DNA after which was air dried. Micro-Ultraviolet Spectrophotometer Nanodrop 2 000 was used for DNA analysis and the concentration was adjusted to 60 ng/μl.
(3) dCAPS reaction and analysis.Primers of 4451_fon closely linked with resistance gene of Fusarium wilt race 1 reported by Zhang et al.[2]were chosen for amplification. The forward sequence (5’→3’)was “AAATGGGTACTGGTGGTCGCTC” and the reverse sequence(5’→3’)was “TTCCTCTTCTTCTGTTTCTCCACAA”,which were synthesized by Boshang Biological Technology co.,LTD(Shanghai).
Amplification reaction system was 25 μl including 12.5 μl 2×Taq Master Mix,2 μl template DNA,2 μl 10 μmol/μl mixed primers and 8.5 μl ddH2O. Amplification reaction process was as follows: initial denaturation at 94 ℃for 5 min;94 ℃20 s,55 ℃20 s,72 ℃30 s for 34 cycle;extend at 72 ℃for 5 min;stored at 4 ℃. The 2×Taq Master Mix was bought from Beijing Baitaike Biotechnology Co.,LTD.
The product of dCAPS was undergone enzyme cleavage wirh Taq I.The recognition site for enzyme cleavage was T’CGA. The enzymatic system was 15 μl including 1.5 μl 10×Buffer, 1.0 μl restriction enzyme, 5 μl PCR product and 7.5 μl H2O.The process of enzyme cleavage was as follows:enzyme cleavage at 65 ℃for 16 h, degeneration at 80 ℃for 20 min and stored at 4 ℃.The restriction enzyme was bought from Fermentas Company.
8.0% polyacrylamide gel electrophoresis was used for electrophoresis of the products from amplification/enzyme cleavage and the electrophoresis was performed at 280V constant voltage for 50 min. After that,the products were stained with silver staining and photographed for analysis. The genotype was resistant and homozygous when 233 bp band was occurred after the products of enzyme cleavage underwent electrophoresis. The genotype was susceptible and homozygous when 244 bp band was occurred. The genotype was resistant and heterozygous when bands of 233 bp and 244 bp were occurred and the plant was heterozygote.
Results and analysis
Artificial inoculation identification for Fusarium wilt resistance
Identification of Fusarium wilt resistance by artificial inoculation was conducted on twenty two diploid and tetraploid monophyletic parents and their hybrid generations with the resistant parent “Sugarlee”and susceptible parent “14CB11” as references.The results showed that all the diploid monophyletic groups (B61-68), most tetraploid monophyletic groups (D5-D13)and the resistant parent “Sugarlee” were expressed as highly resistance(HR), which were resistant materials.Tetraploid monophyletic grousp of D16,D17 and “14CB11”were susceptible, which were susceptible materials. The F1 were shown as moderate resistance (MR), which were resistant material as well(Table 1).
Analysis on the dCAPS marker of Fusarium wilt-resistant gene of diploid watermelon
The dCAPS marker 4451_fon was used for the analysis on individual plants of eight monophyletic groups(B61 -B68) of diploid watermelon.Thirty plants in each monophyletic group were selected and a total of 240 plants were analyzed. The results showed that after amplification and enzyme cleavage of the DNA extracted from individual plants of the diploid watermelon,233 bp bands were stably occurred, which was consistent with that of the resistant parent “Sugarlee”.The result indicated that the eight resistant lines were materials with resistant genotype for Fusarium wilt race,which were resistant monophyletic groups (Fig.1). The analysis results were accorded with the results of artificial inoculation identification(Table 1).
Analysis on the dCAPS marker of Fusarium wilt-resistant gene of tetraploid watermelon
The dCAPS marker 4451_fonwas used for the analysis on individual plant of nine tetraploid monophyletic groups,D5-D13 and D16-D17. Thirty plants in each monophyletic group were selected and a total of 330 plants were analyzed. The results showed that after amplification and enzyme cleavage of the DNA extracted from individual plants of the tetrappoid monophyletic groups, D5-D13, 233bp bands were stably occurred, which was consistent with that of the resistant parent “Sugarlee”.The result indicated that the nine tetraploid monophyletic groups were materials with resistant genotype for Fusarium wilt race, which were resistant materials(Fig.2). Only 244bp bands were occurred after amplification and enzyme cleavage of the DNA extracted from individual plants of the tetrappoid monophyletic groups, D16 and D17,which were accorded with that of the susceptible parent “14CB11”.The result indicated that the two tetraploid monophyletic groups were materials with susceptible genotype for Fusarium wilt race and belonged to susceptible monophyletic group (Fig.2). The above results were accorded with the results of artificial inoculation identification(Table 1).
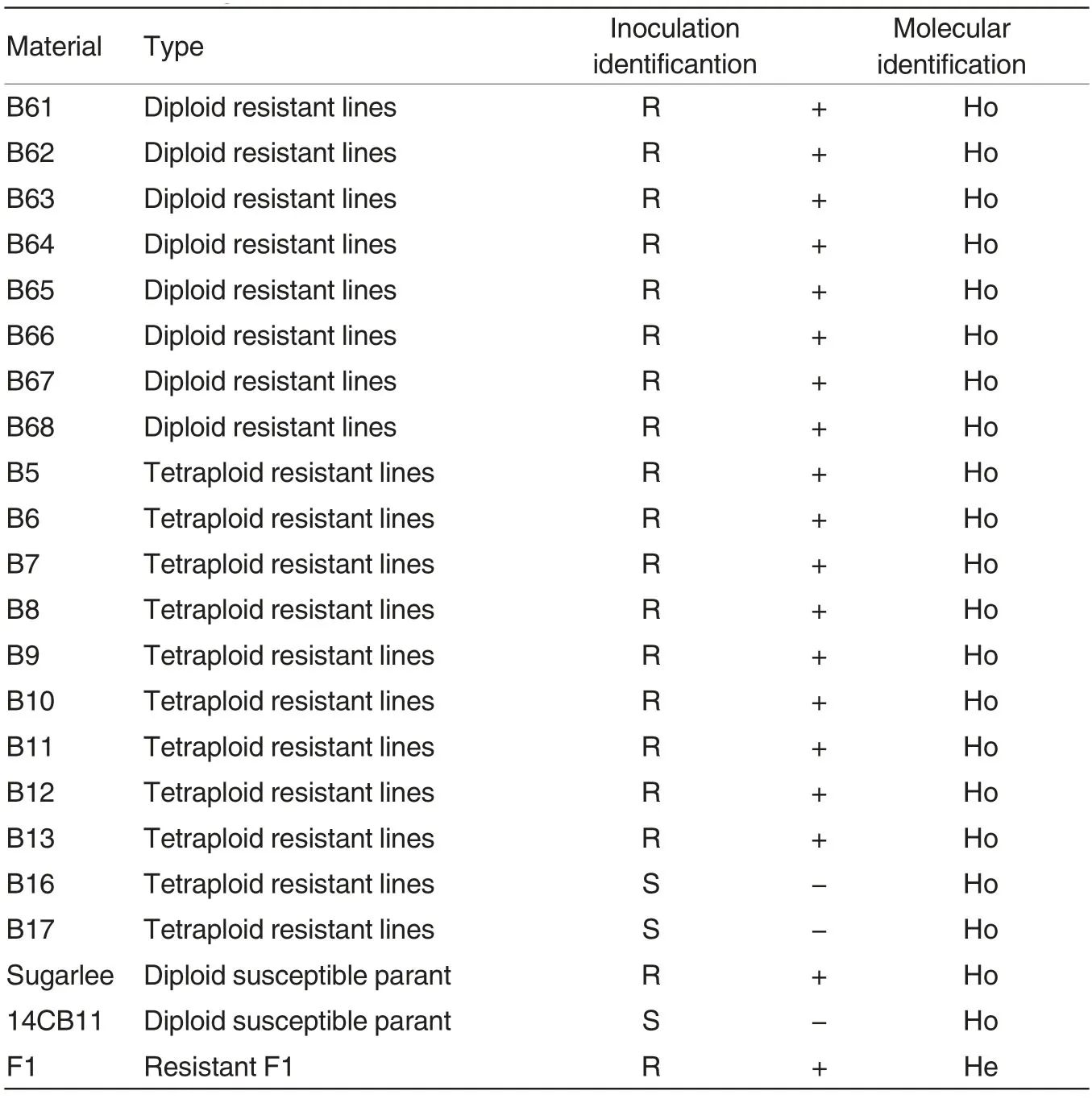
Table 1 Artificial inoculation identification and molecular identification of watermelon selecting lines with resistance to Fusarium wilt
Analysis on homozygous/heterozygous genotypes with Fusarium wilt resistance
The dCAPS marker 4451_fon is a co-dominant marker, which can be used for the distinguishing of homozygous and heterozygous genotypes.According to the analysis on the detection results of diploid and tetraploid monophyletic groups, 233bp bands could stably occurred after the amplification products from the DNA of resistant monophyletic groups were undergone enzyme cleavage while the 244 bp bands occurred before enzyme cleavage were disappeared. The phenomenon indicated that the resistance gene of diploid and tetraploid resistant lines had been become homozygote after years of selfing, and the resistance gene in its inbred progenies would not segregate. Resistance selection was needless in subsequent selection of agronomic traits by selfcrossed purification. 244bp and 233bp bands (Fig.3) occurred from individual plants of the F1of resistant parent“Sugarlee” and susceptible parent“Black Diamond”. The F1expressed as heterozygous resistant genotype.Resistance identification and selection needs to be continued in its progenies if it is used for Fusarium wilt-resistant breeding.
Discussions
Selection is one of the important parts in crop breeding.For watermelon resistant breeding, resistant materials are hard to be selected according to the comparison of phenotypic traits during the breeding process. Disease nursery planting and observation, and artificial inoculation identification were usually used for selection, which are easily affected by pathogenic environment with bad selection accuracy and low efficiency. Molecular marker-assisted selection is a selection technol-ogy based on the differential analysis of DNA sequence, which can reveal differences among breeding materials from molecular genetics.The selection result is reliable because the method is not easily affected by environment.Selection can be conducted accurately in early-generation that breeding process will be accelerated and breeding efficiency will be improved with the utilization of gene pyramiding[6].
Molecular marker-assisted selection was conducted on the monophyletic groups with the published dCAPS marker “4451_fon”which was closely linked with resistance gene of Fusarium wilt race 1. The results showed that the molecular identification results were completely consistent with that of artificial inoculation identification. Genotypes of the resistant monophyletic groups can be easily distinguished because dCAPS marker 4451-fon is a co-dominant marker.Resistant selection is needless in progenies according to the reservation of individual plants with homozygous resistance gene that the breeding cycle will be extremely shortened. However,artificial inoculation identification cannot distinguish resistant heterozygote and homozygote, which cannot provide selection information in this aspect.Therefore,dCAPS marker 4451-fon can be used as an important tool for the molecular marker assisted selection of watermelon with resistance to Fusarium wilt.
[1]ZHANG Y (张 屹),WEI L (魏 林),XU Y(许 勇), et al. Advances in watermelon Fusarium wilt (西瓜枯萎病的研究进展)[J].Hunan Agricultural Sciences(湖南农业科学),2013,(05):67-70.
[2]ZHANG Y(张 屹),ZHANG HY(张海英),GUO SG (郭绍贵),et al.Developments of molecular markers tightly linked to Fon-1 fro resistance to Fusarium oxysporum f. sp. niveum Race 1 in Watermelon(西瓜枯萎病菌生理小种1 抗性基因连锁标记开发)[J]. Scientia Agricultura Sinica (中国农业科学), 2013, 46(10):2085-2093.
[3]JI JB (吉加兵).Identification method for Fusarium wilt resistance at seedling stage of watermelon(西瓜枯萎病苗期抗性鉴定方法的探讨)[J].China Cucurbits and Vegetables (中国瓜菜), 1992(1):35-39
[4]ZHANG XW(张学炜),HUANG XS(黄学森),GU QS(古勤生),et al.A preliminary report of resistance identification for Fusarium wilt of watermelon varieties(西瓜品种对枯萎病抗性鉴定研究初报)[J].China Cucurbits and Vegetables(中国瓜菜),1991(1):22-24
[5]MURRAY MG,THOMPSON WF.Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research[J]. 1980,8(19):4321-4325
[6]HE GM(何光明),SUN CQ(孙传清), FU YC (付永彩), et al. Clustering analysis on rice senility-resistant ipt gene and bacterial blight-resistant gene Xa23 (水稻抗衰老ipt 基因与抗白叶枯病基因Xa23 的聚合研究)[J].Journal of Genetics and Genomics(遗传学报),2004,31(8):836-841.
 Agricultural Science & Technology2015年10期
Agricultural Science & Technology2015年10期
- Agricultural Science & Technology的其它文章
- Research Advances in Gene Regulation and Genetic Improvement of Fish Feeding
- Instrucions for Authors
- Cambridge Scientific Abstracts (CSA)
- Overview of Pharmaceutical Research on the Poria with Hostwood of Traditional Chinese Medicine
- Study on Relative Soil and Water Conservation Benefits of Ridge Tillage in Different Terrain Conditions in the Black Soil Area of Northeast China
- Applicability of Biodegradable Mulch in Maize Production in Northeastern Yunnan
