Mapping and validation of a dominant salt tolerance gene in the cultivated soybean(Glycine max) variety Tiefeng 8
Rongxia Guan,Jiangang Chen,Jinghan Jiang,Guangyu Liu,Ying Liu,Lei Tian,Lili Yu,Ruzhen Chang,Li-juan Qiu*
The National Key Facility for Crop Gene Resources and Genetic Improvement(NFCRI),Key Laboratory of Germplasm&Biotechnology(MOA),Institute of Crop Sciences,Chinese Academy of Agricultural Sciences,Beijing 100081,China
1.Introduction
Soybean(Glycine max[L.]Merr.)is classified as a salt-sensitive glycophyte [1].In soybean,salinity stress inhibits seed germination and seedling growth.Several salt-tolerant soybean accessions have been identified[2,3].To characterize the inheritance of salt tolerance,Abel [4] performed crosses of parents differing in chloride accumulation tolerance.The F2population segregated in a 3:1 ratio of non-necrotic plants(very low in chloride)to necrotic plants(very high in chloride),and the segregation ratio from a test cross was 1:1.From these results,it was concluded that exclusion and inclusion of chloride in soybean leaves and stems were controlled by a single dominant gene Ncl and a recessive gene ncl,respectively.Lee et al.[5]evaluated the salt tolerance of a F2:5population derived from the cross of salt-tolerant parent S-100 with salt-sensitive parent Tokyo.Salt tolerance was scored from 0(all plants dead)to 5(plants with normal green leaves).As a result,a major quantitative trait locus (QTL) for salt tolerance was mapped near the simple sequence repeat(SSR) marker Sat_091 on soybean linkage group N (chromosome 3),accounting for 41%,60%,and 79% variation of total genetic variation under three different conditions.The authors speculated that this QTL was the dominant gene Ncl reported by Abel [4].The major QTL in this genomic region was confirmed in the cultivated soybeans FT-Abyra and Jindou No.6 as well as a wild-type soybean,JWS156-1 [6,7].Another salt tolerance-associated QTL was identified in a Chinese cultivar Nannong 1138-2 and mapped between SSR markers Sat_164 and Sat_358 on chromosome 18 [8].Besides these QTLs,allelism testing showed that a wild soybean accession (Glycine soja) PI483463 carried a single dominant salt-tolerance gene,different from the one found in G.max S-100,but also mapping to soybean chromosome 3 [9,10].Recently,an ion transporter gene GmCHX1,located on soybean chromosome 3,was cloned from wild soybean by a combination of genome sequencing and QTL mapping[11].
Thousands of Chinese soybean accessions have been screened for salt tolerance,and several Chinese cultivated soybeans cultivars have been identified as sources of salt tolerance at seedling and adult stages [3,12].Genetic analysis using F1hybrids and F2and F2:3populations constructed from crosses between tolerant varieties Wenfeng 7,Teifeng 8,and Jindou 33 and sensitive cultivars Union,Hark,and Zaoshu 6 revealed that salt tolerance in soybean at the seedling stage was controlled by a single dominant gene[13].Using the same crosses as those used by Shao and Chang [13],Guo et al.[14]found a codominant RAPD marker (a 600 bp product in the sensitive parent and a 700 bp product in the tolerant parent)closely linked to the salt-tolerance gene,but its location was not determined.In the present study,we mapped the dominant salt-tolerance gene on chromosome 3 and used salt tolerance-linked markers for determining the salt tolerance of soybean accessions.
2.Materials and methods
2.1.Development of mapping populations
Soybean variety Tiefeng 8 had been determined to be salt-tolerant,whereas 85-140 was relatively sensitive[12,13,15].A segregating F2population (n = 392) was developed from the cross 85-140 × Tiefeng 8 in 2005.The F1plant was verified using SSR markers polymorphic between the parents [16],and a F2:3family (n = 239) with sufficient seed was used for mapping the salt tolerance gene.To confirm the gene detected in the F2:3population,an F5:6RIL mapping population (n = 230)developed by single-seed descent from the F2:3families was employed.
2.2.Evaluation of salt tolerance
Twelve seeds of each F2:3line were planted in a 100 ×80 × 80-mm pot filled with vermiculite and watered with tap water from the bottom every three days.The experiment was conducted under a rainout shelter under ambient weather conditions at the Chinese Academy of Agricultural Sciences in July 2009.The daily air temperature ranged from 33 (daily highest) to 21 °C (night lowest) with an average daily temperature of approximately 25 °C.Each of these experiments was conducted with two replications,and each pot had eight to ten plants.Twenty-two F2:3lines and the two parents were placed in 600 × 400 × 80-mm tanks.Ten days after planting,when the true leaves of plants were fully expanded,2 L of 200 mmol L-1NaCl was added twice within six days to each tank.Thereafter,2 L of tap water was added to each tank every three days.When the salt-sensitive parent 85-140 appeared salt-toxicity symptoms,about 2 weeks after treatment,the genotype of each family was scored for salt tolerance.Lines with no obvious salt toxicity were considered tolerant,whereas lines with obvious leaf scorch were considered sensitive.Lines appearing both symptomatic and asymptomatic were considered heterzygous.
Salt-tolerance evaluation of the F5:6RIL population and 58 cultivated soybean accessions was performed in July of both 2011 and 2012,respectively,in two replications as described for F2:3families.After treatment for 2 weeks,the RILs(n = 8–10)were scored for salt tolerance.A salt-tolerance rating for each of the 58 soybean accessions was assigned according to the level of leaf chlorosis (Table S1).The salt-tolerance ratings ranged on a scale from 1 to 5,as follows:1)plants with normal green leaves; 2) less than 25% leaf area showing chlorosis; 3)less than 50% leaf area showing chlorosis; 4) more than 50%leaf area showing chlorosis,but plants not completely dead;5)all plants dead.The salt-tolerance evaluations of the RIL population and the 58 soybean accessions were both performed twice.
2.3.DNA isolation,RAPD analysis,and SCAR marker development
Leaf samples from all F2and F5individuals and the parents were collected and frozen in liquid nitrogen.For the germplasm used for marker-assisted selection and two parents,leaf samples were collected from ten plants of each accession.DNA was isolated with a Genomic DNA Purification kit(Fermentas,Burlington,Canada) and diluted to a final concentration of 20 ng μL-1.PCR was conducted in a 20-μL reaction mixture containing 40 ng DNA and 1 U Ex Taq(TaKaRa,Japan) using RAPD primer 5′-GGCACGTAAG-3′under the following conditions: 95 °C for 5 min,followed by 35 cycles of 95 °C for 30 s,37 °C for 30 s,and 72 °C for 1 min.The PCR products were separated on 2% agarose gel with 1× TAE buffer at 40 V for 3 h and stained with ethidium bromide.
The RAPD polymorphic fragments present in the salttolerant and-sensitive parents were excised and purified from the agarose gel and then cloned into the pEASY-T1 Cloning Vector (TransGen Biotech,Beijing,China).The positive (white) colonies were verified by PCR and the insert was sequenced on an ABI 3730XL Capillary DNA Sequencer using the fluorescent dye terminator method.A sequencecharacterized amplified region (SCAR) marker was designed from the sequence information of the two fragments.The SCAR marker(QS08064),amplified by using forward 5′-ACGTAAGTGG TTGAAGGCGTT-3′and reverse 5′-GGGCAAGGGATATGAAAA-3′primers,was used to genotype from both parents and the individuals in both populations.PCR was performed in 20 μL reaction mixtures containing 1× PCR buffer,200 μmol L-1MgCl2,150 μmol L-1dNTPs,150 μmol L-1of each primer,40 ng DNA,and 1 U Taq-polymerase (Takara).PCR conditions were as follows: 94 °C for 5 min followed by 35 cycles of 94 °C for 30 s,55 °C for 30 s,72 °C for 45 s,and a final extension of 5 min at 72 °C.The PCR products were separated on 2% agarose gels with 1× Tris-acetate-EDTA(TAE) buffer at 100 V for 40 min and stained with ethidium bromide.
2.4.SSR marker analysis
Forty-two SSR markers from soybean chromosome 3 were tested for polymorphism between the two parents and nine polymorphic markers were identified.The SSR profile was the same as for the SCAR analysis.PCR products were then separated on 6% denaturing gels (19 acrylamide:1N,N′-methylene-bis-acrylamide) containing 7 mol L-1urea with 1× TBE for 1.5 h at 100 W,and the fragments were visualized with silver staining [17].
2.5.Development of InDel markers for gene mapping
The soybean reference sequence of Williams 82(http://www.phytozome.net/soybean) and information about insertions and deletions in wild soybean(ftp://147.46.250.193/gff/)were used for developing InDel markers(Table 1).A total of 9 pairs of primers flanking 3–9-bp insertion/deletions were designed with Primer 3 software (http://primer3.sourceforge.net/),with Tm values of 54–56 °C.Four of them were polymorphic InDel markers and were used for mapping the salt-tolerance locus.
2.6.Genetic mapping of the salt-tolerance gene
Thirteen polymorphic markers were used to screen the DNA from the individuals of the F2and RIL populations.The IciMapping software (v3.3; http://www.isbreeding.net/) was used to calculate genetic linkages and map distances[18].
2.7.Phylogenetic analysis
Phylogenetic analysis of 58 soybean accessions was done based on the neighbor-joining (NJ) method implemented in PowerMarker version 3.25 [19],and the neighbor-joining tree was viewed using MEGA 4.0[20].

Table 1-Sequences of SCAR and InDel primer pairs and the sizes of PCR products generated by each primer pair.
3.Results
3.1.Phenotypic variation in F2:3 and RIL populations
To characterize the variation for salt tolerance in the two populations derived from Tiefeng 8 and 85-140,the 239 and 230 lines of the F2:3segregating and RIL(F5:6)populations were evaluated for salt tolerance.Based on leaf chlorosis,the offspring in the F2:3family showed an expected segregation ratio of 1:2:1(χ2= 7.03 <χ2(2)0.01= 9.21)(tolerant,42;segregating,132; sensitive,65).In the RIL population,95 RILs showed salt tolerance and 114 RILs showed salt sensitivity,fitting a ratio of 1:1(tolerant: sensitive) (χ2= 1.55 <χ2(1)0.01= 3.84).
3.2.Development of a codominant SCAR marker and mapping of the salt-tolerance gene
The two polymorphic fragments amplified by the RAPD primer from cultivated soybeans Tiefeng 8 and 85-140 were cloned and sequenced[14].The allele size of the RAPD marker was 767 bp for the salt tolerance and 655 bp for the salt sensitive,respectively.The DNA sequence identity between tolerance and sensitive alleles was 80.8%,with 4 InDels (10,129,6,and 2 bp) and 2 single-nucleotide polymorphisms(SNPs) (Fig.1).The corresponding soybean sequences were searched with BLAST (Basic Local Alignment Search Tool)against the Phytozome soybean database (http://www.phytozome.net/).The two fragments from Tiefeng 8 and 85-140 showed identities of 92.2% and 88.4%,respectively,with a reference DNA fragment that is part of gene Glyma03g32920.1 between nucleotides 40,650,610 and 40,651,263 on chromosome 3.A pair of SCAR primers was designed according to the insertions and deletions in the sequences of Tiefeng 8 and 85-140,with expected fragments of 383 bp and 263 bp,respectively(Table 1,Fig.1).
To map the salt-tolerance gene,42 SSR markers on soybean chromosome 3 were selected from SoyBase (http://soybase.org/) and used to identify polymorphism between Tiefeng 8 and 85-140,and nine of them showed polymorphism.The 239 F2plants were genotyped with the nine SSR markers and one SCAR marker QS08064.The salt-tolerance gene was closely linked with QS08064 and located between the SSR markers Barcsoyssr_3_1301 and Barcsoyssr_3_1315(Fig.2-A).
For further narrow the mapping region for the salt-tolerance gene of the salt-tolerance locus,an additional four InDel markers(Table 1)between Barcsoyssr_3_1301 and Barcsoyssr_3_1315 were developed based on the insertion/deletion information of a deeply sequenced wild soybean [21] and used to genotype both the F2and RIL populations (Fig.2-A,B).Representative recombinant individuals and lines from F2and RIL populations displaying fragments differing in length between marker QS1112 and Barcsoyssr_3_1301 were shown in Fig.2-C.All lines displaying the fragment from Tiefeng 8 between markers Barcsoyssr_3_1301 and QS08064 were salt-tolerant.Accordingly,the salt-tolerance gene was assigned to the 209-kb interval between Barcsoyssr_3_1301 and QS08064.

Fig.1-Alignment of DNA fragments from salt-tolerant and-sensitive parents.
3.3.Salt tolerance of soybean germplasm and relationship between phenotype and genotype
Under salt treatment,the phenotypes of soybean accessions were scored according to leaf chlorosis on a visual rating scale ranging from 1 to 5.The salt-tolerant variety Tiefeng 8(leaf scorch 1)and the sensitive line 85-140(leaf scorch 4)were used as references.As shown in Table S1,17 and 18 accessions showing superior salinity tolerance had leaf scorch ratings of 1 and 2,respectively,whereas 23 accessions showing different levels of salt sensitivity had leaf scorch ratings from 3 to 5.To evaluate the relationship between salt tolerance and allelic variation of markers associated with the salt tolerance gene,58 soybean accessions were screened with SCAR marker QS08064,two SSR markers,Barcssoysr_3_1306,Barcsoyssr_3_1310,and InDel marker QS080465,which cosegregated with the salt-tolerance gene (Fig.2-B).For SCAR marker QS08064,32 of the 35 tolerant accessions displayed the expected 383-bp allele.For the InDel marker QS080465,most (33 of 35) of the salt-tolerant accessions displayed the expected 152-bp allele(Table 2,Fig.3).Two salt-tolerant accessions,Cutler 71 and Franklin,carried salt sensitivity-related alleles of size 148 bp at locus QS080465 and 263 bp at locus QS08064,whereas the salt-sensitive accession Peking carried tolerance-associated alleles at both loci.Six and four alleles were detected at SSR loci Barcsoyssr_3_1310 and Barcsoyssr_3_1306,respectively,with one allele at locus Barcsoyssr_3_1310 and 3 alleles at locus Barcsoyssr_3_1306 being detected primarily in the salt-tolerant accessions and the other six alleles detected mainly in the salt-sensitive accessions (Table S1).The 58 accessions were clustered into three different major branches on a neighbor-joining tree based on SCAR marker QS08064,SSR markers Barcsoyssr_3_1310 and Barcsoyssr_3_1306 and InDel marker QS080465;two groups contained salt-sensitive soybeans and the third contained tolerant ones (Fig.4).These results indicate that markers closely linked to the salt-tolerance gene can be used for marker-assisted selection of salt-tolerant soybean.
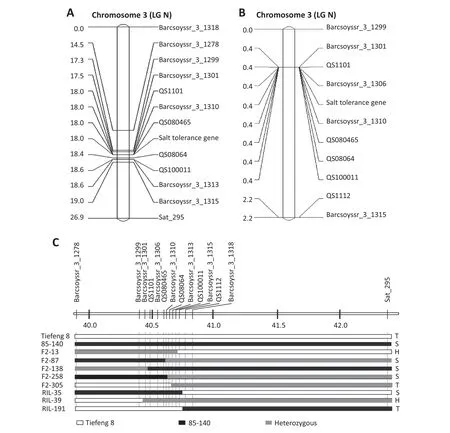
Fig.2-Linkage map of F2 population(A)and RIL population(B) for the cross of 85-140 × Tiefeng 8,showing the positions of molecular markers and salt-tolerance gene.Markers are shown to the right and distances in cM to the left of the chromosome;(C)Graphical genotypes of representative recombinant lines of F2 and RILs produced from 85-140 × Tiefeng 8 in the candidate region of chromosome 3.T,S,and H indicate that the recombinant line is salt-tolerant,salt-sensitive,or heterozygous(segregating),respectively.
4.Discussion
The salt-tolerance phenotype in varieties Tiefeng 8 and Wenfeng 7 is a monogenically controlled trait,and a codominant RAPD marker closely linked to the salt tolerance gene was identified [13,14].In this study,we sequenced specific fragments of RAPD marker from the salt-tolerant and-sensitive parents Tiefeng 8 and 85-140 and converted the RAPD marker into a codominant SCAR marker.BLAST search of the specific fragments showed that it was located on chromosome 3,on which a conserved salt-tolerance QTL has been mapped [5–8,10].Using F2:3and RIL populations derived from the cross of Tiefeng 8 and 85–140,we also mapped the dominant salt-tolerance gene on chromosome 3 in an interval of 209 kb,suggesting coincidence with the QTL located previously [5,6,15].The candidate region detected in PI 483463 was 658 kb,overlapping the interval of this study [10].The consistency of the mapping region of the salt-tolerance gene/QTL in different soybean backgrounds suggests that this gene/QTL is a major locus controlling salt tolerance in soybean.According to the annotation of the Williams 82 genome,22 genes are predicted to be located in the 209 kb region,including sodium/hydrogen exchanger (CHX) families (Glyma03g32890.1 and Glyma03g32900.1).In the Arabidopsis CHX family,several members including AtCHX13,AtCHX17,and AtCHX20 have been shown to play a role in potassium transport [22–24],and AtCHX21,expressed in root endodermal cells,has been shown to regulate Na+concentration in the xylem [25].Recently,a major QTL for salt tolerance in G.soja,W05,was mapped to a 388 kb region on soybean chromosome 3,and GmCHX1(Glyma03g32900.1) was confirmed as a novel gene conferring salt tolerance by use of a root-hair transformation system[11].In our previous study,the salt-tolerant parent Tiefeng 8 was found to accumulate significantly less Na+in the shoot than the salt sensitive parent 85-140[12,26].Valencia et al.observed that salt-sensitive soybean cultivars accumulated 2.64 and 1.96 times more Na+and Cl-in leaves than tolerant accessions[27].However,no DNA markers associated with Na+or Cl-content have been mapped on soybean chromosome 3 where the salt-tolerance gene has been mapped.Thus,further studies should be conducted to understand the mechanisms underlying salt tolerance in soybean.
The identification of the salt-tolerance QTL/gene on chromosome 3 in different backgrounds suggests the possibility of marker-assisted selection for salt-tolerant soybean varieties using the markers associated with the salt-tolerance gene.We screened 58 diverse soybean accessions with three DNA markers,Barcsoyssr_3_1306,Barcsoyssr_3_1310 and QS080465,which cosegregated with the salt-tolerance locus.The InDel marker QS080465 was developed from the promoter region of a sodium/hydrogen exchanger gene,Glyma03g32890.1,whereas SSR marker Barcsoyssr_3_1310 was located in the intron region of Glyma03g32900.1.Of the 35 soybean accessions carrying the 152-bp tolerance allele at locus QS080465,33 were salt-tolerant,indicating a selection efficiency of 94.3%.The selection efficiencies of Barcsoyssr_3_1306 and Barcsoyssr_3_1310 were 76.2%and 90.1%,respectively (Table 2).We also tested the selection efficiency of SCAR marker QS08064 and found that 32 of 40 accessions with the 383-bp tolerance allele were salt-tolerant,corresponding to a selection efficiency of 80.0%.
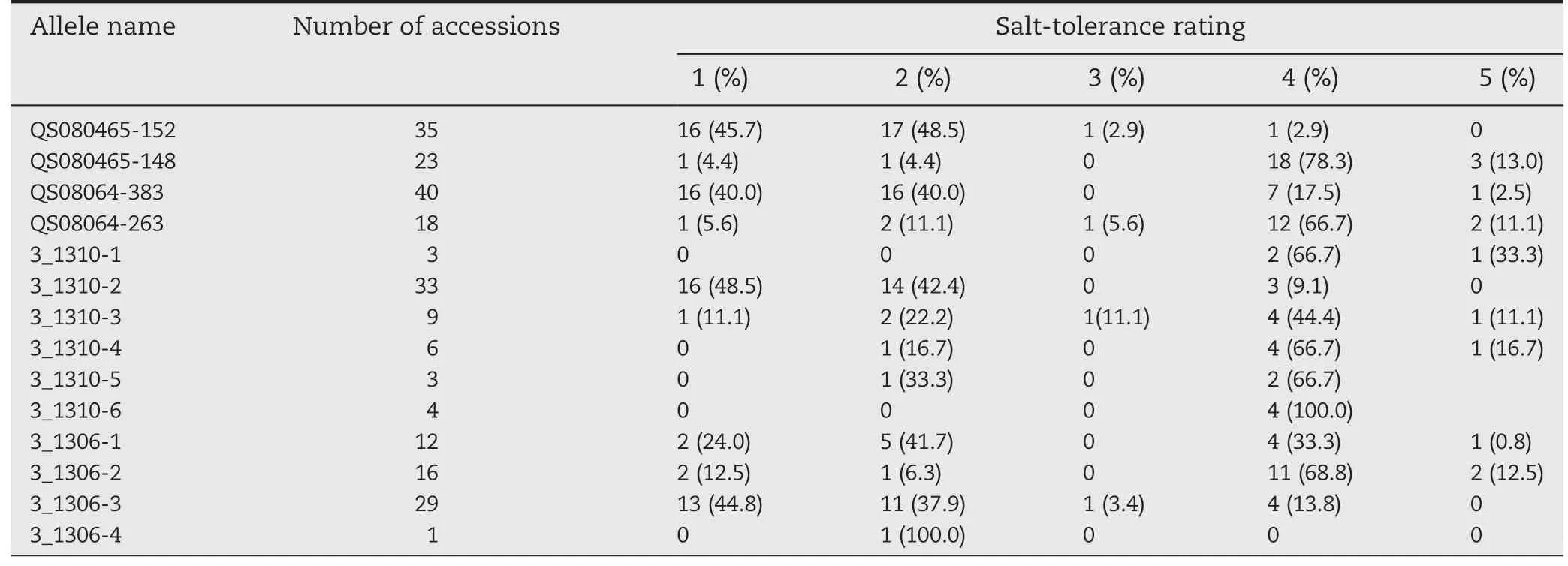
Table 2-Distribution of alleles of QS080465,QS08064,Barcsoyssr_3_1310,and Barcsoyssr_3_1306 in 58 soybean accessions with different salt-tolerance rating.
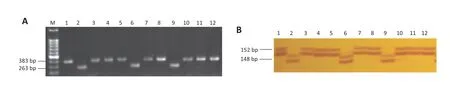
Fig.3-Allelic patterns of SCAR marker QS08064(A)and InDel marker QS0465(B) in 12 of 58 soybean accessions.M,molecular-weight marker(100-bp ladder);Numbers above images are the codes of individuals in Table S1.
In previous studies,Lee et al.[5]and Hamwieh et al.[7]tried to study the association between the salt-tolerance QTL and linked SSR markers (Sat_091 and Satt237) by tracing the pedigrees of S-100 and FT-Abyara,the parents used in QTL mapping.They found in both studies that the salt-tolerance alleles were always associated with the salt-tolerance phenotype in the descendants.However,they found different SSR marker alleles in S-100 and FT-Abyara.This finding may reflect the nature of SSR markers,which can amplify up to 34 alleles at one locus in different backgrounds [28–30],while the same allele may not always be associated with salt tolerance in different backgrounds,as we observed for Barcsoyssr_3_1306 and Barcsoyssr_3_1310 in the present study.Only two alleles for InDel marker QS080465 and SCAR marker QS08064 were identified among the 58 soybean accessions.QS080465 appeared to have a higher selection efficiency than did QS08064.The salt-sensitive variety Williams 82 carried the salt-sensitive allele at both QS08064 and QS080465(Table S1).Although six and four alleles were identified within the 58 soybean accessions at the SSR loci Barcsoyssr_3_1310 and Barcsoyssr_3_1306,an association between alleles and salt tolerance was observed.Most of the salt-tolerant and-sensitive soybeans were observed to cluster in different clades based on the data from the four markers linked with the salt-tolerance gene(Fig.4).Taking together the mapping of a salt-tolerance QTL/gene in the same interval of chromosome 3 in both wild and cultivated soybean and the high selection efficiency of markers located in the interval suggests that the salt-tolerance gene is relatively common in soybean germplasm and that all these QTLs or genes correspond to the Ncl locus initially reported by Abel [4].However,reliable evidence is needed to confirm that the same gene is responsible for salt tolerance in both wild and cultivated soybean.
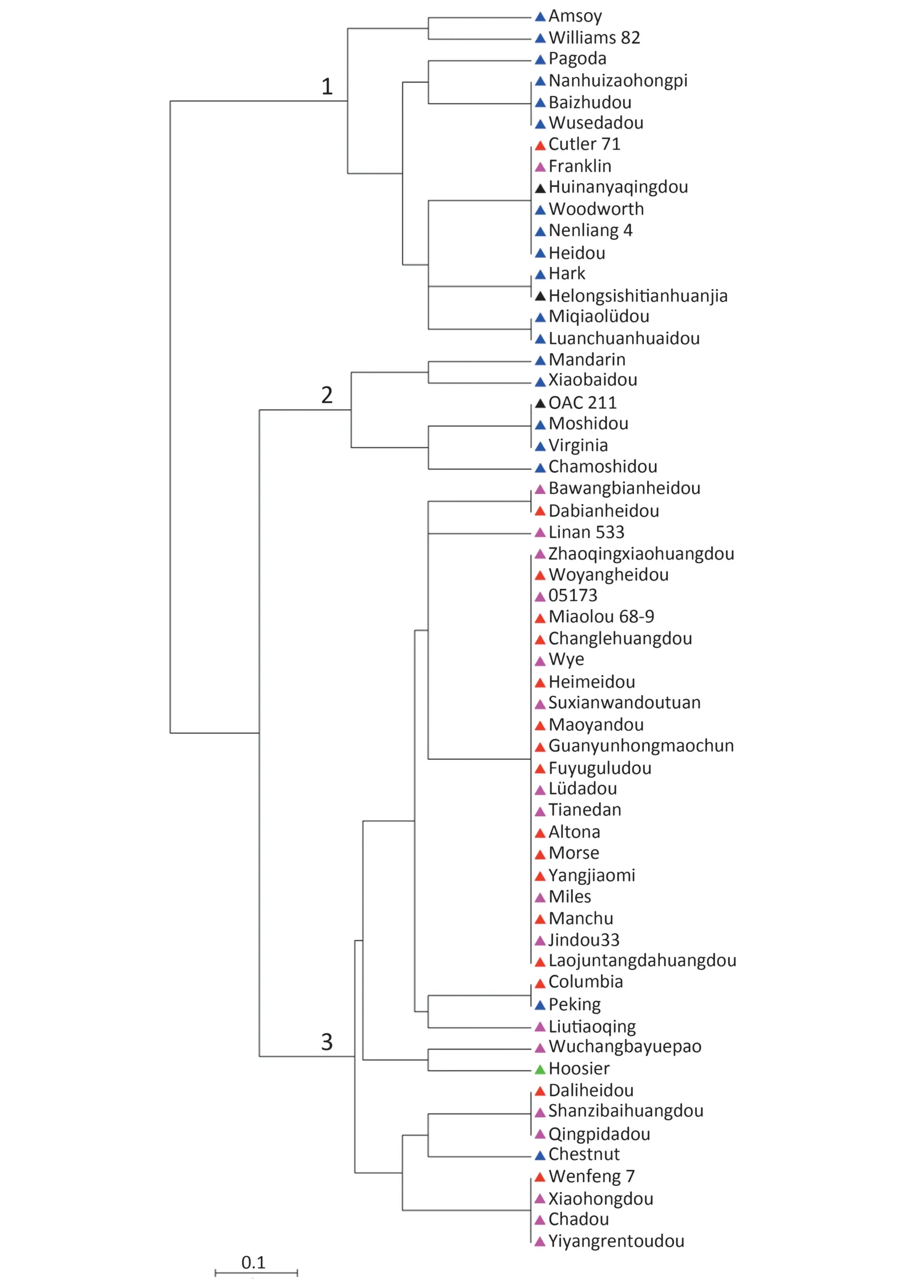
Fig.4-Neighbor-joining tree of 58 soybean accessions based on SCAR marker QS08064,InDel marker QS080465,and SSR markers Barcsoyssr_3_1310 and Barcsoyssr_3_1306.Taxon labels correspond to the accession names shown in Table S1.Salt-tolerance ratings are depicted as triangles color-coded as follows:red:rating 1,purple:rating 2,green:rating 3,blue:rating 4,and black:rating 5.Numbers at nodes represent subgroups.
This work was financially supported by the National Natural Science Foundation of China (30971801,31271752,30490250-1),the National Key Technologies R&D Program in the 12th Five-Year Plan (2012AA101106),the National Foundation for Transgenic Species (2009ZX08009-088B),and the Agricultural Science and Technology Innovation Program.We thank Ben Averitt,Department of Crop and Soil Environmental Sciences at Virginia Tech,for editorial support.
Supplementary material
Supplementary table to this article can be found online at http://dx.doi.org/10.1016/j.cj.2014.09.001.
[1] A.Lauchli,Salt exclusion:an adaptation of legumes for crops and pastures under saline conditions,in:R.C.Stapes (Ed.),Salinity Tolerance in Plants,John Wiley &Sons,New York,1984,pp.171–187.
[2] G.H.Abel,A.J.MacKenzie,Salt tolerance of soybean varieties(Glycine max L.Merrill) during germination and later growth,Crop Sci.4(1964) 157–161.
[3] G.Shao,J.Song,H.Liu,Preliminary studies on the evaluation of salt tolerance in soybean varieties,Sci.Agric.Sin.6 (1986)30–35(in Chinese with English abstract).
[4] G.H.Abel,Inheritance of the capacity for chloride inclusion and chloride exclusion by soybeans,Crop Sci.9(1969)697–698.
[5] G.J.Lee,T.E.Carter Jr.,M.R.Villagarcia,Z.Li,X.Zhou,M.O.Gibbs,H.R.Boerma,A major QTL conditioning salt tolerance in S-100 soybean and descendent cultivars,Theor.Appl.Genet.109 (2004) 1610–1619.
[6] A.Hamwieh,D.H.Xu,Conserved salt tolerance quantitative trait locus(QTL) in wild and cultivated soybeans,Breed.Sci.58 (2008) 355–359.
[7] A.Hamwieh,D.D.Tuyen,H.Cong,E.R.Benitez,R.Takahashi,D.H.Xu,Identification and validation of a major QTL for salt tolerance in soybean,Euphytica 179(2011)451–459.
[8] H.T.Chen,S.Y.Cui,S.X.Fu,J.Y.Gai,D.Y.Yu,Identification of quantitative trait loci associated with salt tolerance during seedling growth in soybean(Glycine max L.),Aust.J.Agric.Res.59 (2008) 1086–1091.
[9] J.D.Lee,J.G.Shannon,T.D.Vuong,H.T.Nguyen,Inheritance of salt tolerance in wild soybean(Glycine soja Sieb.and Zucc.)accession PI483463,J.Hered.(2009) esp027.
[10] B.K.Ha,T.D.Vuong,V.Velusamy,H.T.Nguyen,J.G.Shannon,J.D.Lee,Genetic mapping of quantitative trait loci conditioning salt tolerance in wild soybean (Glycine soja) PI 483463,Euphytica 193 (2013) 79–88.
[11] X.Qi,M.-W.Li,M.Xie,X.Liu,M.Ni,G.Shao,C.Song,A.K.-Y.Yim,Y.Tao,F.L.Wong,Identification of a novel salt tolerance gene in wild soybean by whole-genome sequencing,Nat.Commun.5 (2014) 4340.
[12] G.Liu,R.Guan,R.Chang,L.Qiu,Correlation between Na+contents in different organs of soybean and salt tolerance at the seedling stage,Acta Agron.Sin.37(2011) 1266–1273(in Chinese with English abstract).
[13] G.Shao,R.Chang,Study on inheritance of salt tolerance in soybean,Acta Agron.Sin.20(1994) 721–726 (in Chinese with English abstract).
[14] B.Guo,L.Qiu,G.Shao,R.Chang,L.Liu,Z.Xu,X.Li,J.Sun,Tagging a salt tolerant gene using PCR markers in soybean,Sci.Agric.Sin.33(2000)10–16(in Chinese with English abstract).
[15] J.Jiang,R.Guan,Y.Guo,R.Chang,L.Qiu,Simple evaluation method of tolerance to salt at seedling stage in soybean,Acta Agron.Sin.39(2013)1248–1256(in Chinese with English abstract).
[16] L.Tian,R.X.Guan,Z.X.Liu,R.Z.Chang,L.J.Qiu,Verity identification of soybean hybrids using SSR markers,J.Plant Genet.Resour.9 (2009) 443–447.
[17] B.J.Bassam,G.Caetano-Anollés,P.M.Gresshoff,Fast and sensitive silver staining of DNA in polyacrylamide gels,Anal.Biochem.196 (1991) 80–83.
[18] J.Wang,H.Li,L.Zhang,L.Meng,Users' manual of QTL IciMapping.The Quantitative Genetics Group,Institute of Crop Science,Chinese Academy of Agricultural Sciences(CAAS),Beijing,China,and Genetic Resources Program,International Maize and Wheat Improvement Center(CIMMYT),Apdo,Mexico.,(2013).
[19] K.Liu,S.V.Muse,PowerMarker: an integrated analysis environment for genetic marker analysis,Soybean Sci.21(2005) 2128–2129.
[20] K.Tamura,J.Dudley,M.Nei,S.Kumar,MEGA4: molecular evolutionary genetics analysis (MEGA)software version 4.0,Mol.Biol.Evol.24 (2007) 1596–1599.
[21] M.Y.Kim,S.Lee,K.Van,T.H.Kim,S.C.Jeong,I.Y.Choi,D.S.Kim,Y.S.Lee,D.Park,J.Ma,W.Y.Kim,B.C.Kim,S.Park,K.A.Lee,D.H.Kim,K.H.Kim,J.H.Shin,Y.E.Jang,K.D.Kim,W.X.Liu,T.Chaisan,Y.J.Kang,Y.H.Lee,J.K.Moon,J.Schmutz,S.A.Jackson,J.Bhak,S.H.Lee,Whole-genome sequencing and intensive analysis of the undomesticated soybean (Glycine soja Sieb.and Zucc.)genome,Proc.Natl.Acad.Sci.U.S.A.107(2010) 22032–22037.
[22] J.Zhao,N.H.Cheng,C.M.Motes,E.B.Blancaflor,M.Moore,N.Gonzales,S.Padmanaban,H.Sze,J.M.Ward,K.D.Hirschi,AtCHX13 is a plasma membrane K+transporter,Plant Physiol.148 (2008) 796–807.
[23] F.Cellier,G.Conejero,L.Ricaud,D.T.Luu,M.Lepetit,F.Gosti,F.Casse,Characterization of AtCHX17,a member of the cation/H+exchangers,CHX family,from Arabidopsis thaliana suggests a role in K+homeostasis,Plant J.39(2004) 834–846.
[24] S.Padmanaban,S.Chanroj,J.M.Kwak,X.Li,J.M.Ward,H.Sze,Participation of endomembrane cation/H+exchanger AtCHX20 in osmoregulation of guard cells,Plant Physiol.144(2007) 82–93.
[25] D.Hall,A.R.Evans,H.J.Newbury,J.Pritchard,Functional analysis of CHX21: a putative sodium transporter in Arabidopsis,J.Exp.Bot.57(2006) 1201–1210.
[26] J.Jiang,Y.Guo,R.Chang,R.Guan,L.Qiu,Simple evaluation method of tolerance to salt at seedling stage in soybean,Acta Agron.Sin.39(2013)1–9(in Chinese with English abstract).
[27] R.Valencia,P.Y.Chen,T.Ishibashi,M.Conatser,A rapid and effective method for screening salt tolerance in soybean,Crop Sci.48(2008) 1773–1779.
[28] M.S.Akkaya,A.A.Bhagwat,P.B.Cregan,Length polymorphisms of simple sequence repeat DNA in soybean,Genetics 132 (1992) 1131–1139.
[29] N.Diwan,J.Bouton,G.Kochert,P.Cregan,Mapping of simple sequence repeat(SSR)DNA markers in diploid and tetraploid alfalfa,Theor.Appl.Genet.101 (2000) 165–172.
[30] R.X.Guan,R.Z.Chang,Y.H.Li,L.X.Wang,Z.X.Liu,L.J.Qiu,Genetic diversity comparison between Chinese and Japanese soybeans (Glycine max (L.)Merr.) revealed by nuclear SSRs,Genet.Resour.Crop.Evol.57(2010) 229–242.
- The Crop Journal的其它文章
- Maize forage aptitude: Combining ability of inbred lines and stability of hybrids
- Molecular detection of Xanthomonas oryzae pv.oryzae,Xanthomonas oryzae pv.oryzicola,and Burkholderia glumae in infected rice seeds and leaves
- Growth,photosynthesis and nitrogen metabolism in soybean varieties after exclusion of the UV-B and UV-A/B components of solar radiation
- Differences between soybean genotypes in physiological response to sequential soil drying and rewetting
- Genetic background effects on QTL and QTL × environment interaction for yield and its component traits as revealed by reciprocal introgression lines in rice
- Brief Guide for Authors

