Effect of siRNA interference on nerve growth factor in intervertebral disc inflammation rats
1Department of Orthopedics,Qilu Hospital Affiliated to Shandong University, No.107 of Wenhua Xi Road, Jinan 250012, China
2Department of Orthopedics, Gaomi People’s Hospital, Gaomi 261500, China
3Shengli Oilfield Central Hospital, No.31 of Jinan Road of Dongying, Dongying 257034, China
Effect of siRNA interference on nerve growth factor in intervertebral disc inflammation rats
Ming-Lei Lang1,3, Ai-Lin Qin2, Jian-Min Li1, Peng Fu3*
1Department of Orthopedics,Qilu Hospital Affiliated to Shandong University, No.107 of Wenhua Xi Road, Jinan 250012, China
2Department of Orthopedics, Gaomi People’s Hospital, Gaomi 261500, China
3Shengli Oilfield Central Hospital, No.31 of Jinan Road of Dongying, Dongying 257034, China
Objective:To investigate the inhibition effect of siRNA interference on NGF induced by inflammatory factor IL-6, and IL-1 so as to provide novel targets for clinical treatment of discogenic low back pain.Methods:The intervertebral disc nucleus and annulus fibrosus cells of rats were separated .The cells were co-cultured with different concentrations (10 nmol/L, 20 nmol/L, 50 nmol/L, 100 nmol/L) of IL-6 and IL-1β. The NGF- siRNA was leaded into the cocultured cells with its import ability assessed by flow cytometry instrument tests,before and after which the NGF mRNA expression was detected by real-time Q-PCR and the NGF content was detected by ELISA.Results:Flow cytometry instrument test results showed that the NGF-siRNA cell conversion rate was 99.8%. Real-time Q-PCR detection results showed that compared with negative control group, the NGF mRNA expression of co-cultured cells treated by 10 nmol/ L, 20 nmol/L, 50 nmol/L, 100 nmol/L IL-6 and IL-1β were respectively raised 3.4, 3.7, 4.7, 3.7 times which were all significantly down-regulated after the import of NGF- siRNA. EILSA detection results showed that compared with negative control group, the NGF content of cocultured medium treated by 10 nmol/L, 20 nmol/L, 50 nmol/L, 100 nmol/L I-L6 and IL-1β were respectively raised 2.9, 3.3, 4.5, 7.4 times which were all significantly decreased after the import of NGF- siRNA.Conclusions:These molecular biological results suggest that inflammatory factor IL-6 and IL-1β could stimulate NGF on intervertebral disc cells in vitro culture model and its efficiency is concentration dependent, while siRNA interference can inhibit the stimulation effect of IL-6 and IL-1β on intervertebral disc cell, which provides a new targets for the clinical treatment of discogenic low back pain.
ARTICLE INFO
Article history:
Received 10 April 2014
Received in revised form 15 May 2014
Accepted 15 July 2014
Available online 20 September 2014
RNA
Small interfering
Nerve growth factor
Low back pain
Intervertebral disk displacemento
1. Introduction
Discogenic low back pain (DLBP) is the hot spot of modern orthopedic research with its complex etiology, unclear pathogenesis and absence of effective clinical treatment[1]. Epidemiological survey results showed an increasing incidence of DLBP as the upgrading of people’s life pace and work intensity[2,3]. Electrophysiological studies indicated that, under normal physiological conditions, nerve fiber only exists at the outer layer of the fibrous ring of intervertebral disc, while constitutively distributed in the nucleus pulposus and the annulus inner layer in DLBP patients. This evidence suggests that the ingrowth of nerve fibers is the important pathogenesis mechanism associated with DLBP[4,5], which provide theoretical bases for the clinical treatment at molecular biology level. Nerve growth factor (NGF) overexpression and excessive secretion of inflammatory factors are the key in the DLBP pathogenesis[6,7]. This study aims to investigate the effect of siRNA interference on NGF in rats with inflammatory disc.
2. Materials and methods
2.1. Experimental animals
Sixty Sprague-Dawley (SD) rats of specific pathogen free level were selected, which were half male and half female, weighing 180-240 g, with mean weight as (223±12.4) g, aged 2.5-3.5 months, with mean age of (2.94±0.73) months. They were provided by the Animal Center of Shandong University School of Medicine, China with license No. SYXK (Lu) 2007-0081.
2.2. Main instruments and reagents
Main instruments: PCR instrument (C1000 Thermal cycler, BIO-RAD); fluorescence quantitative instrument (IQ5, BIORAD); ice machine (AF100, Scotsman); cryogenic overweight centrifuge (Scanspeed 1730, Labogene); micro-oscillator (QL-901, Haimen Kirin Medical Instrument Factor, Haimen, China); flow cytometry (EPICS®ALTRA™, Beckman). Reagents: LipofectamineTM2000 (Invitrogen); Trizol total RNA extraction kit (Invitrogen); ELISA measurement kit (Chemicon); Oligo dT Primer, Random Primer, 5× PrimeScriptTM Buffer and other PCR reagents (TAKARA); ethanol and chloroform (domestic analytical pure); interleukin-6 and interleukin-1β (Peprotechec Company); DMEM/F12 media and fetal bovine serum (Gibco).
2.3. Methods
2.3.1. In vitro culture of cells
Cells were cultured in vitro as the previously described methods[8]. The intervertebral disc was rinsed with icecold PBS three times immediately after removal, the nucleus pulposus tissue was isolated and cut into pieces in DMEM/ F12 medium, then the pieces were centrifuged at 4 000 r/min, 4 ℃ and resuspended, incubated at 37 ℃ with 5% CO2for 8 h. The supernatant was removed after centrifugation at 4 000 r/min, 4 ℃, the cells were rinsed with PBS and digested with 0.25 % trypsin and 2% Ⅱ collagenase. After another 4 ℃centrifugation at 4 000 r/min, the supernatant was removed and the cells was resuspended in DMEM/F12 medium, then the resuspended cells were counted to determine the number of cells up to 104/L. Subsequently the cells at 1× 104/L were transferred to 5% CO2incubator and cultured 37 ℃. After cells were cultured for 7 d, the cells were transferred to serum-free medium for additional 24 h. The cells were divided into a control group and an experimental group, culturing in DMEM/F12 medium containing 0.3% fetal bovine serum, experimental group was added with 10 nmol/L, 20 nmol/L, 50 nmol/L, 100 nmol/L of interleukin-6 and interleukin-1β for 48 h.
2.3.2. Import and identification of NGF-siRNA
Cells at the logarithmic growth phase were incubated onto the six-well culture plate at the density of 2.5×105/mL. In accordance with the kit instructions, DEPC H2O and LipofectamineTM2000 were diluted and then mixed with NGF-siRNA, the mixture was incubated for 10 minutes and added to the culture plate (2.5×105/hole) in 37 ℃, 5% CO2incubator. The culture medium was changed promptly. The conversion rate of the cells was determined with flow cytometry to assess transfer efficiency.
2.3.3. Determination of NGF mRNA
Q-PCR primer sequence of NGF mRNA was as follows. Forward: 5’-ACT TCA GCA TTC CCT TGA CAC A-3’, Reverse: 5’-ACG GGC AGC TAT TGG TTC AG-3’, Probe: 5’-FAM-CCC TCC GCA GAG CCC GCA-TAMRA-3’; GAPDH, the reference gene, primer sequence: Forward: 5’-AGG GCT GCC TTC TCT TGT GA-3’, reverse 5’-AAC TTG CCG TGG GTA GAG TCA-3’, probe: 5’-FAM-CCA TCA ACG ACC CCT TCA TTG ACC TC-TAMRA-3’. Reverse transcription and PCR amplification was automatically performed with by PCR instrument, and paired using a conventional system. Reaction conditions: Reverse transcription: 7 ℃ for 15 min, 85 ℃ for 5 s; amplification: 93 ℃ for 2 min; 93 ℃ for 15 s, 55 ℃ for 25 s, 72 ℃ for 25 s, totally 40 cycles; 72 ℃ for 10 min. Ct value (the number of copies/µL cDNA) refers to the number of cycles when fluorescence signal reaches the preset threshold value, Ct values for each template has a log-linear relationship with the initial copy number,ie., the higher the initial copy number is, the smaller Ct value is. Due to the difference of total RNA concentration in different samples, NGF mRNA expression is calculated according to the following formula: Cttargetgene/Ctreferencegene.
2.3.4. Determination of NGF
Cell culture medium 100 µL were collected to measure NGF levels in strict accordance with the ELISA kit instructions.
2.4. Statistical analysis
Data were analyzed using SPSS 17.0 software. NGF mRNA expression and NGF levels were normally distributed and expressed as mean±SD. The difference before and after the introduction NGF-siRNA was compared with pairedt-test. Multiple-group comparison was performed using one-wayanalysis of variance. α = 0.05 was considered as significant difference.
3. Results
3.1. Identification of cells
Toluidine blue staining showed that the nucleus pulposus cells stained were strongly positive (Figure 1A); fibrous ring cells were purple in nuclei and dark blue in cytoplasm, most of the stained cells were fusiform shaped, a large number of vacuoles were visible in the cytoplasm (Figure 1B). Safranin O staining showed that, nucleus pulposus cells stained were positive, the cytoplasm was pink, and brownish red perinuclear granules were visible (Figure 2A); fibrous ring cells were lightly stained (Figure 2B). Cells were successfully cultured in vitro, nucleus pulposus cells and fibrous annulus cells were obtained.
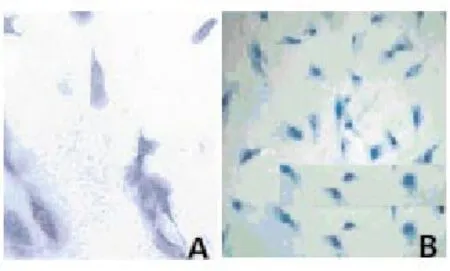
Figure 1. Dyeing appraisal results of intervertebral disc nucleus pulposus cells and annulus fibrosus cells of rats.NOTE: A, nucleus pulposus cells, B, annulus fibrosus cells; dyed with toluidine blue, ×200.
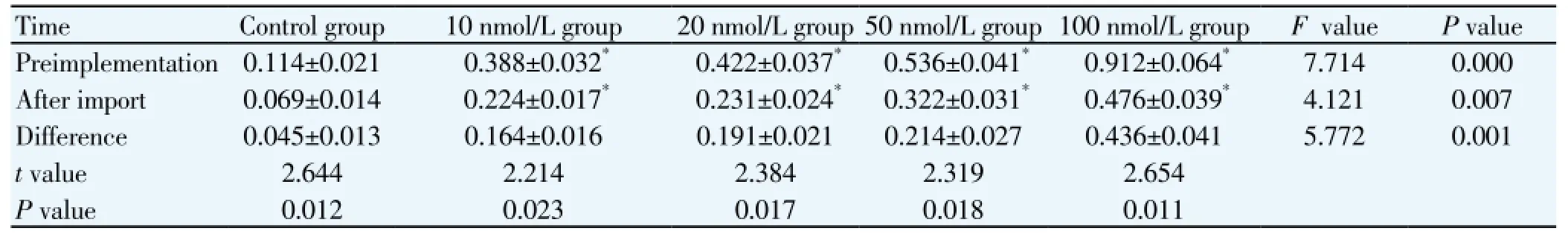
Table 1 Compare of NGF mRNA expression before and after NGF-siRNA interference (mean±sd, nmol/L).
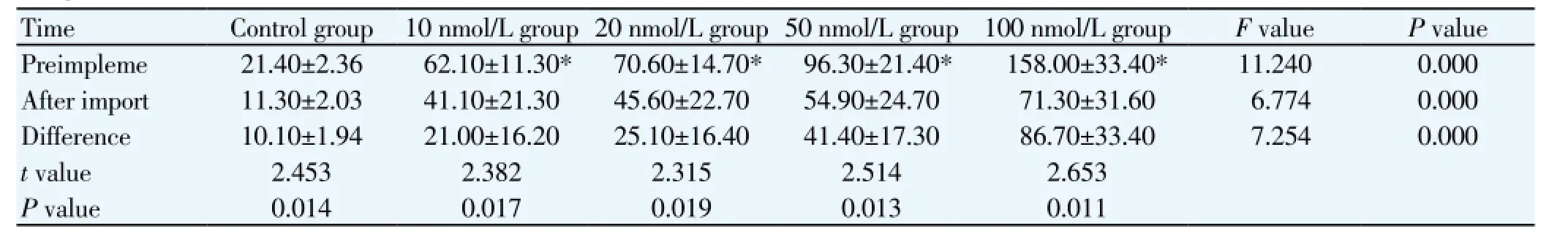
Table 2 Compare of NGF contents before and after NGF-siRNA interference (man±sd, ng/L)
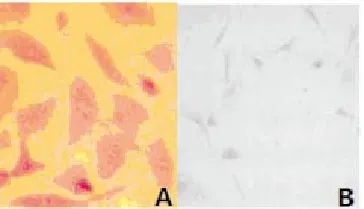
Figure 2. Dyeing appraisal results of intervertebral disc nucleus pulposus cells and annulus fibrosus cells of rats.NOTE: A, nucleus pulposus cells, B, annulus fibrosus cells; dyed with sarranine, ×200.
3.2. Conversion rate of cells
Flow cytometry analysis showed that, NGF-siRNA cell conversion rate was up to 99.8% (Figure 3).
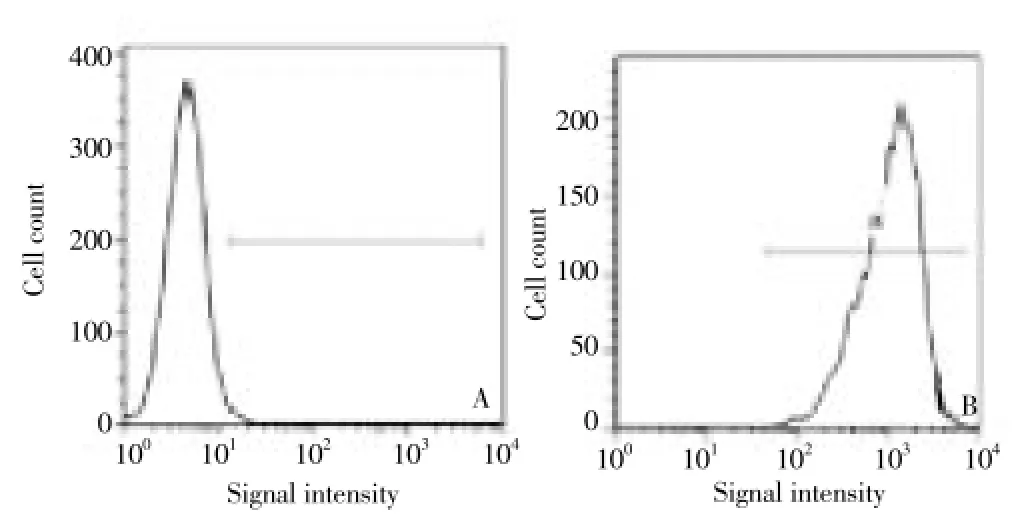
Figure 3. Flow cytometry results of cultured cells before and after NGF-siRNA.(A) Before NGF-siRNA; (B) After NGF-siRNA; marked by Cy5.
3.3. NGF mRNA relative expression
Q-PCR results of GAPDH and NFG mRNA are shown inFigure 4. A and B are respectively the standard amplification curve of GAPDH and NFG mRNA. The standard amplification curve at different concentrations showed good parallelism and surface smooth, indicating that the amplification conditions were accurate with a high specificity. C and D are respectively the standard linear regression graphs of GAPDH and NFG mRNA. The linear regression graphs have a good regression and high reproducibility, and the regression coefficientR2was>0.99. E and F are respectively the specimen amplification curve of GAPDH and NFG mRNA. Before the NGF-siRNA intervention, NGF mRNA expression in 10 nmol/L, 20 nmol/L, 50 nmol/L, 100 nmol/L groups was increased 3.4, 3.7, 4.7, 8.0 times compared with the control group. After NGF-siRNA intervention, NGF-mRNA relative expression in each group was decreased 39.5%, 45.5%, 45.3%, 39.9%, 47.8% compared with before intervention (P<0.05; Table 1).

Figure 4. Q-PCR results of GAPDH and NFG mRNA.A and B represent standard amplification curve for GAPDH and NFG mRNA; C and D represent standard regression line for GAPDH and NFG mRNA; E and F represent sample amplification curve for GAPDH and NFG mRNA.
3.4. Determination of NGF levels
Before NGF-siRNA intervention, NGF levels in 10 nmol/L, 20 nmol/L, 50 nmol/L, 100 nmol/L groups were increased 2.9, 3.3, 4.5, 7.4 times compared with the control group; after NGF-siRNA intervention, NGF levels in each group were decreased 47.2% , 33.8% , 35.4%, 43.0%, 54.9% compared with before intervention (P<0.05; Table 2).
4. Discussion
DLBP is also known as intradiscal disorders, showing a progressive degeneration. As the progress of the disease, adjacent segments may be affected, eventually involving the whole vertebral system[9]. From the view of morphological pathology, DLBP patients are accompanied with multiple fractures and damage in intervertebral disc and nucleus pulposus, as well as loss of bone density, leading to abnormal lordosis, motion segment instability and neurological mechanical injury; from the view of neuropathology, increased innervation density, lower nociceptor threshold value, and a wide range of painsensitive reaction are detected in DLBP patients; from the view of biological pathology, the mechanical injury and pain-sensitive response in DLBP patients are mediated by inflammatory reaction, they are mutually complementary and interdetermined.
Previous studies[11,12] have shown that, a large number of inflammatory mediators such as tumor necrosis factorα, interleukin-1β, interleukin-6 are involved in the process of intervertebral disc damage, and their contents in intervertebral disc are greatly changed at acute onset period. This evidence suggests that inflammatory mediators are an important incentive for the exacerbations of DLBP, but the underlying mechanism remains unclear. In this study, inflammatory intervertebral disc cells were treated with interleukin-1β and interleukin-6. The results showed that, before the intervention of interleukin-1β and interleukin-6, there were low NGF levels and weakly positive mRNA expression in the model of intervertebral disc inflammation established using Freund’s adjuvant; after the intervention, NGF levels and NGF mRNA expression were significantly up-regulated, in a dose-dependent manner. Along with the increase of interleukin-1β and interleukin-6 concentrations, the NGF levels and NGF mRNA expression was gradually increased. This evidence suggested that secretion of inflammatory cytokines interleukin-1β and interleukin-6 is the important cause of NGF activation in the rat intervertebral disc. NGF supplies one of the basic nutrients for autologous neurons and may participate in a series of fundamental physiological functions of the nerve cells, such as growth, development, repair, and regeneration. NGF is ubiquitously distributed in the rat embryo intervertebral discs and can maintain the spinal nerve cells differentiation and proliferation[13]. In healthy adult rats, NGF is mainly located at the outer edge of intervertebral disc, and scarcely seen in nucleus pulposus; their abnormal expression is closely related to the initiation of self-repair of damaged nerve cells, thus having been widely concerned in bone and neurosurgery injury research[14]. In addition, NGF abnormal expression is considered the major pathological mechanism of nerveingrowth in DLBP, and inflammatory pain signaling in DLBP is mainly mediated by NGF-dependent neurons[15]. NGF can not only induce the nerve ingrowth in DLBP, but also involves inflammatory pain sensitization. Therefore, we hypothesized that NGF abnormal expression caused by interleukin-1β and interleukin-6 is an important mechanism of intervertebral disc injury. Based on the above speculation, we preliminarily determined the NGF abnormal expression using siRNA interference, the results showed that NGF content and mRNA expression levels were significantly reduced after the intervention of NGF-siRNA. This finding indicates that, NGF siRNA interference can inhibit the stimulation effect of proinflammatory cytokines interleukin-6 and interleukin-1β on NGF in intervertebral disc cells, which provide a novel approach for the treatment of DLBP.
The present study revealed the possible mechanism of proinflammatory cytokines interleukin-6 and interleukin-1 β-induced DLBP from the in vitro cell levels, and provided crucial data for the feasibility of NGF-siRNA interference in the treatment of DLBP. However, the limitations of this study lies on the use of cultured rat intervertebral disc cells in vitro. On the one hand, we prepared the models of inflammatory intervertebral disc using Freund’s adjuvant according to the principle of that, direct stimulation of 65 kD heat shock protein, an arthritis-induced antigen in Mycobacterium tuberculosis, could trigger inflammatory reaction within 10-20 days and reach the peal at 20 days. The exact mechanism associated with DLBP is very complex, and most of affected patients suffer from a long-term, chronic lesion. Therefore Freund’s adjuvant-caused model of inflammatory disc only partially mimics inflammatory reaction in the DLBP, and the establishment of models needs further improvement. On the other hand, the present study only confirmed the in vitro applicability of NGF-siRNA interference in the treatment of DLBP, and further investigations are needed for in vivo tests.
Conflict of interest statement
We declare that we have no conflict of interest.
[1] Tang SJ, Huang QS. Diagnosis and treatment progress of intervertebral disc source sex lumbago. Chin J Orthopaedic Surg 2012; 17(17): 1307-1310.
[2] Wang HH. The diagnosis of intervertebral disc source sex lumbago progress. J Clinical Pract Hospital 2012; 9(6): 205-208.
[3] Hurri H, Karppinen J. Discogenic pain. Pain 2013; 112(3): 225-228.
[4] Aoki Y, Takahashi Y, Ohtori S, Moriya H, Takahashi K. Distribution and immunocytochemical characterization of dorsal root ganglion neurons innervating the lumbar intervertebral disc in rats: a review. Life Sci 2011; 74(2): 2627-2642.
[5] Guan Y, Ni JX. Cells of the intervertebral disc source sex lumbago pathology research progress. J Reh Med Chin 2012; 27(7): 683-686.
[6] Ozawa T, Ohtori S, Inoue G, Doya H, Saito T, Iti T, et al. The degenerated lumbar intervertebral disc is innervated primarily by peptide-containing sensory nerve fibers in humans. Spine 2012; 31(21): 2418-2422.
[7] Kakutani K, Nishida K, Uno K, Takada T, Shimomura T, Maeno K, et al. Prolonged down regulation of specific gene expression in nucleus pulposus cell mediated by RNA interference in vitro. J Orthop Res 2012; 24(6): 1271-1278.
[8] Wang G, Han DF, Shi YZ, Chen ZW, Li YQ, Liu SL. IL - 1 beta and IL-6 on the influence of the intervertebral disc nucleus pulposus cells model NGF expression. Chin J Pathol Physiol 2010; 26(7): 1265-1269.
[9] Freemont AJ, Peacock TE, Goupille P, Hoyland JA, O’Brien J, Jayson MI. Nerve ingrowth into diseased intervertebral disc in chronic back pain. Lancet 2011; 350(9072): 178-181.
[10] Coppes MH, Marani E, Thomeer RT, Oudega M, Groen GJ. Innervation of “painful” lumbar discs. Spine 2012; 22(20): 2342-2350.
[11] Wang G, Liu SL, Cheng ZA, Shi YZ, Han DF, Huang DS. The influence of lumbago discectomy for lumbar intervertebral disc protrusion. Chin J Orthop Surg 2009; 17(6): 23-26.
[12] Roberts S, Eisenstain SM, Menage Y, Evans EH, Ashton IK. Mechanoreceptors in intervertebral discs. Morphology, distribution and neuropeptides. Spine 2012; 20(24): 2645-2651.
[13] Abe Y, Akeda K, An HS, Aoki Y, Pichika R, Muehleman C, et al. Proinflammatory cytokines stimulate the expression of nerve growth factor by human intervertebral disc cells. Spine 2011; 32(6): 635-642.
[14] Han D, Ding Y, Liu SL, Wang G, Si IC, Wang X, et al. Double role of Fas ligand in the apoptosis of intervertebral disc cells in vitro. Acta Biochim Biophys Sin (Shanghgai) 2009; 41(11): 938-947.
[15] Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 2013; 391(6669): 806-811.
ment heading
10.1016/S1995-7645(14)60127-9
*Corresponding author: Peng Fu, Professor, M.D., Supervisor of Doctoral Students, Shengli Oilfield Central Hospital, No.31 of Jinan Road of Dongying, Dongying 257034, China.
E-mail: Lala0815@126.com
Foundation project: It is supported by Shandong Province Natural Science Foundation (28172a2).
 Asian Pacific Journal of Tropical Medicine2014年9期
Asian Pacific Journal of Tropical Medicine2014年9期
- Asian Pacific Journal of Tropical Medicine的其它文章
- Sonic Hedgehog signaling pathway in primary liver cancer cells
- Runx3 might participate in regulating dendriti cell function in patients with irritable bowel syndrome
- Protection effect of Emodin pretreatment on intestinal I - RI damage of intestinal mucosa in ratsa
- Protective effect and mechanism of lithium chloride pretreatment on myocardial ischemia-reperfusion injury in rats
- Protective effects of Ginseng mixture on myocardial fibrosis in rats
- Nerve protective effect of rhTPO and G-CSF on hypoxic ischemic brain damage in rats
