miRNA在植物种子发育过程中的作用
龚淑敏,丁艳菲,朱诚
中国计量学院生命科学学院,浙江省海洋食品品质及危害物控制技术重点实验室,杭州 310018
miRNA在植物种子发育过程中的作用
龚淑敏,丁艳菲,朱诚
中国计量学院生命科学学院,浙江省海洋食品品质及危害物控制技术重点实验室,杭州 310018
MicroRNA(miRNA)是一类小分子非编码RNA,通过降解靶基因途径在转录后水平调控基因表达,参与植物生长、发育以及逆境胁迫应答等多种细胞代谢活动。种子是植物生长的基础要素,是农业生产的重要资料。与种子发育相关的miRNA已在多种植物中得到鉴定。文章综述了参与植物种子发育过程的miRNA及其在种子发育中的具体调控机制,旨在为利用miRNA提高种子遗传特性提供研究思路。
种子;miRNA;发育;调控机制;植物
MicroRNA(miRNA)是一类内源性的、长度为20~25nt、单链的非编码调控小分子RNA,在真核生物的基因转录和转录后调控中发挥重要作用[1,2]。在秀丽隐杆线虫(Caenorhabditis elegans)中首次发现miRNA后[3],几乎所有真核生物中都被证明存在miRNA。2002年,植物miRNA在拟南芥(Arabidopsis thaliana)中被发现[4,5],此后植物miRNA的研究开始得到研究人员的普遍重视。研究表明,miRNA在植物中广泛存在,并且参与植物生长发育、细胞分化、生物及非生物胁迫应答等过程[6~8]。
种子是植物生长的基础要素,是农业生产中重要的生产资料,对种子中miRNA的研究将对调控种子发育成熟及胁迫响应具有重要意义。本文总结了近年来国内外研究发现的植物种子中的miRNA,探讨了miRNA如何通过调节激素信号转导、抗氧化作用、糖转化、细胞生长等途径,参与种子发育的调控。
1 miRNA在植物中的成熟及作用机制
植物中miRNA在RNA聚合酶Ⅱ作用下,转录成几百个核苷酸的原初miRNA转录子(primary miRNA,pri-miRNA)[9]。接着,在RNA聚合酶Ⅲ的介导下,DCL1-HYL1复合体在pri-miRNA的颈端近基部剪切形成3¢末端有二核苷酸突出、5¢末端有磷酸基、长度约为70~300nt具有茎环结构的miRNA前体(miRNA precursor,pre-miRNA)。在DCLl酶的介导下[4,10,11],pre-miRNA被剪切成miRNA/miRNA*双体[12],miRNA/miRNA*双链的 3¢端被 HUA ENHANCER1(HEN1)甲基转移酶甲基化[13]。大多数甲基化的miRNA/miRNA*在Exportin25的同源物HASTY(HST)转运蛋白的帮助下从细胞核转移到细胞质中[14,15]。成熟的miRNA结合Argonaute(AGO)形成 RNA诱导的沉默复合体(RNA-induced silencing complex,RISC)[16~18],从而实现对靶基因的调控,miRNA*则被降解。
miRNA的主要功能是进行转录后调控。植物中miRNA的作用机制包括mRNA剪切[5]和蛋白翻译抑制[19],通过这两种沉默机制调控目的基因的表达。如果miRNA与mRNA完全互补,miRNA就引导mRNA特异性切割;如果两者没有足够的互补性,则引起翻译抑制[9,20]。大多数植物miRNA与靶序列的开放阅读框(ORF)完全匹配,因而,植物中miRNA主要以降解靶 mRNA途径下调靶基因的表达[7,20]。
2 参与种子发育的miRNA
miRNA参与植物生长发育过程,并对逆境胁迫应答具有调控作用[20~22]。植物中miRNA的发现,为种子生物学提供了新的研究方向。与种子发育相关的miRNA已经在拟南芥、水稻(Oryza sativa L.)、玉米(Zea mays L.)、油菜(Brassica napus)中得到鉴定。Körbes等[23]利用高通量测序技术鉴定了甘蓝型油菜发育种子中的miRNA,共得到35个miRNA家族。Zhao等[24]检测高含油量和低含油量甘蓝型油菜种子中miRNA表达模式的差异,共鉴定到50个保守的miRNA、37个未报道的保守miRNA及9个新的miRNA,其中miR156、miR167、miR390、miR2111、miR6028和miR6029在两种种子中差异表达;猜测miR156、miR167和miR6029通过调控靶基因SPL、ARF和 VIP1转录因子及 miR390通过靶基因ta-siRNA介导的生长素信号通路调控早期胚胎发育,从而影响了甘蓝型油菜种子的含油量。Xue等[25]利用大规模平行测序技术(Massively parallel signature sequencing,MPSS)和生物信息学技术鉴定水稻种子中的miRNA及靶基因,发现miR167、miR397、miR398、miR408、miR528、miR1866-3p和miRc11在水稻的种子中优先表达,暗示这些miRNA在水稻种子发育中的重要功能。Kang等[26]通过深度测序和miRNA微阵列芯片结合的方法研究miRNA在玉米发育种子中的作用机制,发现125个保守miRNA和54个新的miRNA;其中miR166、miR167和miR319表达量很高。Han等[27]鉴定了小麦(Triticum aestivum L.)幼苗、旗叶、发育种子中的miRNA,发现4个已知miRNA、22个未知miRNA在种子中高表达,miR164和miR160在种子发育过程中表达量增加,而miR169的表达量降低,种子特异miRNA的预测靶基因包括转录因子、酶、核/染色质装配因子、核糖体再生因子、细胞成分等。
植物中miRNA的靶基因多为转录因子,miRNA通过调控这些关键因子在植物生长过程中发挥重要作用[28~30]。种子的发育主要包括胚和胚乳的发育,及其种子贮藏物质的积累[31]。miRNA通过信号转导(ABA、生长素、油菜素甾醇等)、淀粉合成、抗氧化作用、糖转化、细胞生长等作用途径调控种子的发育过程。表1和图1分别列举了近年来发现的种子发育相关的miRNA种类及它们参与种子发育的具体作用途径。
3 miRNA参与种子发育的具体途径
3.1 脱落酸信号转导
脱落酸(ABA)是植物种子休眠、成熟的重要调节因子,许多ABA信号转导蛋白都参与了种子发育过程[32,33]。ABA含量对水稻种子细胞分裂、淀粉沉积、灌浆率等具有正向调控作用[34],在水稻灌浆期对水稻进行ABA处理可以加快细胞分裂、增加细胞数量、提高灌浆率,从而增加劣势籽粒的重量[35]。但ABA含量升高,种子发育会被抑制。miR159的靶基因是转录因子MYB33和MYB101,是ABA信号的正向调控因子。过表达miR159可以抑制MYB33和MYB101的转录水平,使种子对ABA信号不敏感[36]。研究发现,miR159在水稻劣质籽粒中的表达量高于优势籽粒,暗示miR159通过调控种子对ABA信号的转导,影响了种子的灌浆[37]。miR169通过靶基因核转录因子NF-YA调控拟南芥种子对ABA信号的转导,过表达NF-YA可以降低种子对ABA信号的敏感性[32,38,39]。
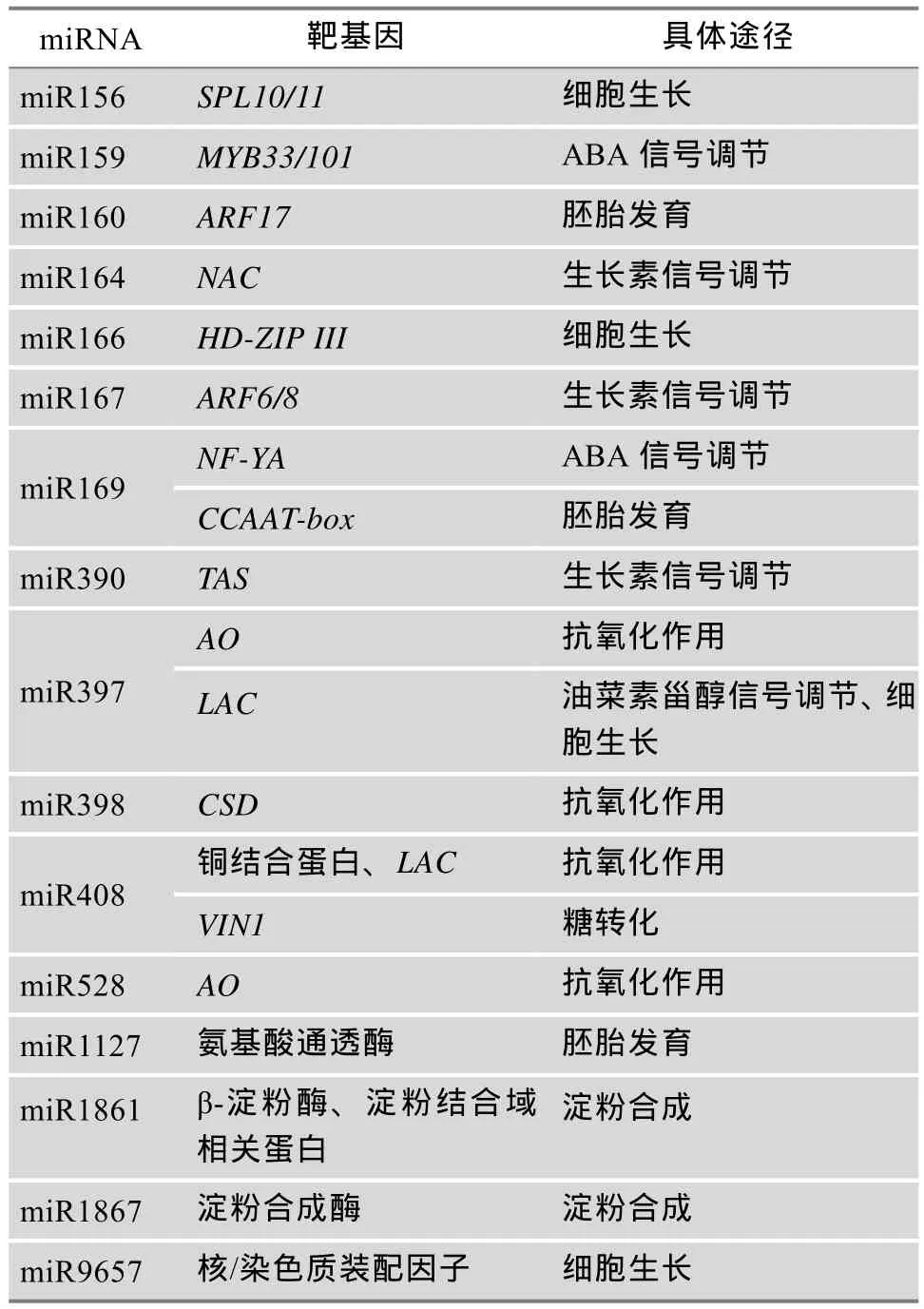
表1 参与种子发育miRNA的种类和具体作用途径
3.2 生长素信号转导
生长素是种子发育过程中的关键因素,参与了种子形态形成、细胞分裂和细胞膨胀等过程[40]。生长素响应因子ARF(AUXIN RESPONSIVE FACTOR)是生长素转导通路上的重要组分,通过结合生长素诱导基因的上游特异顺式作用元件,参与生长素信号转导途径。Kang等[26]利用高通量测序和表达谱芯片技术,发现miR167在发育的玉米种子中表达量很高。miR167的靶基因是ARF6和ARF8,已在拟南芥和水稻中得到鉴定[41,42],因此推测miR167通过调控生长素的感应和转导在种子发育过程中发挥重要作用。
miR390通过调控靶基因反干扰小RNA(ta-siRNA, TAS)参与种子的发育过程。Zhao等[24]通过对油菜早期胚胎中小RNA的表达分析,发现miR390通过反式作用反干扰小RNA介导的生长素信号转导途径,参与了油菜早期胚胎的发育过程。研究发现,miR164介导的生长素调控机制能够调控种子灌浆[43]。miR164是生长素相关的miRNA,在水稻优质穗和劣质穗中都存在,并且在优质穗中表达量更高[37]。miR164通过靶向NAC转录因子,调节植物内部生长素平衡。NAC基因在植物发育中具有生长素信号转导、防御等功能,通过调节衰老增加小麦种子中蛋白、锌、铁等的含量[44]。Han等[27]在发育的小麦种子中发现miR164的表达量呈现上升趋势,而干扰miR164调控则会导致胚胎发育异常,说明miR164通过其靶基因NAC对生长素信号转导的调控,参与了种子发育过程。

图1 典型的miRNA在种子发育过程中的作用途径NAC:NAC转录因子;ARF:生长素响应因子(Auxin response factor);TAS:反干扰小RNA(ta-siRNA);CSD:铜超氧化物歧化酶(Copper superoxide dismutase);AO:L-抗坏血酸氧化酶;LAC:漆酶(Laccase);BR:油菜素甾醇(Brassinosteroid)。
3.3 油菜素甾醇信号转导
油菜素甾醇(Brassinosteroid,BR)是一类在植物生长发育中起重要作用的植物激素。研究发现,miR397通过靶向漆酶(Laccase,LAC)调控水稻种子的大小和产量。miR397的靶基因为LAC,其产物是类漆酶蛋白,参与油菜素甾醇信号转导。过表达miR397能够增加种子大小、促进圆锥花序分支和增加主穗粒数,从而增加产量[45]。miR397作为一类在不同作物中高度保守的miRNA,其对油菜素甾醇信号转导的调控作用有待在其他作物中进一步验证。
3.4 抗氧化作用
在植物中,铜是进行光合作用的必需微量元素,铜的稳态在种子发育中发挥着重要作用。而铜/锌超氧化物歧化酶是叶绿体中异常丰富的铜相关蛋白,能够特异地清除超氧阴离子自由基,维持氧自由基的平衡[46]。miR408的保守靶基因是铜离子结合蛋白及漆酶[47]。漆酶是一种结合多个铜离子的蛋白质,属于铜蓝氧化酶,具有催化氧化的作用。由此可见,植物miR408通过调节铜离子的浓度和催化氧化作用维持体内的稳态和维持氧自由基的平衡,为种子的发育维持一个稳定的内环境。
miR397、miR398和miR528在水稻种子胚中表达量很高,它们的靶基因编码了铜结合蛋白[43,48,49]。蔗糖介导的miR398通过降低两个铜超氧化物歧化酶(Copper superoxide dismutase,CSD)基因的表达,从而参与铜的稳态调控[50]。miR398的这一调控作用在水稻、拟南芥和大豆中是保守的。水稻中miR397和miR528靶向L-抗坏血酸氧化酶(AO),AO通过调控信号转导来调节植物的氧化还原状态[51]。AO能够促进抗坏血酸(AA)的氧化从而获得脱氢抗坏血酸(DHA),DHA的积累可以阻止细胞分裂[52]。而种子在早期发育阶段需要具备很高的细胞分裂能力,过表达miR397和miR528能够使AO和DHA维持在很低的水平,促进细胞分裂。以上研究结果揭示了miRNA通过抗氧化途径调控种子的发育过程。
3.5 糖转化
miR408是一种广泛存在于拟南芥、水稻、玉米、甘蔗等植物中的高度保守的miRNA。最近的研究表明,水稻miR408通过介导VIN1基因表达对种子发育具有重要的影响。VIN1是一种液泡转化酶,其主要功能是将蔗糖水解成葡萄糖和果糖。通过对水稻miR408转基因株系种子胚中糖含量的测定,发现miR408表达水平与种子蔗糖含量呈正相关性,而其靶基因表达则与蔗糖含量呈负相关性[46]。由此可知,miR408通过糖转化途径参与了水稻种子的发育过程。
3.6 细胞生长
研究发现,miR166是一类在种子发育过程中调控细胞生长的miRNA。Zhang等[53~55]研究发现,玉米中 miR166的靶基因是 HD-ZIP III(Class-III homeodomain-leucine zipper)基因,HD-ZIP III能够调控种子成熟及侧根生长。miR156是另一类调控细胞生长的miRNA,它靶向SPL10和SPL11基因,能够导致不正常的细胞分裂[56],控制种子的发育[57,58]。植物漆酶能够聚合木质素单体,形成木质素。在拟南芥中发现,miR397b通过靶基因LAC4调控木质素的合成和种子产量[59]。过表达miR397b减少了木质素的沉积,纤维束二层壁厚减少,同时使植物发育出两个以上的花芽序,从而得到更多的种子数量。这与水稻中miR397a和miR397b过表达的结果相似
[43]。在开花植物中,miR397通过木质素的调节,影响种子产量的调控机制可能普遍存在。在小麦发育种子中,miR9657a的靶基因是核/染色质装配因子,是细胞增殖中DNA复制所必需的。miR9661靶向F-box蛋白结构域。研究发现,F-box蛋白编码基因在种子发育的不同阶段表现出不同的转录水平,暗示F-box蛋白可能参与了种子发育过程[60,61]。
3.7 淀粉合成
谷类作物如水稻、玉米、小麦等种子主要由淀粉组成,种子的发育主要是淀粉沉积的过程。淀粉合成酶是淀粉合成的关键酶。水稻中淀粉合成酶的活性与水稻灌浆率正相关。降解组测序和5¢-RACE实验发现,miR1867靶向淀粉合成酶蛋白,参与淀粉合成通路,从而调控水稻灌浆率[62]。miR1861被证明调控β-淀粉酶和淀粉结合域相关蛋白,从而在水稻种子发育中发挥重要作用[63]。
3.8 胚胎发育
胚胎发育是种子发育中的重要过程。在小麦发育种子中,miR169的表达量较高,miR169通过靶基因CCAAT-box转录因子参与胚胎发育过程[27,64]。 miR1127b的靶基因为氨基酸通透酶基因,在早期种子发育中发挥重要作用。在早期胚胎形成阶段,氨基酸通透酶摄取氨基酸进入胚乳并向发育中的胚胎提供氨基酸[65]。miR160通过调控靶基因ARF17,造成异常胚胎对称[66]。
4 展 望
基因表达变化在种子发育过程中发挥着重要作用,miRNA作为关键的后转录调控因子,调控众多与种子发育相关基因的表达。近年来,越来越多与种子发育相关的miRNA通过高通量测序、基因芯片、大规模平行测序、生物信息学、降解组测序等技术在多种植物中得到鉴定。这些miRNA同它们的靶基因对种子的发育构成了庞大的调控网络。参与种子发育的miRNA及其靶基因目前已得到广泛的研究,本文总结了与种子发育相关的miRNA及其靶基因,重点综述了miRNA在种子发育过程中发挥作用的具体调控机制。
目前已在多种植物种子中鉴定到miRNA,并预测了它们的靶基因及功能,但真正明确其具体调控机制的miRNA还很少。如miRNA对种子活力的调控机制还不明确。有研究发现,miRl64、miRl68、miR395、miR399、miR817等在水稻不同活力种子胚中的表达量呈规律性变化。其中miR817的靶蛋白是ATP合成酶的构成蛋白及膜蛋白,参与种子的活力调控[25,67]。但是这些miRNA究竟是如何调控种子活力的?其具体功能与作用机制是什么?这些都还未知,还需进一步深入研究。miRNA对种子发育的交叉调控作用也是未解之谜。miRNA对生长素、脱落酸、赤霉素等植物激素信号转导的调控是否具有交叉作用?仍然需要进一步探索来找寻答案并明确其具体的调控方式。
因此,种子发育相关miRNA的靶基因和具体调控机制研究应该成为今后研究的重点。miRNA对谷类作物种子的灌浆、油菜类种子的脂质存储、种子活力的提高等均极有利于农业生产,相信对miRNA的研究将为更好的发挥其在调控种子生长发育中的作用起到极大的帮助。
[1]Bartel DP.MicroRNAs:Genomics,biogenesis,mechanism, and function.Cell,2004,116(2):281–297.
[2]Zamore PD,Haley B.Ribo-gnome:The big world of small RNAs.Science,2005,309(5740):1519–1524.
[3]Lee RC,Feinbaum RL,Ambros V.The C.elegans heterochronicgenelin-4encodessmallRNAswith antisense complementarity to lin-14.Cell,1993,75(5):843–854.
[4]Park W,Li JJ,Song RT,Messing J,Chen XM.CARPEL FACTORY,a Dicer homolog,and HEN1,a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol,2002,12(17):1484–1495.
[5]Llave C,Kasschau KD,Rector MA,Carrington JC. Endogenousand silencing-associated smallRNAsin plants.Plant Cell,2002,14(7):1605–1619.
[6]Jones-Rhoades MW,Bartel DP,Bartel B.MicroRNAs and their regulatory roles in plants.Annu Rev Plant Biol, 2006,57:19–53.
[7]Teotia S,Tang GL.To bloom or not to bloom:role of MicroRNAs in plant flowering.Mol Plant,2015,8(3): 359–377.
[8]Khraiwesh B,Zhu JK,Zhu JH.Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim Biophys Acta,2012,1819(2):137–148.
[9]Jia L,Zhang DY,Qi XW,Ma B,Xiang ZH,He NJ. Identification of the conserved and novel miRNAs in mulberry by high-throughput sequencing.PLoS One,2014, 9(8):e104409.
[10]Reinhart BJ,Weinstein EG,Rhoades MW,Bartel B,Bartel DP.MicroRNAs in plants.Genes Dev,2002,16(13): 1616–1626.
[11]Xie ZX,Allen E,Fahlgren N,Calamar A,Givan SA, Carrington JC.Expression of Arabidopsis MIRNA genes. Plant Physiol,2005,138(4):2145–2154.
[12]魏强,梁永宏,李广林.植物miRNA的进化.遗传, 2003,35(3):315–323.
[13]BaranauskėS,MickutėM,Plotnikova A,Finke A, Venclovas Č,Klimašauskas S,Vilkaitis G.Functional mapping of the plant small RNA methyltransferase:HEN1 physically interactswith HYL1andDICER-LIKE 1 proteins.Nucleic Acids Res,2015,43(5):2802–2812.
[14]BollmanKM,AukermanMJ,ParkMY,HunterC, BerardiniTZ,PoethigRS.HASTY,theArabidopsis ortholog of exportin 5/MSN5,regulates phase change and morphogenesis.Development,2003,130(8):1493–1504.
[15]Park MY,Wu G,Gonzalez-Sulser A,Vaucheret H,Poethig RS.Nuclear processing and exportof microRNAs in Arabidopsis.Proc Natl Acad Sci USA,2005,102(10): 3691–3696.
[16]Xie M,Zhang SX,Yu B.microRNA biogenesis, degradation and activity in plants.Cell Mol Life Sci, 2015,72(1):87–99.
[17]Bonnet E,Van de Peer Y,Rouzé P.The small RNA world of plants.New Phytol,2006,171(3):451–468.
[18]Khan Y,Yadav A,Bonthala VS,Muthamilarasan M,Yadav CB, Prasad M. Comprehensive genome-wide identification and expression profiling of foxtail millet [Setaria italica(L.)]miRNAs in response to abiotic stress and development of miRNA database.Plant Cell,Tissue and Organ Culture(PCTOC),2014,118(2):279–292.
[19]ReinhartBJ,Slack FJ,Basson M,PasquinelliAE, Bettinger JC,Rougvie AE,Horvitz HR,Ruvkun G.The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans.Nature,2000,403(6772): 901–906.
[20]Ding YF,Tao YL,Zhu C.Emerging roles of microRNAs in the mediation of drought stress response in plants.J Exp Bot,2013,64(11):3077–3086.
[21]Carrington JC,Ambros V.Role of microRNAs in plant and animal development.Science,2003,301(5631):336–338.
[22]Zhang BH.MicroRNA:a new target for improving plant tolerance to abiotic stress.J Exp Bot,2015,66(7): 1749–1761.
[23]Körbes AP,Machado RD,Guzman F,Almerão MP,de Oliveira LFV,Loss-Morais G,Turchetto-ZoletAC, Cagliari A,Maraschin FS,Margis-Pinheiro M,Margis R. Identifying conserved and novel micrornas in developing seeds of Brassica napus using deep sequencing.PLoS One, 2012,7(11):e50663.
[24]Zhao YT,Wang M,Fu SX,Yang WC,Qi CK,Wang X J. Small RNA Profiling in Two Brassica napus Cultivars Identifies MicroRNAs with Oil Production-and Development-Correlated Expression and New Small RNA Classes.Plant Physiol,2012,158(2):813–823.
[25]Xue LJ,Zhang JJ,Xue HW.Characterization and expression profiles of miRNAs in rice seeds.Nucleic Acids Res,2009,37(3):916–930.
[26]Kang MM,Zhao Q,Zhu DY,Yu JJ.Characterization of microRNAs expression during maize seed development. BMC Genomics,2012,13:360.
[27]Han R,Jian C,Lv JY,Yan Y,Chi Q,Li ZJ,Wang Q,Zhang J,Liu XL,Zhao HX.Identification and characterization of microRNAs in the flag leaf and developing seed of wheat (Triticum aestivum L.).BMC Genomics,2014,15:289.
[28]Rhoades MW,Reinhart BJ,Lim LP,Burge CB,Bartel B, Bartel DP.Prediction of plant microRNA targets.Cell, 2002,110(4):513–520.
[29]Sunkar R,Chinnusamy V,Zhu JH,Zhu JK.Small RNAs as big players in plant abiotic stress responses and nutrient deprivation.Trends Plant Sci,2007,12(7):301–309.
[30]MitsudaN,Ohme-TakagiM.Functionalanalysisof transcription factors in Arabidopsis.Plant Cell Physiol, 2009,50(7):1232–1248.
[31]朱诚.植物生物学.北京:北京师范大学出版社,2012: 254–258.
[32]Li DT,Wang LW,Liu X,Cui DZ,Chen TT,Zhang H, Jiang C,Xu CY,Li P,Li S,Zhao L,Chen HB.Deep sequencing of maize small RNAs reveals a diverse set of microRNA in dry and imbibed seeds.PLoS One,2013, 8(1):e55107.
[33]Finkelstein R,Reeves W,Ariizumi T,Steber C.Molecular aspects of seed dormancy.Annu Rev Plant Biol,2008,59: 387–415.
[34]YangJC,ZhangJH,Wang ZQ,LiuK,Wang P. Post-anthesis development of inferior and superior spikelets in rice in relation to abscisic acid and ethylene.J Exp Bot,2006,57(1):149–160.
[35]Zhang ZX,Chen J,Lin SS,Li Z,Cheng RH,Fang CX, Chen HF,Lin WX.Proteomic and phosphoproteomic determination of ABA's effects on grain-filling of Oryza sativa L.inferior spikelets.Plant Sci,2012,185–186: 259–273.
[36]Reyes JL,Chua NH.ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination.Plant J,2007,49(4):592–606.
[37]Peng T,Sun HZ,Qiao MM,Zhao YF,Du YX,Zhang J,Li JZ, Tang GL, Zhao QZ. Differentially expressed microRNA cohorts in seed development may contribute to poor grain filling of inferior spikelets in rice.BMC Plant Biol,2014,14:196.
[38]Li WX,Oono Y,Zhu JH,He XJ,Wu JM,Iida K,Lu XY, CuiX,JinH,Zhu JK.TheArabidopsisNF-YA5 transcription factorisregulated transcriptionally and posttranscriptionally to promote drought resistance.Plant Cell,2008,20(8):2238–2251.
[39]Mu JY,Tan HL,Hong SL,Liang Y,Zuo JR.Arabidopsis transcription factor genes NF-YA1,5,6 and 9 play redundant roles in male gametogenesis,embryogenesis, and seed development.Mol Plant,2013,6(1):188–201.
[40]Schruff MC,Spielman M,Tiwari S,Adams S,Fenby N, Scott RJ.The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling,cell division,and the size of seeds and other organs.Development,2006,133(2): 251–261.
[41]Yang JH,Han SJ,Yoon EK,Lee WS.Evidence of an auxin signal pathway,microRNA167-ARF8-GH3, and its response to exogenous auxin in cultured rice cells.Nucleic Acids Res,2006,34(6):1892–1899.
[42]Jones-Rhoades MW, Bartel DP. Computational identification ofplantmicroRNAsand theirtargets, including a stress induced miRNA.Mol Cell,2004, 14(6):787–799.
[43]Yi R,Zhu ZX,Hu JH,Qian Q,Dai JC,Ding Y. Identification and expression analysis of microRNAs at the grain filling stage in rice(Oryza sativa L.)via deep sequencing.PLoS One,2013,8(3):e57863.
[44]Uauy C,Distelfeld A,Fahima T,Blechl A,Dubcovsky J.A NAC gene regulating senescence improves grain protein, zinc,and iron content in wheat.Science,2006,314(5803): 1298–1301.
[45]Zhang YC,Yu Y,Wang CY,Li ZY,Liu Q,Xu J,Liao JY, Wang XJ,Qu LH,Chen F,Xin P,Yan C,Chu J,Li HQ, ChenYQ.OverexpressionofmicroRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching.Nat Biotechnol,2013,31(9):848–852.
[46]马圣运.os-miR408的表达模式及其在水稻种子发育中的功能[学位论文].杭州:浙江大学,2012.
[47]Sun GL.MicroRNAs and their diverse functions in plants. Plant Mol Biol,2011,80(1):17–36.
[48]Sunkar R,Zhu JK.Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis.Plant Cell,2004, 16(8),2001–2019.
[49]Liu B,Li PC,Li X,Liu CY,Cao SY,Chu CC,Cao XF. Loss of function of OsDCL1 affects microRNA accumulation and causes developmental defects in rice. Plant Physiol,2005,139(1):296–305.
[50]Song QX,Liu YF,Hu XY,Zhang WK,Ma B,Chen SY, Zhang JS.Identification of miRNAs and their target genes in developing soybean seeds by deep sequencing.BMC Plant Biol,2011,11:5.
[51]Pignocchi C,Kiddle G,Hernández I,Foster SJ,Asensi A, Taybi T, Barnes J, Foyer CH. Ascorbate oxidase-dependent changes in the redox state of the apoplast modulate gene transcript accumulation leading to modified hormone signaling and orchestration of defense processes in tobacco.PlantPhysiol,2006,141(2): 423–435.
[52]Potters G,Horemans N,Caubergs RJ,Asard H.Ascorbate and dehydroascorbate influence cell cycle progression in a tobacco cell suspension.Plant Physiol,2000,124(1):17–20.
[53]Zhang LF,ChiaJM,KumariS,Stein JC,Liu ZJ, Narechania A,Maher CA,Guill K,McMullen MD,Ware D.A genome-wide characterization of microRNA genes in maize.PLoS Genet,2009,5(11):e1000716.
[54]Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J,Tiedemann J,Kroj T,Parcy F.bZIP transcription factors in Arabidopsis.Trends Plant Sci, 2002,7(3):106–111.
[55]Hawker NP,Bowman JL.Roles for class III HD-Zip and KANADI genes in Arabidopsis root development.Plant Physiol,2004,135(4):2261–2270.
[56]Nodine MD,Bartel DP.MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis.Genes Dev,2010,24(23):2678–2692.
[57]Palatnik JF,Allen E,Wu X,Schommer C,Schwab R, Carrington JC, Weigel D. Control of leaf morphogenesis by microRNAs.Nature,2003,425(6955): 257–263.
[58]Wang SK,Wu K,Yuan QB,Liu XY,Liu B,Lin XY,Zeng RZ,Zhu HT,Dong GJ,Qian Q,Zhang GQ,Fu XD. Control of grain size,shape and quality by OsSPL16 in rice.Nat Genet,2012,44(8):950–954.
[59]Wang CY,Zhang SC,Yu Y,Luo YC,Liu Q,Ju CL,Zhang YC,Qu LH,Lucas WJ,Wang XJ,Chen YQ.MiR397b regulates both lignin contentand seed numberin Arabidopsis via modulating a laccase involved in lignin biosynthesis.Plant Biotechnol J,2014,12(8): 1132–1142.
[60]Jain M,Nijhawan A,Arora R,Agarwal P,Ray S,Sharma P, Kapoor S,Tyagi AK,Khurana JP.F-box proteins in rice. Genome-wide analysis,classification,temporal and spatial gene expression during panicle and seed development,and regulation by light and abiotic stress.Plant Physiol,2007, 143(4):1467–1483.
[61]Galli V,Guzman F,de Oliveira LFV,Loss-Morais G, Körbes AP,Silva SDA,Margis-Pinheiro MMAN,Margis R.Identifying MicroRNAs and Transcript Targets in Jatropha Seeds.PLoS One,2014,9(2):e83727.
[62]Zhang H,Li HW,Yuan LM,Wang ZQ,Yang JC,Zhang JH. Post-anthesis alternate wetting and moderate soil drying enhances activities of key enzymes in sucrose-to-starch conversion in inferior spikelets of rice.J Exp Bot,2012,63(1):215–227.
[63]Peng T,Sun HZ,Du YX,Zhang J,Li JZ,Liu YX,Zhao YF, Zhao QZ.Characterization and expression patterns of microRNAs involved in rice grain filling.PLoS One,2013, 8(1):e54148.
[64]Siefers N,Dang KK,Kumimoto RW,Bynum WE,Tayrose G,HoltBF.Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity.Plant Physiol, 2009,149(2):625–641.
[65]Schmidt R,Stransky H,Koch W.The amino acid permease AAP8 is importantforearly seed developmentin Arabidopsis thaliana.Planta,2007,226(4):805–813.
[66]Mallory AC,Bartel DP,Bartel B.MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essentialfor properdevelopmentand modulates expression of early auxin response genes.Plant Cell,2005, 17(5):1360–1375.
[67]成海兰.miRNA与水稻种子活力的相关性研究[学位论文].长沙:湖南师范大学,2011.
(责任编委:张宪省)
Role of miRNAin plant seed development
Shumin Gong,Yanfei Ding,Cheng Zhu
Key Laboratory of Marine Food Quality and Hazard Controlling Technology of Zhejiang Province,College of Life Sciences,China Jiliang University,Hangzhou 310018,China
MicroRNA(miRNA),a class of non-coding small RNAs,has been reported to be involved in a broad range of metabolic and physiological processes in plants,such as plant growth,development and responses to stresses. They participate in gene expression by degrading target genes at post-transcriptional levels.Seeds are the basic elements of plant growth and important materials for agriculture.miRNAs have been identified to be involved in seed development in many plants.Herein we review recent progresses on the miRNAs involved in seed development and their regulatory mechanisms,which will help to provide insights into further research to improve seed quality.
seed;miRNA;development;regulatory mechanism;plant
2015-01-08;
2015-03-18
国家自然科学基金项目(编号:31401299,31170251,31470368)和浙江省自然科学基金项目(编号:LY13C020002,LZ14C020001)资助
龚淑敏,在读硕士研究生,专业方向:植物逆境生理与分子生物学。E-mail:gongshumin1029@126.com
丁艳菲,副教授,研究方向:植物逆境生理与分子生物学。E-mail:dingyanfei1984@126.com
龚淑敏和丁艳菲为并列第一作者。
朱诚,博士,教授,研究方向:植物逆境生理与分子生物学。E-mail:pzhch@cjlu.edu.cn
10.16288/j.yczz.15-020
时间:2015-5-13 13:57:35
URL:http://www.cnki.net/kcms/detail/11.1913.R.20150513.1357.001.html

