中甸刺玫的叶绿体基因组特征及种内变异



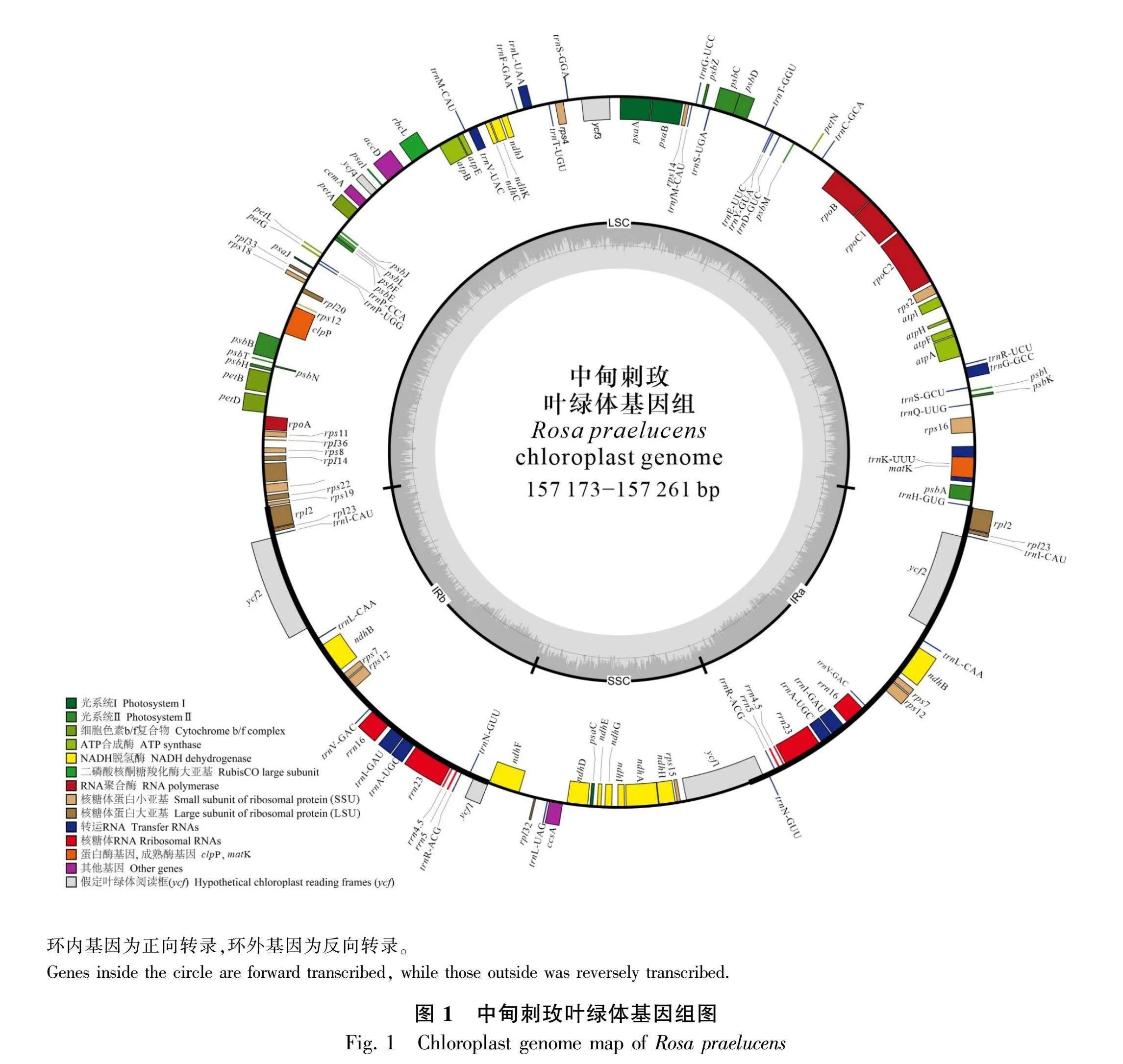

摘 要: "中甸刺玫(Rosa praelucens)是云南香格里拉市的特有“极危”植物和国家二级重点保护植物,也是著名的高山花卉和重要的十倍体月季种质资源,种内存在丰富的表型多样性。为了澄清中甸刺玫种内表型变异的遗传背景,该文利用二代测序技术对40个不同表型的中甸刺玫代表性个体的叶绿体基因组进行测序、组装注释和比较分析。结果表明:(1)中甸刺玫的基因组序列长157 173~157 261 bp,植株间仅相差88 bp,共编码132个功能基因,主要为与光合作用和自我复制相关的基因。全部基因共由27 155个密码子编码,以A-和U-为末端的密码子较常见。(2)中甸刺玫的叶绿体基因组共鉴定出36个重复序列和73个简单重复序列(SSRs),后者大部分为单核苷酸SSRs,主要位于大单拷贝(LSC)区的基因间隔区。(3)中甸刺玫种内叶绿体全基因组的单倍型多样性为0.928±0.027,核酸多态性(Pi)为0.000 12;位于LSC的petN-trnD、psaA-ycf3等基因间隔区,以及rps16和ycf1等基因的核酸多态性相对较高;不同表型的代表性个体的叶绿体基因组间在结构上不存在大片段或基因的逆转或者丢失。该研究结果表明,中甸刺玫种内在叶绿体基因组大小、序列和结构等方面均高度保守,其种内丰富的表型多样性并非由叶绿体基因组变异而引起。
关键词: 中甸刺玫, 叶绿体基因组, 比较基因组, 简单重复序列, 核酸多态性, 密码子偏好
中图分类号: "Q943
文献标识码: "A
文章编号: nbsp;1000-3142(2025)01-0015-16
基金项目: "国家自然科学基金(31972443); 云南省高层次科技人才及创新团队选拔专项(202305AS350002)。
第一作者: 王其刚(1977—),硕士,研究员,研究方向为月季遗传育种,(E-mail)171068976@qq.com。
*通信作者: "蹇洪英,博士,研究员,研究方向为月季种质资源研究与利用,(E-mail)ynwildflower@aliyun.com。
Chloroplast genome features and intraspecific
variation of Rosa praelucens
WANG Qigang"CAO Shirui1,2, WANG Huichun"MA Changle2,
YAN Huijun"QIU Xianqin"JING Weikun"JIAN Hongying1*
( 1. Flower Research Institute, Yunnan Academy of Agricultural Sciences, Kunming 650205, China; 2. College
of Landscape and Horticulture, Southwest Forestry University, Kunming 650224, China )
Abstract: "Rosa praelucens is a critically endangered alpine wild flower endemic to Shangri-La City of Yunnan Province. Rich in phenotypic diversity and with a high ploidy level of decaploid, R. praelucens is a very important rose germplasm resource. In order to clarify the genetic background of its phenotypic variation, the chloroplast genomes of 40 R. praelucens individual plants representing different phenotypes within the species were sequenced by using the Illumina HiSeq 2000 platform, and then assembled, annotated, compared and analyzed. The results were as follows: (1) Chloroplast genomes of R. praelucens were 157 173 - 157 261 bp in length, with a size difference of 88 bp among different individual plants. The genomes encoded 132 function genes, mainly related with photosynthesis and self-replication. A total of 27 155 codons, preferring using codon ending of A or U, were found in all the coding sequences. (2) A total of 36 repeats and 73 simple sequence repeats (SSRs) were detected in the chloroplast genome of R. praelucens. Most of the SSRs were mononucleotide type and located in the intergenic region of large single scale (LSC). (3) The haplotype diversity among the 40 chloroplast genomes was 0.928±0.027, and the nucleotide polymorphism (Pi)"was 0.000 12. The intergenic region of petN-trnD and psaA-ycf gene rps16 and ycf1 were relatively more divergent. No reverse or loss of large DNA fragments and genes were found among the chloroplast genomes of different individuals. These results indicate that the chloroplast genomes are highly conserved in size, sequence and structure within R. praelucens. The rich intraspecific phenotypic diversity is not caused by the variation of chloroplast genomes among different individual plants.
Key words: Rosa praelucens, chloroplast genome, comparative "genome, simple sequence repeats (SSRs), nucleotide polymorphism (Pi), codon preference
叶绿体在植物的生活史中具有重要作用(Wicke et al., 2011)。大多数维管植物的叶绿体基因组长度约为150 kb,为保守的四分体结构,包括一个大单拷贝区(large single copy, LSC)、一个小单拷贝区(small single copy, SSC)和两个反向重复区(inverted repeats, IRs)(Wicke et al., 2011; Shetty et al., 2016; Zhu et al., 2016)。高等植物的叶绿体基因组高度保守,但某些类群的叶绿体基因组中存在大片段的倒置(Sun et al., 2017)、大量重复序列(Guisinger et al., 2011)、基因丢失或假基因化(Ye et al., 2018)以及IR区的扩张或收缩(Li et al., 2017; Liu et al., 2017)。叶绿体基因组在大多数被子植物中为母系遗传(Neale amp; Sederoff, 1989;Daniell et al., 2016)。与核基因组相比,叶绿体基因组具有分子量低、结构简单、保守性强等特点,还包含大量的重复序列,包括简单重复序列(simple sequence repeats, SSRs)(Cavalier-Smith, 2002)。因此,被广泛应用于系统发育、DNA条形码、基因工程和亲缘关系等研究(Dong et al., 2018)。随着二代测序技术(next-generation sequencing, NGS)的发展,越来越多的植物叶绿体全基因组序列被报道,NCBI数据库迄今已公布了超过8 500个植物叶绿体基因组。
中甸刺玫(Rosa praelucens)是云南省香格里拉市的特有“极危”植物(Ku amp; Robertson, 2003;覃海宁等,2017),也是国家二级保护植物(http://www.forestry.gov.cn/main/3954/20210908/163949170374051.html)。中甸刺玫是著名的高山花卉(Li amp; Zhou, 2005)和重要的月季种质资源,既耐寒(邓菊庆等,2013),也高抗蚜虫(范元兰等,2021)。自Jian等(2010)发现其是蔷薇属野生种中唯一的最高倍性——十倍体(2n=10x=70)以来,人们对中甸刺玫的生境和群落特征(关文灵等,2012)、繁育系统(伍翔宇等,2014)、种群现状(周玉泉等,2016)、系统位置(王开锦等,2018)、基于染色体荧光原位杂交的核型特征(方桥等,2020)以及遗传多样性和遗传结构(Jian et al., 2018a)等进行了系统研究,发现中甸刺玫种内存在丰富的表型多样性,其中花色和花型变异尤其显著(李树发等,2013;Jian et al., 2018a)。
理解多倍性如何修饰表型性状是进化生物学的一个研究热点和主要目标(Balao et al., 2011)。大量研究表明自然形成或人工诱导的多倍体植物都会产生遗传和表观遗传的改变,从而改变基因的表达,使其在遗传、生理和形态上产生分化,形成新的表型(Ramsey amp; Schemske, 2002)。对于十倍体的中甸刺玫而言,其种内丰富的表型变异的机制尚不清楚。中甸刺玫的高倍性特征,限制了多种分子技术手段在其遗传背景研究上的应用。Jian等(2017)报道了中甸刺玫的叶绿体基因组大小、各分区的长度和GC含量、编码的基因数量等基本信息,发现中甸刺玫的叶绿体基因组大小为157 186 bp,与同属的单瓣月季花(R. chinensis var. spontanea)等其他植物相比,其叶绿体基因组最大且在LSC区的psbM和trnD之间有一个长为505 bp的插入。在此基础上,为了探究中甸刺玫种内表型变异的遗传背景,本研究利用二代测序技术对40个不同表型的中甸刺玫代表性个体的叶绿体基因组进行了测序、组装和比较分析,探讨以下问题:(1)中甸刺玫叶绿体基因组的序列特征和密码子偏好性;(2)中甸刺玫种内不同表型个体在叶绿体基因组上是否存在较大的变异。以期为中甸刺玫的物种形成和保护提供更多的遗传信息,也为探讨其种内表型变异的分子机制提供叶绿体基因组方面的基础数据。
1 材料与方法
1.1 研究材料
用于叶绿体基因组序列特征分析的中甸刺玫植株来源于香格里拉市小中甸镇和平村塘安培组(99°49′38.1″ E、27°32′16.68″ N, 3 248 m),其原始序列已上传至NCBI,序列号为MG450565.1。其余40个不同表型的代表性植株的基本信息见表1。于2021年6月底在野外采集当年生成熟健康叶片,立即用硅胶进行干燥,-18 ℃低温保存用于后续实验。
1.2 研究方法
1.2.1 基因组总DNA的提取、测序、组装及注释 使用改良的CTAB法进行中甸刺玫叶片的总DNA提取,达到建库测序要求的DNA送到北京诺禾致源科技股份有限公司用Illumina Hiseq 2000测序平台进行建库测序,每个样品得到约3.5 Gb的150 bp短片段原始序列(raw data),用NGSQC Toolkit_v2.3.3软件(Patel amp; Jain, 2012)按照默认参数对原始序列进行过滤筛选,得到高质量的有效序列(clean data)。使用GetOrganelle(https://github.com/Kinggerm/GetOrganelle)进行de novo从头组装,得到叶绿体全基因组序列。将组装好的基因组序列使用CpGAVAS(Liu et al., 2012)自动进行注释,用Genious 9.1(Kearse et al., 2012)进行校对和调整每个注释基因的边界区域,采用OGDRAW(Lohse et al., 2013)绘制叶绿体基因组物理图谱。
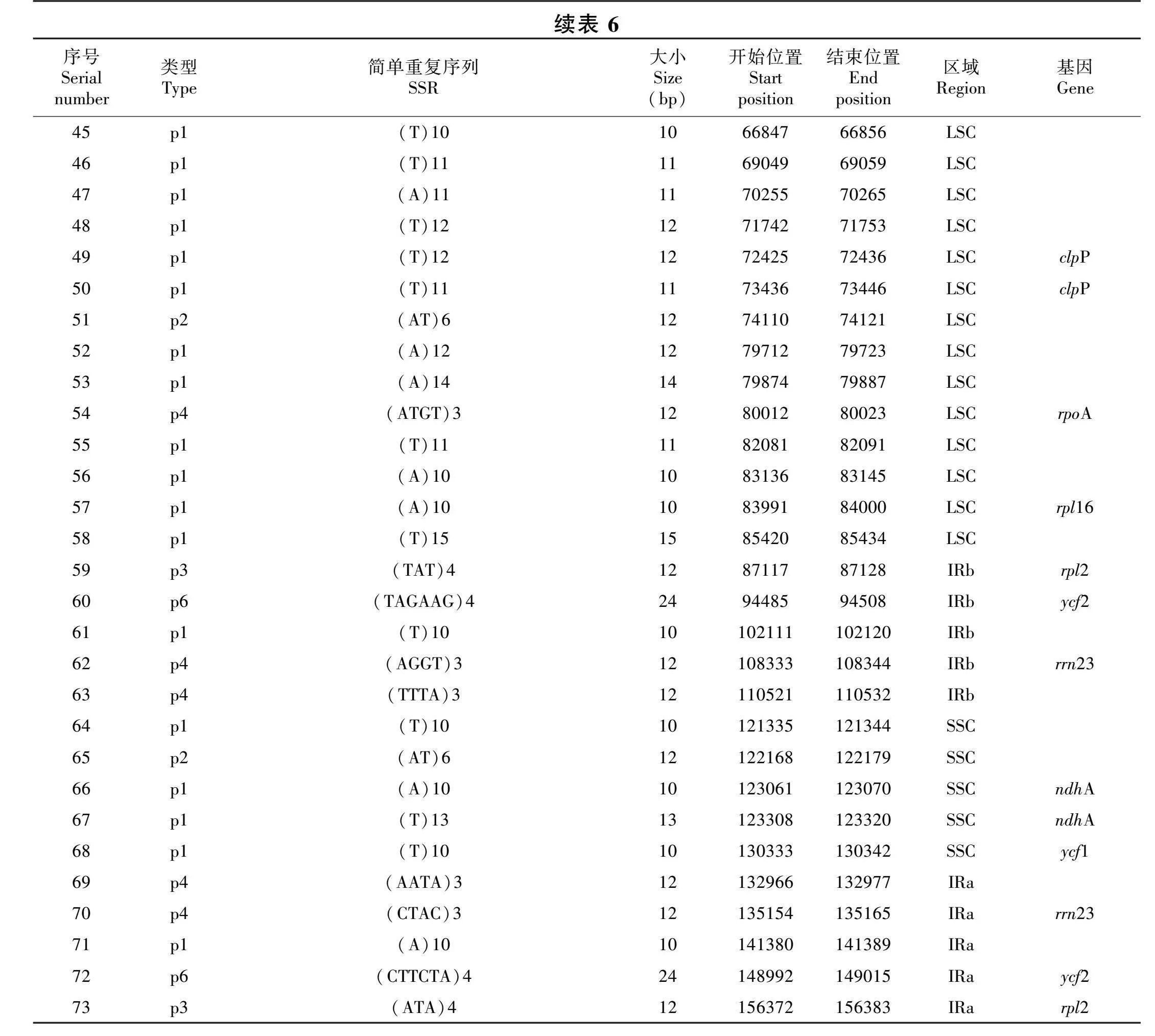
1.2.2 叶绿体基因组结构分析 利用Geneious软件对已上传到NCBI、序列号为MG450565.1的中甸刺玫叶绿体全基因组进行叶绿体全基因组编码基因构成统计。用MEGA6(Tamura et al., 2013)进行密码子偏好分析,计算同义密码子相对使用值(relative synonymous codon usage values; RSCU)并统计AT含量。用重复序列分析软件REPuter软件(https://bibiserv.cebitec.uni-bielefeld.de/reputer)(Kurtz et al., 2001),搜索基因组中的正向重复(forward repeats)和反向重复(reverse repeats)序列。软件运行时,设置搜索的重复序列长度不小于20 bp,序列一致性大于85%。此外,利用MISA(Beier et al.,2017)软件来鉴定简单重复序列,搜索的阈值设置为单核苷酸、二核苷酸、三核苷酸、四核苷酸、五核苷酸和六核苷酸重复次数分别不小于10、5、4、3、3和3。
1.2.3 种内不同个体的叶绿体基因组序列比较 在Geneious软件中调用Mauve程序(Darling et al., 2004)对中甸刺玫40个代表性植株的叶绿体基因组进行比对,分析不同植株的叶绿体基因组间是否存在大片段的逆转或丢失。用DNASP v5.10软件(Librado amp; Rozas, 2009)计算种内叶绿体基因组的单倍型多样性和核苷酸多态性(Pi),筛选叶绿体基因组中的高变区。
2 结果与分析
2.1 中甸刺玫叶绿体基因组的结构特征
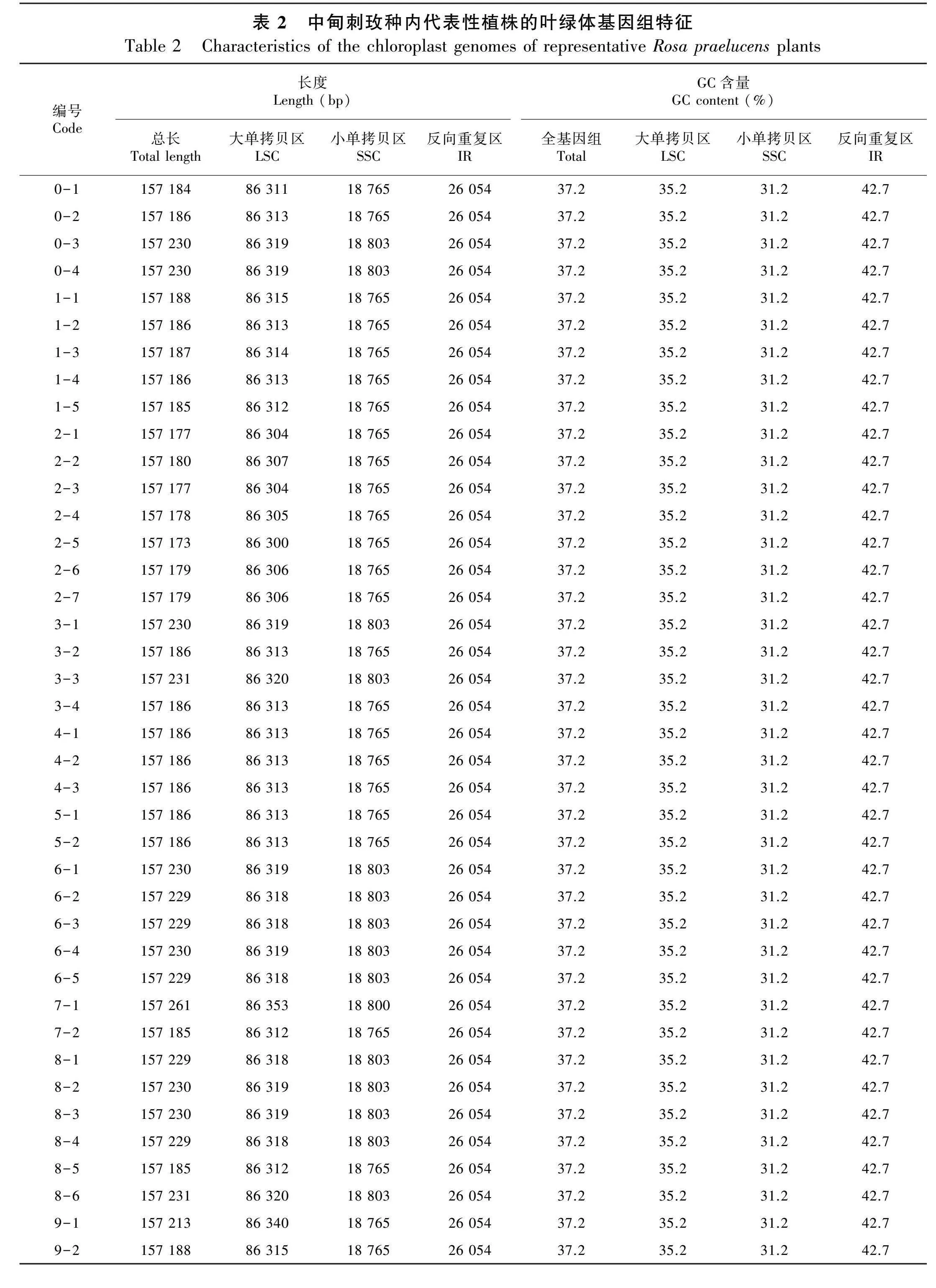
中甸刺玫种内不同表型的40个代表性植株的叶绿体基因组序列的长度、各分区长度、GC含量、编码基因数目等基本信息见图1和表2。中甸刺玫的基因组序列全长157 173~157 261 bp,植株间相差88 bp。基因组最大的是7-1号植株,为157 261 bp,基因组最小的是2-5号植株,为157 173 bp。LSC区长为86 300~86 353 bp,相差53 bp,最长的是7-1号植株,最短的是2-5号植株;SSC区长为18 765~18 803 bp,相差38 bp;反向重复IR区长度均为26 054 bp,说明种内基因组大小的差异主要来源于LSC区和SSC区。基因组的GC含量在不同个体间无显著差异,全基因组的GC含量均为37.2%,其中IR区的GC含量为42.7%,LSC区的GC含量为35.2%,SSC区的GC含量为31.2%。
2.2 中甸刺玫叶绿体基因组的基因构成
由表3可知,中甸刺玫的叶绿体基因组共编码132个功能基因,包括87个蛋白质编码基因,37个tRNA基因和8个rRNA基因。其中,与光合作用有关的基因有45个,与自我复制相关的基因有76个,功能未知的其他基因共11个。6个蛋白编码基因(ndhB、rpl2、rpl23、rps7、rps12、ycf2)、7个tRNAs(trnA-UGC、trnI-CAU、trnI-GAU、trnL-CAA、trnN-GUU、trnR-ACG、trnV-GAC)和4个rRNAs(rrn16、rrn23、rrn 4.5、rrn5)在IR区完全重复。在132个基因中,petB、petD、ndhA、ndhB、rps16、rpl2、rpl16、rpoC1、trnA-UGC、trnI-GAU、trnK-UUU、trnL-UAA、trnV-UAC等13个基因有1个内含子,ycf3和clpP这2个基因各有2个内含子,rps12为反式剪接的基因,其5′端在LSC区,而3′端在IR区重复。
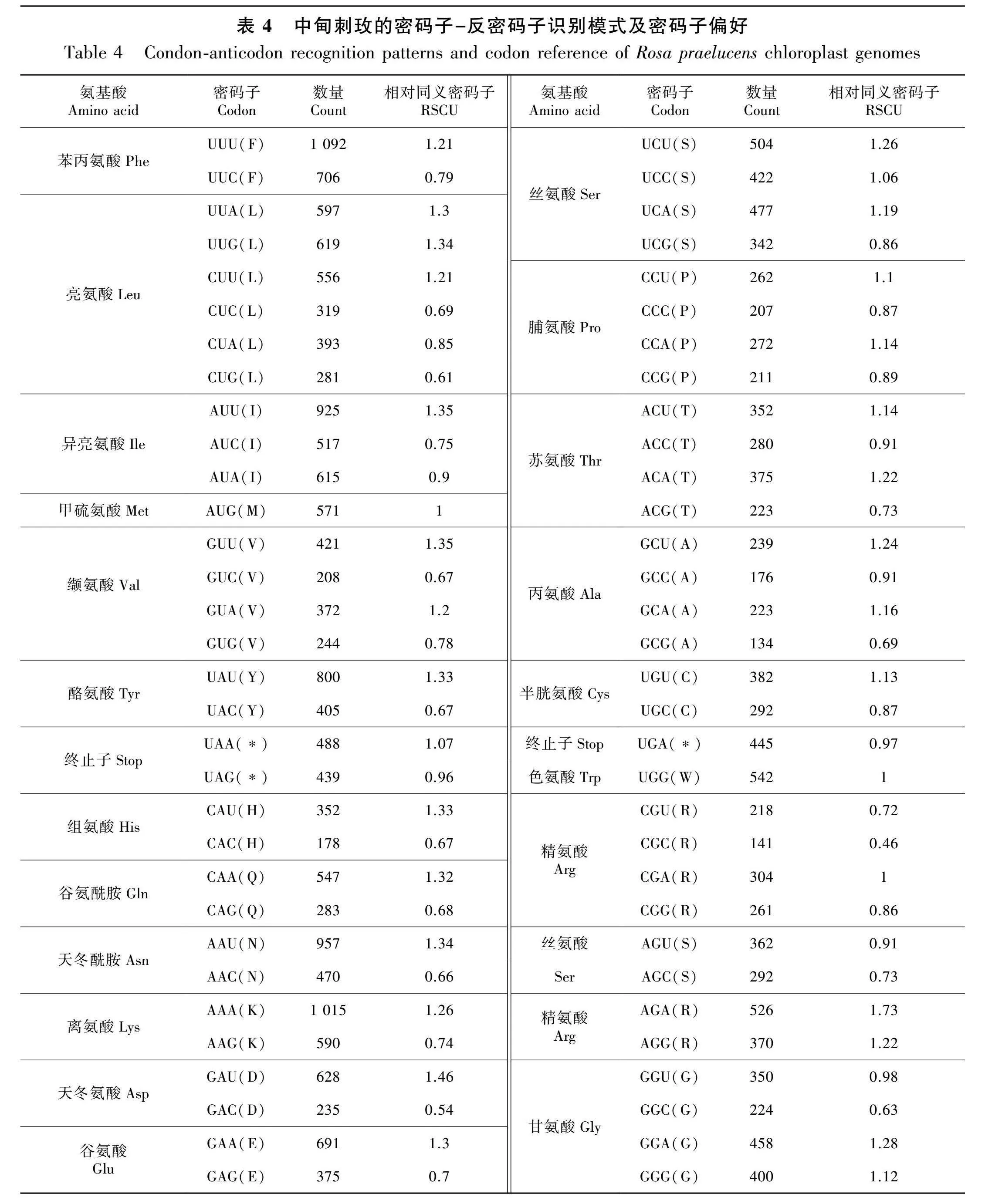
2.3 中甸刺玫叶绿体基因组密码子偏好性
中甸刺玫的叶绿体基因组密码子使用频率(RSCU)如表4。全部基因由27 155个密码子编码,其中亮氨酸(leucine)是使用频率最高的氨基酸,共编码了其中的2 765个密码子,占总数的10.85%,而组氨酸(histidine)是使用频率最低的氨基酸,仅编码了其中的530个密码子,占总数的1.95%。以A-和U-为末端的密码子较常见,除了trnL-CAA、trnS-GGA、精氨酸Arg-AGG和甘氨酸Gly-GGG以外,所有的首选同义密码子(RSCUgt;1)都是以A或U结尾。
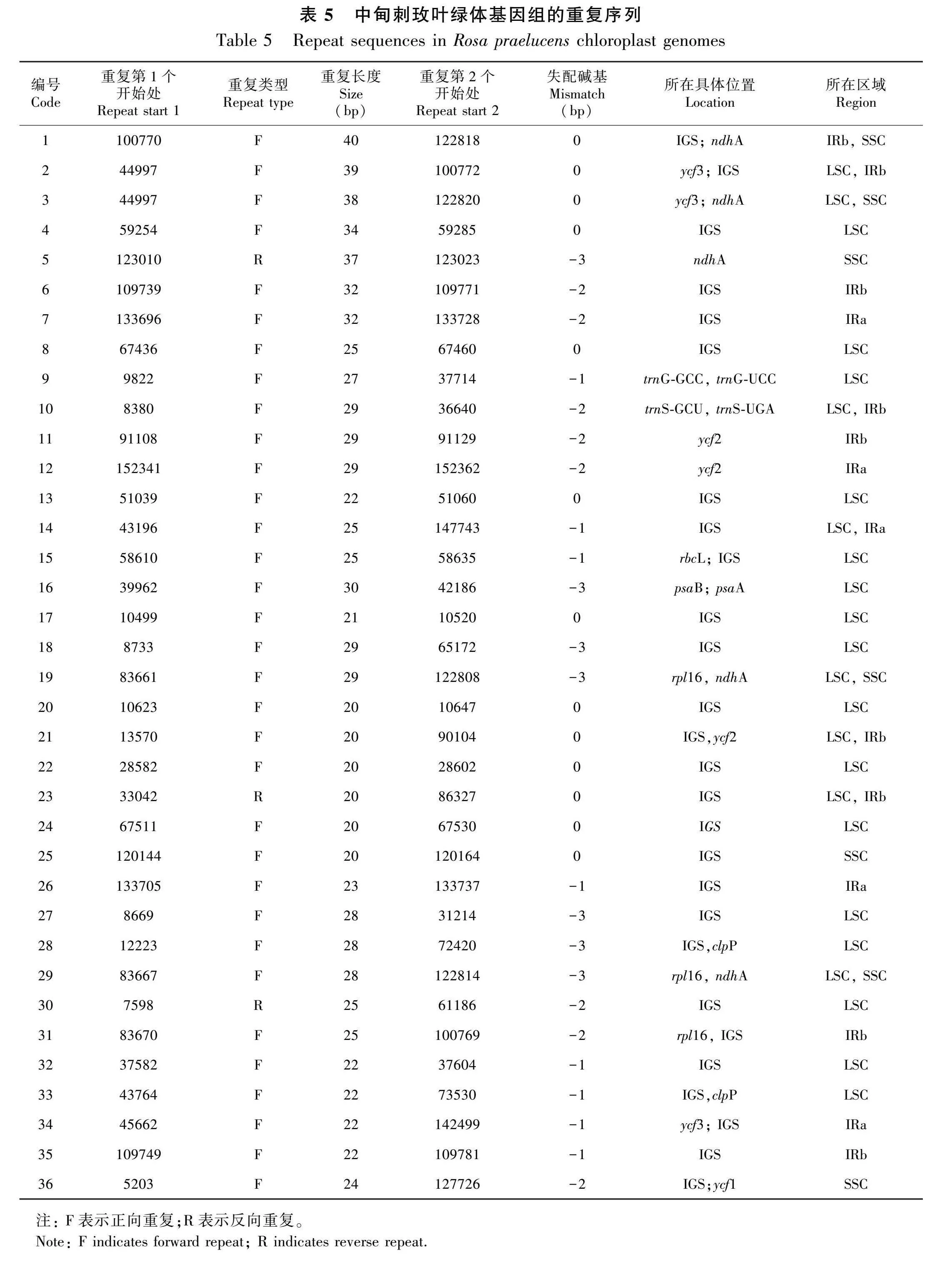
2.4 中甸刺玫叶绿体基因组的重复序列
重复序列分析结果表明,中甸刺玫的叶绿体基因组中共有33个正向重复序列和3个反向重复序列,这些重复序列长度大多为20~30 bp(表5)。最长的重复序列分别位于rps12-trnV(GAC)基因间隔区和ndhA基因的含子区域。大多数重复序列位于LSC区和IR区,还有9个重复序列在不同的区域开始,如第1号重复的2个序列分别开始于IRb区和SSC区。
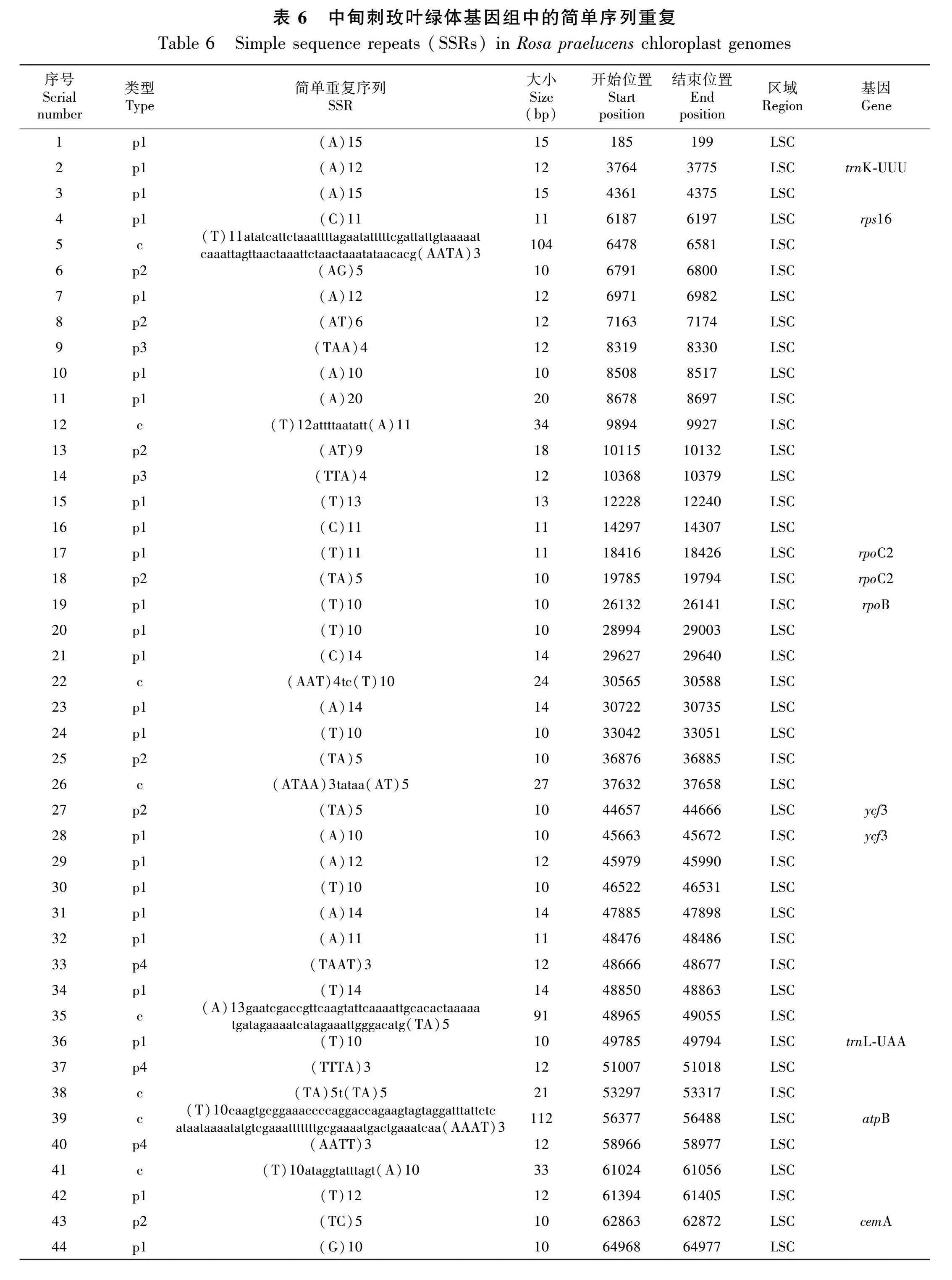
2.5 中甸刺玫叶绿体基因组的简单重复序列
MISA软件在中甸刺玫的叶绿体基因组中共找到73个简单重复序列(SSRs),其中单核苷酸SSRs(A/T/G/C)最多,共有42个;其次是二核苷酸类型(AG/AT/TA/TC),有9个,三核苷酸类型有4个,四核苷酸类型有8个,六核苷酸SSRs有2个,没有五核苷酸SSRs。绝大部分为单纯类型SSRs,复合类型的SSRs较少,二者分别为65个和8个,没有间接型SSRs(表6)。58个SSRs位于LSC区,占全部SSRs的79.5%,5个SSRs位于SSC区,IRa和IRb区各有5个SSRs。只有23个SSRs位于基因中,其他均位于基因间隔区。单核苷酸SSRs中的74%属于A/T,这与SSRs主要由短腺嘌呤(adenine,A)或胸腺嘧啶(thymine, T)重复组成而很少有串联鸟嘌呤(guanine, G)和胞嘧啶(cytosine, C)组成的假说相一致。
2.6 中甸刺玫种内的叶绿体基因组序列差异
中甸刺玫种内不同个体的叶绿体基因组序列差异较小。所有代表性个体的全基因组序列比对分析共检测到58个变异位点共22个单倍型,单倍型多样性为0.928±0.027,核酸多态性为0.000 12。种内叶绿体基因组在基因、基因间隔区的核酸多态性都较低,相对多态性较大的是位于LSC区的psbI-trnS(GCU)、trnS(GCU)-trnG(UCC)、trnG(UCC)-trnfM(CAU)、petN-trnD(GUC)、petA-psbJ、psaA-ycf3等基因间隔区,以及rps16和ycf1等基因(图2)。Mauve比对的结果表明,中甸刺玫种内不同代表性个体的叶绿体基因组在结构上并无显著差异,并且不存在大片段或基因的逆转或者丢失(图3)。
3 讨论与结论
虽然植物的叶绿体基因组在基因组成和排列顺序上具有较高的保守性,但由于长期对不同环境的适应常导致同属植物的叶绿体基因组在大小上发生改变,从而产生结构重排以及IR区的收缩或扩张等(Daniell et al., 2016)。中甸刺玫不同表型40个代表性个体的叶绿体基因组大小为157 173~157 261 bp,整个基因组共编码132个基因,主要与光合作用和自我复制相关。与同属的单瓣月季花(Jian et al., 2018b)、亮叶月季(R. lucidissima)(Zhao et al., 2019)、木香花(R. banksiae)(杨芳,2019),大花香水月季(R. odorata var. gigantea)(Yang et al., 2014)、金樱子(R. laevigata)(Yin et al., 2020)以及其他种(Chen et al., 2019; Cui et al., 2022)的叶绿体基因组相比,中甸刺玫的基因组除在长度上明显多出约500 bp以外,在基因组的GC含量、基因构成和排列顺序上与其他种基本一致,表明蔷薇属植物的叶绿体基因组较保守,种间差异较小,主要在非编码区有序列长度的变化。
密码子是连接核酸和蛋白质的纽带,研究物种的密码子偏好并确定最优密码子,有助于设计基因表达载体来提高目的基因的表达量,在作物遗传育种和品种改良方面具有重要应用价值(Qi et al., 2015)。密码子偏好性分析显示中甸刺玫的叶绿体基因组编码密码子中,亮氨酸是使用频率最高的氨基酸,而组氨酸使用频率最低。此外,中甸刺玫所有的首选同义密码子都是以A-或U-结尾(RSCU gt; 1),由于RSCU大于1则表示实际频率高于其他同义密码子的使用频率(晁岳恩等,2012),因此中甸刺玫的密码子偏好以A-和U-结尾。这与同属的单瓣月季花的密码子偏好性(Jian et al., 2018b)基本一致,为研究相关基因的分子进化和外源表达奠定了基础。
植物的叶绿体简单重复序列为单亲遗传且有较高的种内多态性,是物种进化和多态性的重要遗传标记(Cavalier-Smith, 2002),常被用作野生植物系统演化和居群遗传研究(Provan, 2000; Flannery et al., 2006)以及作物遗传图谱构建的分子标记(Powell et al., 1995; Xue et al., 2012)。由于poly A和poly T相比poly C和poly G可能具有更高的结构稳定性(Gragg et al., 2002),因此大多数植物的叶绿体基因组单核苷酸SSRs多为poly A和poly T结构。中甸刺玫叶绿体基因组中共有73个简单重复序列SSRs,主要为单核苷酸SSRs,大部分由短腺嘌呤(A)或胸腺嘧啶(T)重复组成,主要位于LSC区的基因间隔区,与同属的单瓣月季花(Jian et al., 2018b)基本一致,也与其他多种植物如山茶属(Camellia)(丁祥青等,2022;邓永彪等,2024)、绢蒿属(Seriphidium)(Jin et al., 2023)等类似。
Jian等(2018b)研究表明,蔷薇属内不同物种的叶绿体基因组间的核酸高变区主要位于LSC区的trnK-rps16、ps16-trnQ、trnS-trnG、atpF-atpH、rps2-rpoC2等多个基因隔区、rps19和ycf1等基因的编码区,以及rpl2、rps16、ndhA等基因的内含子区域。中甸刺玫种内不同表型代表性个体的全基因组核酸多态性较低,核酸多态性相对较高的是位于LSC区内的petN-trnD(GUC)、petA-psbJ、psaA-ycf3等少数几个基因间隔区,以及rps16和ycf1等少数几个基因。结合Mauve比对的结果,说明中甸刺玫种内的叶绿体基因组的基因序列和结构均高度保守,不存在大片段序列或基因的逆转或者丢失,其种内不同个体的表型变异并非由叶绿体基因组变异而引起。
综上所述,本研究在对中甸刺玫的叶绿体基因组进行简单报道的基础上,详细地分析了中甸刺玫叶绿体基因组的基因构成、密码子偏好以及简单重复序列等基本特征,并对种内具有不同表型的代表性个体的叶绿体基因组进行了比较基因组分析,结果表明,中甸刺玫种内的叶绿体基因组大小、序列和基因结构等方面均高度保守,不存在大片段序列或基因的逆转或者丢失,为中甸刺玫的保护和开发利用提供了叶绿体基因组方面基础数据。本研究也表明中甸刺玫种内丰富的表型变异并非由叶绿体基因组变异而引起,应结合中甸刺玫的高倍性特征,从染色体的数量和结构、基因表达以及表观遗传等角度进行深入系统的研究。
参考文献:
BALAO F, HERRERA J, TALAVERA S, 2011. Phenotypic consequences of polyploidy and genome size at the microevolutionary scale: A multivariate morphological approach "[J]. The New Phytologist, 192(1): 256-265.
BEIER S, THIEL T, MNCH T, et al., 2017. MISA-web: A web server for microsatellite prediction "[J]. Bioinformatics, 33(16): 2583-2585.
CAVALIER-SMITH ST, 2002. Chloroplast evolution: Secondary symbiogenesis and multiple losses "[J]. Current Biology, 12(2): 62-64.
CHAO YE, CHANG Y, WANG MF, et al., 2012. Codon usage bias and cluster analysis on chloroplastic genes from seven crop species "[J]. Acta Agriculture Boreali-Sinica, 27(4): 60-64. "[晁岳恩, 常阳, 王美芳, 等, 2012. 7种作物叶绿体基因的密码子偏好性及聚类分析 [J]. 华北农学报, 27(4): 60-64.]
CHEN MR, ZHANG C, GAO XF. 2019. The complete chloroplast genome sequence of Rosa pricei (Rosaceae) "[J]. Mitochondrial DNA Part B-Resources, 4(1): 1918-1919.
CUI WH, DU XY, ZHONG MC, et al., 2022. Complex and reticulate origin of edible roses (Rosa, Rosaceae) in China "[J]. Horticulture Research, 9(1): 678-691.
DANIELL H, LIN CS, YU M, et al., 2016. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering "[J]. Genome Biology, 17(1): 1-29.
DARLING ACE, MAU B, BLATTNER FR, et al., 2004. Mauve: Multiple alignment of conserved genomic sequence with rearrangements "[J]. Genome Research, 14(7): 1394-1403.
DENG JQ, JIAN HY, LI SB, et al., 2013. Cold tolerance of several wild Rosa resources endemic of Yunnan "[J]. Southwest China Journal of Agricultural Sciences, 26(2): 273-277. "[邓菊庆, 蹇洪英, 李淑斌, 等, 2013. 几种云南特有蔷薇资源的抗寒性研究 [J]. 西南农业学报, 26(2): 273-277.]
DENG YB, ZHANG J, LAN LL, et al., 2024. Analysis of chloroplast genome features of endangered and rare plant Camellia minima "[J]. Guihaia, 44(1):30-42. "[邓永彪, 张进, 蓝伦礼, 等, 2024. 珍稀濒危植物越南小花金花茶的叶绿体基因组特征分析 [J]. 广西植物, 44(1): 30-42.]
DING XQ, LI WF, WU JL, et al., 2022. Chloroplast genome characteristics and genetic relationship of yellow Camellia "[J]. Journal Fujian Agriculturae Forestry University (Natural Science Edition), 52(3): 1-11. "[丁祥青, 李文芳, 吴丽君, 等, 2022. 4种金花茶叶绿体基因组的比较分析 [J]. 福建农林大学学报(自然科学版), 52(3): 1-11.]
DONG WP, XU C, WU P, et al., 2018. Resolving the systematic positions of enigmatic taxa: Manipulating the chloroplast genome data of Saxifragales "[J]. Molecular Phylogenetics and Evolution, 126(9): 321-330.
FAN YL, CHEN YC, JIAN HY, et al., 2021. Screening of Rosa germplasm resources with resistance to aphids "[J]. Journal of "Yunnan University (Natural Science Edition), 43(3): 619-628. "[范元兰, 陈宇春, 蹇洪英, 等, 2021. 蔷薇属抗蚜种质资源的筛选 [J]. 云南大学学报(自然科学版), 43(3): 619-628.]
FANG Q, TIAN M, ZHANG T, et al., 2020. Karyotype analysis of Rosa praelucens and its closely related congeneric species based on FISH "[J]. Acta Horticulturae Sinica, 47(3): 503-516. "[方桥, 田敏, 张婷, 等, 2020. 中甸刺玫及其近缘种基于FISH的核型分析 [J]. 园艺学报, 47(3): 503-516.]
FLANNERY ML, MITCHELL FJ, COYNE S, et al., 2006. Plastid genome characterisation in Brassica and Brassicaceae using a new set of nine SSRs "[J]. Theoretical and Applied Genetics, 113(7): 1221-1231.
GRAGG H, HARFE BD, JINKS-ROBERTSON S, 2002. Base composition of mononucleotide runs affects DNA polymerase slippage and removal of frame shift intermediates by mismatch repair in Saccharomyces cerevisiae "[J]. Molecular and Cellular Biology, 22(24): 8756-8762.
GUAN WL, LI SF, SONG J, et al., 2012. Study on geographic distribution of Rosa praelucens endemic to Yunnan "[J]. Journal of West China Forestry Science, 41(1): 88-93. "[关文灵, 李世峰, 宋杰, 等, 2012. 云南特有濒危植物中甸刺玫的分布特征研究 [J]. 西部林业科学, 41(1): 88-93.]
GUISINGER MM, KUEHL JV, BOORE JL, et al., 2011. Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: rearrangements, repeats, and codon usage "[J]. Molcular Biology and Evolution, 28(1): 1543-1543.
JIAN HY, LI SF, GUO JL,et al., 2018a. High genetic diversity and differentiation of an extremely narrowly distributed and critically endangered decaploid rose (Rosa praelucens): implications for its conservation "[J]. Conservation Genetics, 19(4): 761-776.
JIAN HY, ZHANG H, TANG KX, et al., 2010. Decaploidy in Rosa praelucens Byhouwer (Rosaceae) endemic to zhongdian plateau, Yunnan, China "[J]. Caryologia, 63(2): 162-167.
JIAN HY, ZHANG SD, ZHANG T, et al., 2017. Characterization of the complete chloroplast genome of a critically endangered decaploid rose species, Rosa praelucens (Rosaceae) "[J]. Conservation Genetics Resources, 10: 851-854.
JIAN HY, ZHANG YH, YAN HJ, et al., 2018b. The complete chloroplast genome of a key ancestor of modern roses, Rosa chinensis var. spontanea, and a comparison with congeneric species "[J]. Molecules, 23: 389.
JIN GZ, LI WJ, SONG F, et al., 2023. Comparative analysis of completeArtemisia subgenus Seriphidium (Asteraceae: Anthemideae) chloroplast genomes: insights into structural divergence and phylogenetic relationships "[J]. BMC Plant Biology, 23(1): 136-150.
KEARSE M,MOIR R, WILSON A, et al., 2012. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data "[J]. Bioinformatics, 28(12): 1647-1649.
KU TC, ROBERTSON KR, 2003. Rosa (Rosaceae) "[M] // WU ZY, RAVEN PH. Flora of China: Vol. 9. Beijing: Science Press; St. Louis: Missouri Botanical Garden Press: 339-381.
KURTZ S, CHOUDHURI JV, OHLEBUSCH E, et al., 2001. REPuter: The manifold applications of repeat analysis on a genomic scale "[J]. Nucleic Acids Research, "29(22): 4633-4642.
LI P, LU RS, XU WQ, et al., 2017. Comparative genomics and phylogenomics of East Asian tulips (Amana, Liliaceae) "[J]. Frontiers in Plant Science, 8: 451.
LI SF, LI CJ, JIAN HY, et al., 2013. Studies on phenotypic diversity of vulnerable Rosa praelucens endemic to Shangrila, Yunnan "[J]. Acta Horticiculturae Sinica, 40(5): 924-932. "[李树发, 李纯佳, 蹇洪英, 等, 2013. 云南香格里拉特有易危植物中甸刺玫的表型多样性 [J]. 园艺学报, 40(5): 924-932.]
LI XX, ZHOU ZK, 2005. Endemic wild ornamental plants from North Western Yunnan "[J]. HortScience, 40(6): 1612-1619.
LIBRADO P, ROZAS J, 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data "[J]. Bioinformatics, 25(11): 1451-1452.
LIU C, SHI L, ZHU Y, et al., 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences "[J]. BMC Genomics, 13: 715.
LIU LX, LI R, WORTH JRP, et al., 2017. The complete chloroplast genome of Chinese bayberry (Morella rubra, Myricaceae): Implications for understanding the evolution of Fagales "[J]. Frontiers in Plant Science, 8: 968.
LOHSE M, DRECHSEL O, KAHLAU S, et al., 2013. Organellar Genome-DRAW — A suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets "[J]. Nucleic Acids Research, 41(W1): 575-581.
NEALE DB, SEDEROFF RR, 1989. Paternal inheritance of chloroplast DNA and maternal inheritance of mitochondrial DNA in loblolly pine "[J]. Theoretical Applied Genetics, 77(2): 212-216.
PATEL RK, JAIN M, 2017. NGS QC toolkit: A toolkit for quality control of next generation sequencing data "[J]. PLoS ONE, 7: e30619.
POWELL W, MORGANTE M, MCDEVITT R, et al., 1995. Polymorphic simple sequence repeat regions in chloroplast genomes: Applications to the population genetics of pines "[J]. Proceedings of the National Academy of Sciences of the United States of America, 92(17): 7759-7763.
PROVAN J, 2000. Novel chloroplast microsatellites reveal cytoplasmic variation in Arabidopsis thaliana "[J]. Molecular Ecology, 9(12): 2183-2185.
QI YY, XU WJ, XING T, et al., 2015. Synonymous codon usage bias in the plastid genome is unrelated to gene structure and shows evolutionary heterogeneity "[J]. Evolutionary Bioinformatics Online, 11: 65-77.
QIN HN, YANG Y, DONG SY, et al., 2017. List of threatened species of higher plants in China "[J]. Biodiversity Science, 25(7): 696-744. "[覃海宁, 杨永, 董仕勇, 等, 2017. 中国高等植物受威胁物种名录 [J]. 生物多样性, 25(7): 696-744.]
RAMSEY J, SCHEMSKE DW, 2002. Neopolyploidy in flowering plants "[J]. Annual Review Ecology and Systematics, 33(1): 589-639.
SHETTY SM, SHAH MUM, MAKALE K, et al., 2016. Complete chloroplast genome sequence of Musa balbisiana corroborates structural heterogeneity of inverted repeats in wild progenitors of cultivated bananas and plantains "[J]. Plant Genome, 9(2): 1-14.
SUN YX, MOORE MJ, LIN N, et al., 2017. Complete plastome sequencing of both living species of Circaeasteraceae (Ranunculales) reveals unusual rearrangements and the loss of the ndh gene family "[J]. BMC Genomics, 18: 592.
TAMURA K, STECHER G, PETERSON D, et al., 2013. MEGA6: molecular evolutionary genetics analysis version 6.0 "[J]. Molecular Biology and Evolution, 30(12): 2725-2733.
WANG KJ, ZHANG T, WANG QG, et al., 2018. The phylogenetic position and hybrid origination of Rosa Praelucens Byhouwer "[J]. Journal of Plant Genetic Resources, 19(5): 1006-1015. "[王开锦, 张婷, 王其刚, 等, 2018. 中甸刺玫的系统位置及杂交起源研究 [J]. 植物遗传资源学报, 19(5): 1006-1015.]
WICKE S, SCHNEEWEISS GM,DE PAMPHILIS CW, et al., 2011. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function "[J]. Plant Molecular Biology, 76(3-5): 273-297.
WU XY, CHEN M, WANG QG, et al., 2014. Comparative study on the breeding systems of Rosa praelucens and Rosa soulieana "[J]. Acta Horticulturae Sinica, 41(10): 2075-2084. "[伍翔宇, 陈敏, 王其刚, 等, 2014. 中甸刺玫和川滇蔷薇的繁育系统比较研究 [J]. 园艺学报, 41(10): 2075-2084.]
XUE J, WANG S, ZHOU SL, 2012. Polymorphic chloroplast microsatellite loci in Nelumbo (Nelumbonaceae) "[J]. American Journal of Botany, 99(6): 240-244.
YANG F, 2019. Sequencing and structural analysis of chloroplast genome in Rosa banksiae "[J]. Genomics and Applied Biology, 38(8): 3586-3594. "[杨芳, 2019. 七里香蔷薇叶绿体基因组测序及结构分析 [J]. 基因组学与应用生物学, 38(8): 3586-3594.]
YANG JB, LI DZ, LI HT, 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs "[J]. Molecular Ecology Resources, 14(5): 1024-1031.
YE WQ, YAP ZY, LI P, et al., 2018. Plastome organization, genome-based phylogeny and evolution of plastid genes in Podophylloideae (Berberidaceae) "[J]. Molecular Phylogenetics and Evolution, 127: 978-987.
YIN XM, LIAO BS, GUO S, et al., 2020. The chloroplasts genomic analyses of Rosa laevigata, R. rugosa and R. canina "[J]. Chinese Medicine, 15: 18.
ZHAO L, ZHANG H, WANG QG, et al., 2019. The complete chloroplast genome of Rosa lucidissima, a critically endangered wild rose endemic to China "[J]. Mitochondrial DNA Part B-Resources, 4(1): 1826-1827.
ZHOU YQ, SU Q, ZHANG H, et al., 2016. Distribution and population quantitative dynamics of critically risked Rosa praelucens Byhouwer "[J]. Journal of "Plant Genetic Resources, 17(4): 649-654. "[周玉泉, 苏群, 张颢, 等, 2016. 极危植物中甸刺玫的分布及种群数量动态 [J]. 植物遗传资源学报, 17(4): 649-654.]
ZHU A, GUO W, GUPTA S, et al., 2016. Evolutionary dynamics of the plastid inverted repeat: the effects of expansion, contraction, and loss on substitution rates "[J]. The New Phytologist, 209(4): 1747-1756.
(责任编辑 李 莉 王登惠)

