利用微卫星标记的魁蚶混交家系鉴定❋
孙 楠, 李 琪, 于 红, 孔令锋
(中国海洋大学水产学院,山东 青岛 266003)
研究简报
利用微卫星标记的魁蚶混交家系鉴定❋
孙 楠, 李 琪❋❋, 于 红, 孔令锋
(中国海洋大学水产学院,山东 青岛 266003)
本文使用6个具有多态性的微卫星位点,对30个魁蚶亲本及其300个子代(共330个个体)进行家系分析,评估苗种生产中亲本与子代间的遗传变异。研究表明,家系鉴定成功率与模拟结果一致,使用6个微卫星位点可以实现超过99%的鉴定率。魁蚶亲本与子代间等位基因多样性相似,子代群体的期望杂合度显著低于亲本群体(P<0.05),观测杂合度和亲本群体相比略有降低,但无显著差异。所有魁蚶父母本都参与了繁殖过程,并具有多重父权和多重母权的现象。研究显示,微卫星作为一种分子标记可以有效应用到家系鉴定中,同时对于养殖魁蚶群体的遗传多样性变化仍需要进一步的监测。本研究为魁蚶人工育苗、养殖生产制定合理的管理方针提供帮助,同时为微卫星作为分子标记进行魁蚶增殖放流溯源提供科学支持。
魁蚶;微卫星;家系分析
魁蚶(Scapharcabroughtonii)属于瓣鳃纲(Lamellibranchia)蚶科(Arcidae),又称大毛蚶、赤贝、血贝,是一种大型底栖经济贝类,广泛分布在中国、日本、韩国及菲律宾海沿岸。其成体个大体肥, 肉质鲜美, 经济价值很高。近年来,由于需求旺盛, 而自然资源远不能满足市场的需要, 因此魁蚶的人工育苗及养殖生产得到快速发展[1-2]。但由于魁蚶具有大部分海洋双壳贝类高繁殖力的特点,因此在其苗种生产中往往使用有限的亲本,而易发生近亲交配。较高的近交率会引起养殖群体遗传多样性下降,这成为魁蚶养殖生产中一个备受关注的问题。
微卫星标记具有多态性高、遵循孟德尔分离定律、共显性遗传等特点,已成为家系鉴定、遗传图谱构建、种群遗传学研究的重要工具[3-5]。利用微卫星标记进行家系鉴定已成功应用于许多水产动物,例如:苏天凤等[6]使用16个微卫星标记鉴定了斑节对虾(Penaeusmonodon)7个全同胞家系的亲缘关系;Norris等[7]在混养的大西洋鲑鱼中,证明了在没有物理标记及背景信息的情况下,微卫星分子标记可以有效的进行家系鉴定和亲缘关系分析。近几年,已先后开发出魁蚶微卫星标记100多个[8-10],本研究从已开发的魁蚶微卫星标记中筛选出6个多态性高、特异性强的微卫星标记,对30个亲本及其混交产生的300个子代进行微卫星分型,分析家系鉴定效率,比较亲本与子代间的遗传变异,以期为魁蚶养殖群体的遗传多样性保护提供基础资料。
1 材料与方法
1.1 样品采集
实验用魁蚶亲贝于2012年6月采自山东省日照海区。随机选择30个魁蚶亲本(14个雄性和16个雌性)进行催产。受精24 h后,D形幼虫用25μm筛绢滤出,移入1.5μL的无菌离心管中,轻微离心后倒出海水,取幼虫和30个亲本组织于100%酒精固定。
1.2 DNA提取及微卫星分析
取亲贝的闭壳肌组织约100mg,采用传统的苯酚-氯仿法来提取基因组DNA[11]。幼虫DNA参照Li等[12]使用Chelex-100树脂提取。实验中选取Sb51[13],ScBr04、 ScBr03、 ScBr20、 ScBr14和ScBr09[8]6个特异性强、多态性较高的微卫星位点进行分析。
聚合酶链式反应(PCR)的总体积为10μL,内含模板DNA 1.0μL,1μmol/L 的正反引物, 0.2mmol/L dNTPs, 1×PCR buffer, 1.5mmol/L MgCl2, 0.25U TaqDNA聚合酶(宝生物工程有限公司)。亲本的PCR扩增程序为:94℃预变性3min,然后进行35个扩增循环,每一循环包括94℃变性30s,退火温度45s,72℃延伸30s,最后72℃延伸10min,4℃保存备用。幼虫的PCR扩增程序为:94℃预变性3min,然后进行7个扩增循环,每一循环包括94℃变性1min,退火温度30s,72℃延伸30s;然后再进行33个循环,每一循环包括94℃变性30s,退火温度30s,72℃延伸30s,最后72℃延伸5min。扩增产物通过6%变性聚丙烯酰胺凝胶进行电泳分离。在1×TBE电泳缓冲液, 220 V恒压下,电泳2.5h,电泳完毕后,银染检测,利用10 bp DNA ladder(Invitrogen公司)作为marker检测等位基因位置。为防止每张板胶孔的差异及读数偏差,每张板中加入若干亲本的PCR产物作为对照。
1.3 统计分析
首先,使用软件CERVUS 3.0模拟估算魁蚶家系鉴定所需要的微卫星位点数及其鉴定效率。具体参数如下:模拟子代数目为10000(循环重复数),候选亲本数为30,亲本检测率100%,位点检测率100%,分型误差率1%,置信水平95%。然后根据实验结果,计算真实的鉴定效率。多态信息含量(PIC)及位点的非排除概率(NE-1P, NE-2P,NE-PP)也由CERVUS 3.0计算,NE-1P、NE-2P和NE-PP分别指位点对第一个亲本、第二个亲本和亲本对的非排除概率。根据位点的PIC值,由高到低逐一增加位点数来计算位点的累积鉴定能力。利用MICRO-CHECKER[14]评估6个微卫星位点的无效等位基因情况。亲本和子代的遗传多样性参数:等位基因数(Na)、期望杂合度(He)和观测杂合度(Ho)由CERVUS 3.0计算。差异显著性由SPSS 19.0(美国SPSS公司)进行非参数两配对样本Wilcoxon符号秩检验[15]。
2 结果
2.1 家系鉴定成功率
用于家系模拟鉴定的6个微卫星位点的等位基因数为6~13,平均值为10.8,平均多态信息含量(PIC)为0.746;对第一个亲本的非排除概率(NE-1P)为0.311~0.839,对第二个亲本的非排除概率(NE-2P)为0.183~0.658,亲本对的非排除概率(NE-PP)为0.055~0.452(见表1)。MICRO-CHECKER分析结果表明,位点Sb51、ScBr04、ScBr20、ScBr14、ScBr09纯合子过剩,可能存在无效等位基因,但都没有大片段基因丢失的现象。

表1 6个微卫星位点的综合信息Table 1 The information of 6 microsatellite loci
注:①Annealing Temperature;②No. of GenBank;③Number of alleles;④Polymorphism information content;⑤Average non-exclusion probability to first parent;⑥Average non-exclusion probability to second parent;⑦Average non-exclusion probability to parent pair;⑧Average
根据位点的PIC值由高到低逐一累加计算6个位点的累积家系鉴定成功率。模拟结果表明:采用4个位点的鉴定成功率为96%;采用5个位点的鉴定成功率能达到99%;使用6个位点,则获得超过99%的鉴定成功率(见图1)。实际家系鉴定在330个个体中进行(包括300个子代和30个亲本)。在95%置信度下,运用5个位点成功鉴定了97%的子代个体,当运用6个位点后,子代的鉴定率达到100%。实际家系鉴定中的累积鉴定成功率与模拟家系基本一致(见图1)。
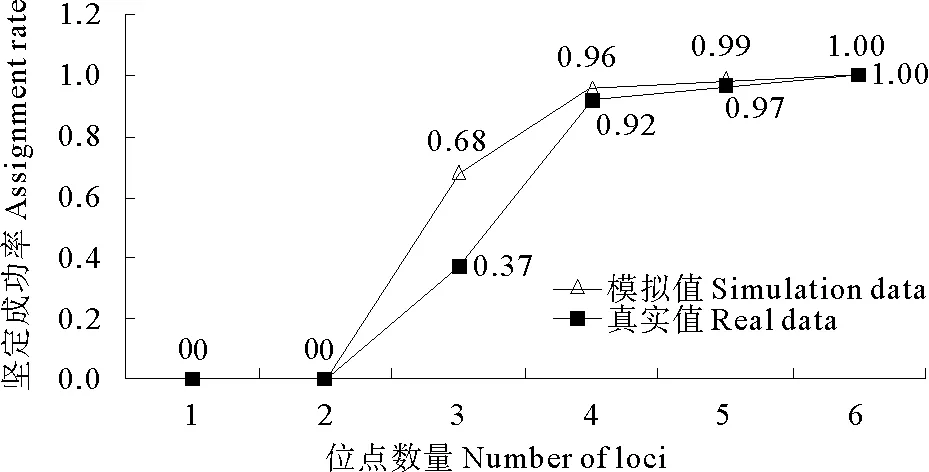
图1 6个微卫星位点模拟和实际鉴定的累积鉴定成功率
2.2 亲本和子代遗传多样性比较
表2显示了6个微卫星位点在亲本和子代群体中的等位基因数(Na),等位基因长度范围,期望和观测杂合度(He、Ho)。亲本群体中等位基因数为6~13,平均值为10.8。子代中没有出现等位基因丢失现象。子代群体的期望杂合度显著低于亲本(P<0.05)。但子代的观测杂合度和亲本群相比略有降低,但是差异不显著(P=0.345)。
家系鉴定结果成功鉴定出14个父本和16个母本组成的144对亲本组合。对于父本,子代数目从7~40,平均数为21.5;对于母本,子代数目从7~34,平均数为18.8。每个亲本的平均子代比例如表3所示。本研究中,所有亲本参与繁殖,母本的平均子代比例为6.25%,父本的平均比例为7.14%。在所有的16个母本中均发现多重父权现象,每个母本的卵子与5~12个父本的精子受精。对所有母本来说,由不同雄性个体作为父本的子代比例和遗传有效父权频率如表4所示。父本根据所产生子代的比例由高到低排列,平均有9个父本与每个母本个体受精,且平均遗传有效父权频率为3.27。在14个父本中也存在多重母权现象,每个父本的精子与4~14个母本的卵子受精。对所有父本来说,由不同雌性个体作为母本的子代个体比例和遗传有效母权频率如表5所示。母本根据所产生子代的比例由高到低排列,平均有10.29个母本与每个父本个体受精,且平均遗传有效母权频率为3.14。

表2 魁蚶亲本和子代遗传多样性的比较
注:①Sample size;②Alleles range;③No. of alleles;④Expected heterozygosity;⑤Observed heterozygosity
3 讨论
3.1 家系分析
在人工育苗生产中,为防止系谱信息错误的逐级放大,导致群体大规模的近交、退化[17],准确的家系鉴定对于避免近交具有重要意义。在实际鉴定之前使用模拟鉴定,可以调整位点的数量,有利于减少成本、提高效率。本研究的实际鉴定结果与模拟鉴定基本一致。
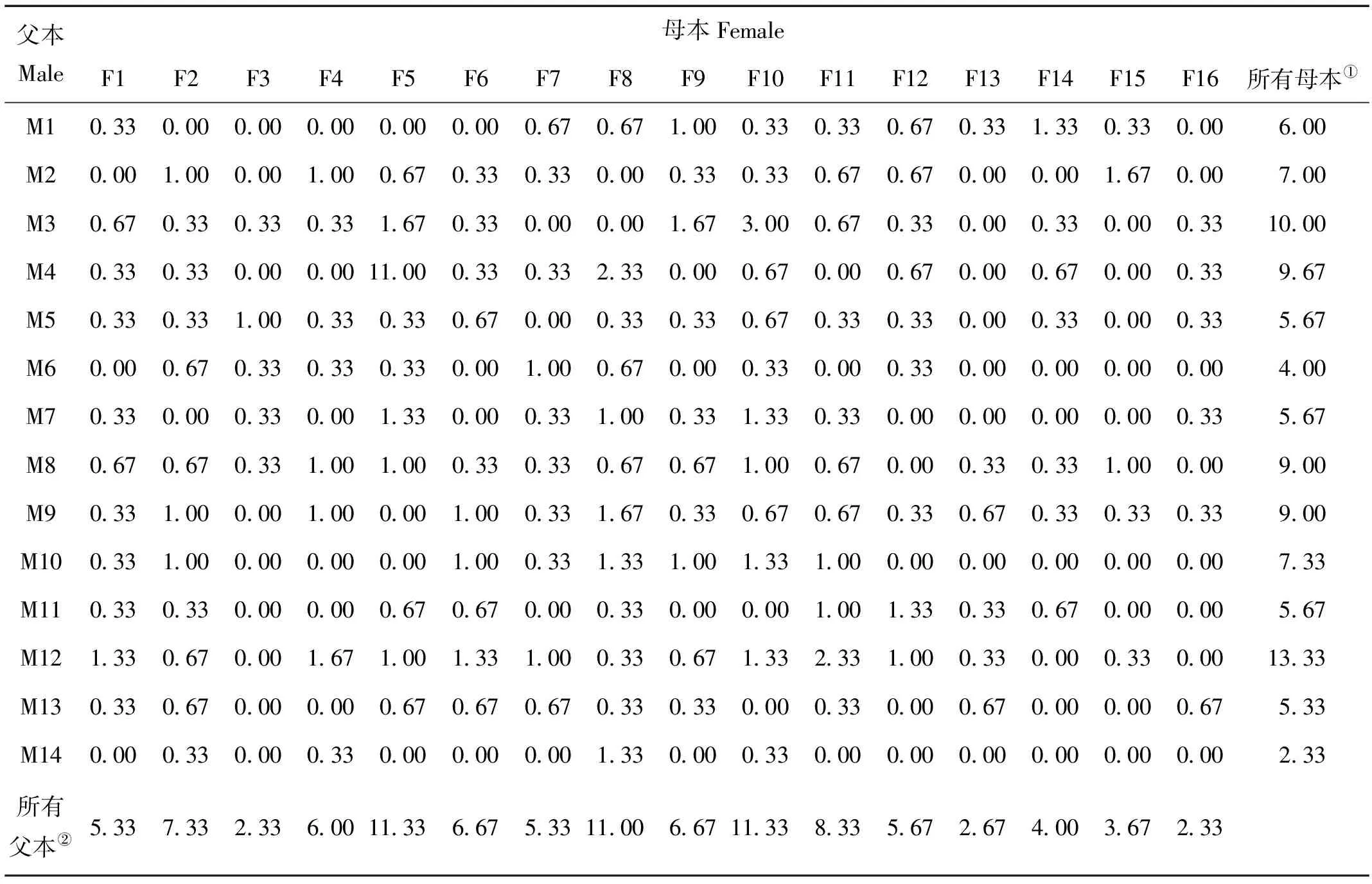
表3 各亲本的繁殖成功率Table 3 Percentage of parental contribution to reproduction
Note:①All females;②All males.
Note:①No. of males participating in clutches;②No. of clutches (% of total);③Paternity proportion of most successful male;④Paternity proportion of second-most successful male;⑤Paternity proportion of third-most successful male;⑥Paternity proportion of fourth-most successful male;⑦Paternity proportion of the least successful male;⑧Genetically effective paternity
基因分型错误、无效等位基因和DNA质量较差是造成家系鉴定不匹配的主要原因[18],其中无效等位基因频率高于5%时,会降低家系鉴定的效率[19]。海洋软体动物的自然种群中,利用微卫星标记经常会观测到杂合子缺失的现象[20-21]。在本研究中,6个微卫星位点中,虽有5个位点(Sb51、ScBr04、ScBr20、ScBr14和ScBr09)由于纯合子过剩,可能存在无效等位基因,但在鉴定过程中,并未降低家系鉴定的置信度。使用这6个魁蚶微卫星位点成功鉴定了30个亲本及其300个子代的亲子关系,表明这6个位点用于家系鉴定的有效性。
3.2 遗传多样性和亲本贡献率
养殖群体和野生群体相比,由于其亲本数量有限,亲本与子代间基因频率经常发生遗传漂变及遗传多样性的降低[22]。遗传多样性的降低表现为等位基因数和杂合度的降低[23-25],会对水产动物生产性状产生不利影响,例如生长率下降、适合度降低[26-27]。本研究中,所有亲本都参与了生产繁殖,避免了等位基因多样性的降低,但由于5个位点可能含有无效等位基因,因此子代和亲本相比,观测杂合度略有降低,但不显著。

表5 交配组母权比例(平均值±标准差)Table 5 Maternity proportions for clutches (Mean ± SD)
Note:①No. of females participating in clutches;②No. of clutches (% of total);③Maternity proportion of most successful female;④Maternity proportion of second-most successful female;⑤Maternity proportion of third-most successful female;⑥Maternity proportion of fourth-most successful female;⑦Maternity proportion of the least successful female;⑧Genetically effective maternity
多重父权或多重母权现象在混交动物中普遍存在[28]。本研究的父权分析表明,在所有的繁殖个体中均存在多重父权现象,每个交配组的遗传有效父权频率显著大于1,说明魁蚶雄性精子的竞争程度比较高。魁蚶的有效母权频率也显著大于1,表明在配子水平上存在多重母权现象,这种现象在大西洋鳕鱼和太平洋牡蛎中也有报道[29-30]。
4 结语
本实验对魁蚶人工养殖群体的研究,证实使用微卫星进行魁蚶家系分析可以鉴定亲本的繁殖成功率,监测生产活动中遗传变化,为今后魁蚶人工育苗、养殖生产制定合理的管理方针提供帮助,同时为微卫星作为分子标记进行魁蚶增殖放流溯源提供科学支持。
[1] 梁超, 杨爱国, 刘志鸿, 等. 4个地理群体魁蚶(Scapharcabroughtonii)的形态差异与判别分析[J]. 海洋科学, 2011, 35(11): 108-113.
[2] Li R, Li Q, Wang C. Sibship reconstruction and effective population size estimation in mass spawning ark shell,Scapharcabroughtoniibased on microsatellite analysis [J]. Genes Genom, 2013, 35(6): 703-708.
[3] Queller D C, Strassmann J E, Hughes C R. Microsatellites and kinship [J]. Trends Ecol Evol, 1993, 8(8): 285-288.
[4] Desvignes J F, Laroche J, Durand J D, et al. Genetic variability in reared stocks of common carp (CyprinuscarpioL.) based on allozymes and microsatellites [J]. Aquaculture, 2001, 194(3): 291-301.
[5] Holland D C. Identity and agency in cultural worlds [M]. Massachusetts: Harvard University Press, 2001.
[6] 苏天凤, 熊小飞, 江世贵, 等. 斑节对虾7个全同胞家系间亲缘关系的微卫星分析 [J]. 南方水产科学, 2010, 6(6): 1-7.
[7] Norris A T, Bradley D G, Cunningham E P. Microsatellite genetic variation between and within farmed and wild Atlantic salmon (Salmosalar) populations [J]. Aquaculture, 1999, 180(3): 247-264.
[8] Sekino M, Kurokawa T, Sasaki K. Multiplex PCR panels of novel microsatellites for the ark shellScapharcabroughtonii(Pteriomorphia, Arcoida) [J]. Conserv Genet Resour, 2010, 2(1): 39-42.
[9] Tian J T, Liu Z H, Zhou L Q, et al. Isolation and characterization of 48 polymorphic microsatellite markers for the blood clamScapharcabroughtonii(Arcidae) [J]. Genet Mol Res, 2012, 11(4): 4501-4507.
[10] Li M, Zhu L, Zhou C Y, et al. Development and characterization of EST-SSR markers fromScapharcabroughtoniiand their transferability inScapharcasubcrenataandTegillarcagranosa[J]. Molecules, 2012, 17(9): 10716-10723.
[11] Li Q, Kijima A. Microsatellite analysis of gynogenetic families in the Pacific oyster,Crassostreagigas[J]. J Exp Mar Biol Ecol, 2006, 331(1): 1-8.
[12] Li Q, Park C, Kijima A. Allelic transmission of microsatellites and application to kinship analysis in newly hatched Pacific abalone larvae [J]. Fisheries Sci, 2003, 69(5): 883-889.
[13] Li N, Li Q, Kong L F, et al. Development of three multiplex PCRs in the ark shellScapharcabroughtoniiand their validation in parentage assignment [J]. Journal of Ocean University of China,(Unpublished).
[14] Oosterhout C V, Hutchinson W F, Wills D P M, et al. Micro-Checker: software for identifying and correcting genotyping errors in microsatellite data [J]. Mol Ecol Notes, 2004, 4(3): 535-538.
[15] Wang J. A new method for estimating effective population sizes from a single sample of multilocus genotypes[J]. Mol Ecol, 2009, 18(10): 2148-2164.
[16] Bekkevold D, Hansen M M, Loeschcke V. Male reproductive competition in spawning aggregations of cod (Gadusmorhua, L.)[J]. Mol Ecol, 2002, 11(1): 91-102.
[17] Hoffman J I, Amos W. Microsatellite genotyping errors: detection approaches, common sources and consequences for paternal exclusion[J]. Mol Ecol, 2005, 14(2): 599-612.
[18] Jerry D R, Preston N P, Crocos P J, et al. Parentage determination of Kuruma shrimpPenaeus(Marsupenaeus)japonicususing microsatellite markers (Bate) [J]. Aquaculture, 2004, 235(1): 237-247.
[19] Marshall T C, Slate J, Kruuk L E B, et al. Statistical confidence for likelihood-based paternity inference in natural populations[J]. Mol Ecol, 1998, 7(5): 639-655.
[20] Prakoon W, Tunkijjanukij S, Nguyen T T, et al. Spatial and temporal genetic variation of green mussel,Pernaviridisin the gulf of Thailand and implication for aquaculture [J]. Mar Biotechnol, 2010, 12(5): 506-515.
[21] Arias-Pérez A, Fernández-Tajes J, Gaspar M B, et al. Isolation of microsatellite markers and analysis of genetic diversity among east atlantic populations of the sword razor shellEnsissiliqua: a tool for population management [J]. Biochemical Genetics, 2012, 50(5-6): 397-415.
[22] Hedgecock D, Sly F. Genetic drift and effective population sizes of hatchery-propagated stocks of the Pacific oyster,Crassostreagigas[J]. Aquaculture, 1990, 88(1): 21-38.
[23] Allendorf F W, Phelps S R. Loss of genetic variation in a hatchery stock of cutthroat trout [J]. T Am Fish Soc, 1980, 109(5): 537-543.
[24] Ryman N, Ståhl G. Genetic changes in hatchery stocks of brown trout (Salmotrutta) [J]. Can J Fish Aquat Sci, 1980, 37(1): 82-87.
[25] Cross T F, King J. Genetic effects of hatchery rearing in Atlantic salmon [J]. Aquaculture, 1983, 33(1): 33-40.
[26] Koehn R K, Diehl W J, Scott T M. The differential contribution by individual enzymes of glycolysis and protein catabolism to the relationship between heterozygosity and growth rate in the coot clam,Mulinialateralis[J]. Genetics, 1988, 118(1): 121-130.
[27] Danzmann R G, Ferguson M M, Allendorf F W. Genetic variability and components of fitness in hatchery strains of rainbow trout [J]. J Fish Biol, 1989, 35(Supplement A): 313-319.
[28] Gibbs H L, Weatherhead P J. Insights into population ecology and sexual selection in snakes through the application of DNA-based genetic markers [J]. J Hered, 2001, 92(2): 173-179.
[29] Li R, Yu R. Parentage determination and effective population size estimation in mass spawning Pacific oyster,Crassostreagigas, based on microsatellite analysis [J]. J World Aquacult Soc, 2009, 40(5): 667-677.
[30] Rakitin A, Ferguson M M, Trippel E A. Sperm competition and fertilization success in Atlantic cod (Gadusmorhua): effect of sire size and condition factor on gamete quality [J]. Can J Fish Aquat Sci, 1999, 56(12): 2315-2323.
责任编辑 朱宝象
Parentage Determination ofScapharcabroughtoniiBased on Microsatellite Analysis
SUN Nan, LI Qi, YU Hong, KONG Ling-Feng
(College of Fisheries, Ocean University of China, Qingdao 266003, China)
In this study, six highly polymorphic microsatellite markers (meanHe= 0.78 andPIC=0.75) were utilized to determine the pedigree and the genetic variability between brood stock and offspring in a mass spawning event of ark shell,Scapharcabroughtonii. Parentage identification was performed on a total of 330 individuals, including 300 offspring and 30 candidate parents. The results showed that the assignment rate was in accordance with simulation. Using 6 microsatellites reached a successful rate of over 99%. The allelic diversity of offspring populations and brood stock was similar wirth each other, but the expected heterozygosity of offspring was less than that of parents significantly (P<0.05). The observed heterozygosity of offspring was a little lower than that of parents, but not significant. All the 30 parents participated in the reproduction. And the existence of multiple paternity and maternity was detected in clutches and dams in these 30 parents. Microsatellite could be effective markers in parentage determination, meanwhile the gentic diversity of hatchery population ofScapharcabroughtoniineed to be monitored. The information obtained in this study was useful for designing suitable management guidelines of cultured stocks in future, and provided a scientific basement for tracing origins in the releasing and enhancement ofScapharcabroughtoniiusing microsatellite as molecular markers.
Scapharcabroughtonii; microsatellites; parentage analysis
国家海洋公益性行业科研专项项目(201205023);国家科技支撑计划项目(2011BAD13B01)资助
2014-07-11;
2014-12-18
孙 楠(1990-),女, 硕士生, 从事水产动物遗传育种学研究。 E-mail: sun-shihan@163.com
❋❋ 通讯作者:E-mail: qili66@ouc.edu.cn
S968.31
A
1672-5174(2015)09-042-07
10.16441/j.cnki.hdxb.20140232

