细菌主导的红肉腐败机制研究进展

摘 要:由细菌主导的红肉腐败是一个涉及多种菌相互竞争、相互作用的复杂动态过程,深入了解细菌引起的红肉腐败机制对于抑制肉类腐败进程、开发红肉及其制品防腐措施、延长肉品货架期有重要意义。本文讨论3 种红肉(牛肉、猪肉和羊肉)屠宰分割过程中腐败微生物的主要污染来源和干预措施,总结不同包装条件下肉类优势腐败菌,重点讨论细菌引起的红肉腐败机制,发现动物皮毛和分割间的接触面是红肉表面微生物污染的主要来源,在各个环节采取2 种或多种抗菌技术能更好地降低细菌初始污染水平;不同红肉中优势菌在相同包装条件下存在一定差异,但大致相同;微生物通过丙酮酸代谢使肉产生异味,同时产生信号分子,调节蛋白酶和脂肪酶分泌,引起肉蛋白降解和脂肪分解,导致肉类产生黏液、软化等腐败现象。本文旨在为控制红肉中的细菌腐败提供理论基础。
关键词:腐败;牛肉;猪肉;羊肉;细菌;优势菌;机制
Advances in Understanding the Mechanism of Red Meat Spoilage Caused by Bacteria
SUN Ge1, WU Tongxuan1, MAO Yanwei1, LI Junling2, ZHU Lixian1, TONG Lin3, ZHANG Xinjun4, CHENG Haijian5, GU Yue6, ZHANG Yimin1,*
(1. College of Food Science and Engineering, Shandong Agricultural University, Tai’an 271018, China;
2. Shandong Animal Product Quality and Safety Center, Jinan 271299, China; 3. Tongliao Comprehensive Experimental Station, National Beef Cattle Industrial Technology System, Tongliao 028000, China; 4. Zhongwei Comprehensive Experimental Station, National Beef Cattle Industrial Technology System, Zhongwei 755000, China; 5. Jinan Comprehensive Experimental Station, National Beef Cattle Industrial Technology System, Jinan 250000, China; 6. Baicheng Comprehensive Experimental Station,
National Beef Cattle Industrial Technology System, Baicheng 137314, China)
Abstract: Bacterial spoilage of red meat is a complex dynamic process involving competition and interaction between different bacterial communities. Deeply understanding the mechanism behind bacterial spoilage of red meat is of great significance for inhibiting the spoilage process, developing preservation measures and prolonging the shelf life of meat products. This review discusses the main sources of and intervention measures for microbial spoilage and contamination in red meat (beef, pork and lamb) during the slaughter and cutting process, and summarizes the dominant spoilage bacteria in red meat under different packaging conditions, with a focus on the mechanism of bacterial spoilage of red meat. It has been found that animal fur and contact surfaces during the segmentation process are the main sources of microbial contamination on the red meat surface. Two or more disinfection techniques can be applied at each step to better reduce the initial microbial contamination level. The dominant bacteria in different red meats under the same packaging conditions are roughly the same with only slight differences. Microorganisms in meat produce off-odors through pyruvate metabolism, while producing signaling molecules that regulate the secretion of proteases and lipases, leading to proteolysis and lipolysis, which in turn results in spoilage phenomena such as getting slimy and softer texture. This review is expected to provide a theoretical basis for the control of bacterial spoilage in red meat.
Keywords: spoilage; beef; pork; lamb; bacteria; dominant bacteria; mechanism
DOI:10.7506/rlyj1001-8123-20240603-134
中图分类号:TS251.5" " " " " " " " " " " " " " " " " " " "文献标志码:A 文章编号:1001-8123(2024)08-0063-09
引文格式:
孙歌, 武桐煊, 毛衍伟, 等. 细菌主导的红肉腐败机制研究进展[J]. 肉类研究, 2024, 38(8): 63-71. DOI:10.7506/rlyj1001-8123-20240603-134." " http://www.rlyj.net.cn
SUN Ge, WU Tongxuan, MAO Yanwei, et al. Advances in understanding the mechanism of red meat spoilage caused by bacteria[J]. Meat Research, 2024, 38(8): 63-71. (in Chinese with English abstract) DOI:10.7506/rlyj1001-8123-20240603-134.
http://www.rlyj.net.cn
肉类在生产、运输、贮藏、分销等任一阶段处理不当均易引起腐败变质,食用腐败肉类是人类感染食源性致病菌的重要途径。我国每年因腐败造成的肉类损失可达9万 t,给肉类行业造成损失达数十亿美元[1]。肉类腐败是一个以微生物为主导的复杂过程。近年来,食品企业及相关研究领域已经采取多种措施减少或灭活肉类产品中的微生物,如建立卫生管理制度、添加防腐剂和采用减菌技术等,但由于微生物污染范围广且极易交叉污染,肉类腐败始终不可避免[2]。在屠宰之前,健康活体动物的肌肉组织通常被认为是无菌的,虽然内在细菌偶尔会以低水平存在于肌肉组织中,但这并非最常见的污染来源。外部污染是肉类和胴体微生物污染的最主要来源[3]。
肉类的初始微生物取决于屠宰时动物的生理状态及污染在屠宰和加工过程中的扩散,而导致肉类腐败的微生物群组成取决于贮藏条件和微生物间的竞争[4]。由于肉类腐败的复杂性和影响肉类腐败各种因素的相互关联性,使得解决这一问题变得非常困难。了解细菌在肉中的腐败机制将有助于制定以经验和证据为基础的缓解策略。目前已有大量研究探索屠宰加工过程中肉类细菌污染来源、细菌在肉中的生长条件及细菌生长竞争和生长活动对肉类货架期和品质的影响,也有一些研究对细菌的腐败机制进行探究。但是在该研究领域还缺乏相关信息和研究成果的总结和归纳。红肉在全球肉类总消费量中占比较大,其中猪肉占比可达36%,牛肉和山羊/绵羊肉占比分别约为24%和5%[5]。本文以红肉中消费量最高的牛肉、猪肉和羊肉为代表,在已有文献基础上对肉类细菌污染来源和腐败机制进行综述,为采取靶向措施延长肉类货架期、控制肉类腐败提供理论支持。
1 红肉中腐败微生物的污染来源和干预措施
在屠宰和加工环节,由于胴体会接触到毛发、兽皮、粪便和加工环境(传送带、生产工具、机器设备、水、操作人员和其他接触材料)等,所以肉类会不可避免地受到微生物污染。在实际生产过程中,胴体间有限的距离和空间还会造成微生物在胴体间的转移。肉牛屠宰过程中,胴体上的菌落总数甚至可达2.7~4.0(lg(CFU/cm2)),分割后生鲜肉的菌落总数也在4.5(lg(CFU/cm2))以上,而生猪屠宰线上菌落总数普遍为3.0~5.3(lg(CFU/cm2)),分割肉的菌落总数甚至超过4.0(lg(CFU/cm2)),绵羊胴体表面的菌落总数则通常为3.1~4.0(lg(CFU/cm2))[6-8]。
屠宰过程中活体动物的皮毛和粪便、泥土及运输、待宰等过程中可转移到皮毛上的污染物是胴体主要的微生物污染源[3]。动物皮毛上通常具有较高的微生物数量和多样性。在肉牛屠宰前,Kang等[9]从皮毛上检出梭菌、拟杆菌、假单胞菌、乳酸菌等对肉腐败有促进作用的微生物。van Ba等[10]在牛皮毛上发现葡萄球菌、芽孢杆菌、埃希氏杆菌。Mohamed等[11]从绵羊皮毛上分离出微球菌、肠球菌、假单胞菌。Sui Lichang等[12]还从生猪皮毛上检测到嗜冷杆菌、气球菌、梭菌、乳杆菌等。不同或相同动物皮毛上的微生物种类差异与农场环境、动物饲养方式、屠宰场环境及季节差异密切相关。动物皮毛上的微生物主要通过剥皮环节转移到胴体表面,Tergney等[13]研究表明,剥皮操作可以使肉牛胴体上的粪便污染率达到30%~60%。肉羊与肉牛屠宰工序相似,经剥皮处理后,胴体表面的菌落总数可达6.95(lg(CFU/100 cm2)),大肠杆菌数量可达2.78(lg(CFU/100 cm2))[14]。Hauge等[14]研究发现,在屠宰前对绵羊进行剪毛处理可以显著降低胴体的细菌负荷。与肉牛和肉羊屠宰流程不同,国内的许多生猪屠宰企业不进行剥皮处理,而采用烫毛和燎毛处理,也可显著降低生猪皮毛上的微生物数量和多样性[15]。但在后续的抛光环节中,从燎毛工序中存活下来的细菌可能会扩散到胴体上,同时抛光设备上附着的微生物也会转移到胴体上[16]。Wheatley等[17]研究发现,抛光后猪胴体表面的菌落总数和肠杆菌数量均有所增加。
在分割环节中,一些研究从刀具、案板、传送带和工人佩戴的手套等物体上均检出较高的菌落总数,且其菌相与生鲜肉初始微生物组成具有较高的相似度,这表明分割过程中大多数细菌通过先污染操作环境再以此为载体接触胴体表面进行传播[16,18]。Wang Hui等[19]指出,假单胞菌是牛肉分割间传送带上的优势微生物群落,其会对冰鲜肉尤其是有氧条件下贮藏产品的货架期产生极大影响。Lavilla Lerma等[20]还从羊肉分割间环境中分离出抗生素耐药性最强的嗜温假单胞菌。还有学者在猪肉分割特定生产线上发现一些丰度较高的乳酸菌、假单胞菌、不动杆菌[21]。微生物污染还可以通过错误的着装、不良的生产规范和卫生规范、不清洁的设备和工作区域引入[22]。
为减少胴体表面的微生物量,国内一些屠宰企业在胴体冷却前普遍采用清水冲洗,或采用高压、热水和蒸汽[6]等物理干预措施。Han Jina等[23]详细报道了这些物理技术的减菌效果,物理干预措施局限性非常显著,例如,清水或高压清水冲洗可将微生物污染扩散到胴体相邻部位,还会促进空气和水中微生物的再污染[23];热水和蒸汽处理需要精准把控处理时间,防止对胴体质量产生不利影响[24]。为此,研究人员开发了应用于胴体表面的化学干预措施。例如,在有机酸喷淋中,乳酸喷淋对胴体表面细菌数量的降低效果最好,其次是醋酸和柠檬酸[25]。Han Jina等[26]研究证明,宰后45 min、9 h和23 h反复喷洒3%乳酸可以将胴体的菌落总数降至2(lg(CFU/cm2))。在冷却过程中反复喷洒300 mg/L过氧乙酸不仅能显著降低牛胴体表面的菌落总数和大肠菌群数,还能改变细菌群落组成。上述几种物理和化学减菌技术均可以减少胴体表面的微生物污染,但仅靠任何单一技术来控制或消除腐败微生物的效果有限,可有效利用干预措施之间的协同作用[23],在屠宰、分割的各个环节针对性地采取2 种或多种减菌措施能更好地降低胴体的初始微生物污染水平,更有效控制生鲜肉类的腐败。
2 红肉中常见优势腐败菌
虽然细菌可以通过多种途径污染胴体,但并不是所有细菌都会引发肉类腐败,环境适应能力强的细菌在肉类中能够占据生长优势,成为优势菌并主导腐败进程。腐败微生物的存活和生长受到肉类包装方式的极大影响。例如,有氧贮藏会促进假单胞菌生长,从而加速肉类腐败进程,而真空包装和气调包装可以促进兼性厌氧细菌(乳酸菌、热杀索丝菌等)占主导地位[4]。表1总结了牛肉、猪肉和羊肉在不同包装方式下冷藏期间的优势腐败菌。即使在低温下贮藏,托盘包装的腐败肉中也经常分离出假单胞菌。研究发现,莓实假单胞菌是腐败肉中最普遍的细菌种类,其次是隆德假单胞菌和荧光假单胞菌[27-28]。
除假单胞菌外,托盘包装红肉的优势菌中还包括一些常见的革兰氏阴性杆菌,包括不动杆菌和气单胞菌。Zhou Cong等[29]还从托盘包装猪肉中发现了库特氏菌。在Mansur等[30]的研究中,黄杆菌在托盘包装牛肉贮藏结束时占主导地位。在真空包装红肉中,与腐败相关的主要菌群为革兰氏阳性菌,包括乳酸菌和热杀索丝菌。但Russo等[31]研究发现,在厌氧条件下热杀索丝菌竞争不过冷藏肉中的乳酸菌。真空包装牛肉中乳酸菌主要以乳杆菌、明串珠菌、肉食杆菌为代表,而真空包装猪肉和羊肉中的乳酸菌均以乳杆菌、肉食杆菌、乳球菌为主。乳杆菌已被证明与肉类酸化、异味和黏液产生的腐败现象有关[32]。明串珠菌与肉类黄油气味与黏液形成、肉色变绿现象相关[33]。除此之外,在3 种红肉的真空包装和牛肉、羊肉的托盘包装中还发现了肉食杆菌,但有学者认为它们对肉类腐败感官品质的影响可以忽略不计[34]。由表1可知,在气调包装肉类冷藏期间,假单胞菌仍然是主要细菌,但包装中高浓度CO2(40%)对假单胞菌有显著抑制作用[35]。Pellissery等[32]称20%~60% CO2还能有效抑制不动杆菌生长。在≥50% O2气调包装条件下,牛肉、猪肉、羊肉中腐败微生物以假单胞菌、热杀索丝菌、乳酸菌为主。乳酸菌在牛肉中主要为明串珠菌、乳杆菌、肉食杆菌,在猪肉中为乳球菌、乳杆菌、明串珠菌,在羊肉中则以乳球菌为代表。气调包装中还值得关注的一类微生物是具有兼性厌氧性的肠杆菌,即使包装中高浓度CO2可以抑制其生长,但在贮藏结束时,肠杆菌仍然是微生物群落的重要组成部分[36]。例如,在≤30% O2气调包装红肉上发现的沙雷氏菌、哈夫尼菌就是2 种常见的肠杆菌,可以代谢肉中的氨基酸,产生胺、氨、甲基硫化物和硫醇,引起肉类产生异味和颜色变化[32]。有研究在60% O2+22%~25% CO2包装猪肉中还发现了特殊腐败微生物发光杆菌[37]。Li Ning等[38]研究发现,发光杆菌在托盘包装猪肉中也可占据主导地位。Yang Xiaoyin等[39]对0.4% CO+30% CO2+69.6% N2包装牛排冷藏过程中微生物群落演替进行研究发现,在CO和高浓度CO2环境中,假单胞菌和环丝菌的生长被完全抑制,乳球菌成为贮藏结束时的主要细菌。通过对牛肉、猪肉、羊肉在不同包装条件下的优势腐败菌分析发现,3 种红肉在相同包装条件下的优势菌虽有一定差异,但是大致相同,有氧包装条件下多以假单胞菌占优势,无氧包装下均以乳酸菌占优势;同时发现,不同研究中同种肉在相同包装条件下的优势菌也存在一定差异,这可能与肉的生产操作和生产环境差异导致的肉初始菌相差异有关,也可能与包装材料、包装条件(如真空度)、贮藏条件(如温度)等有关[3,40]。
3 红肉中微生物的腐败作用机制
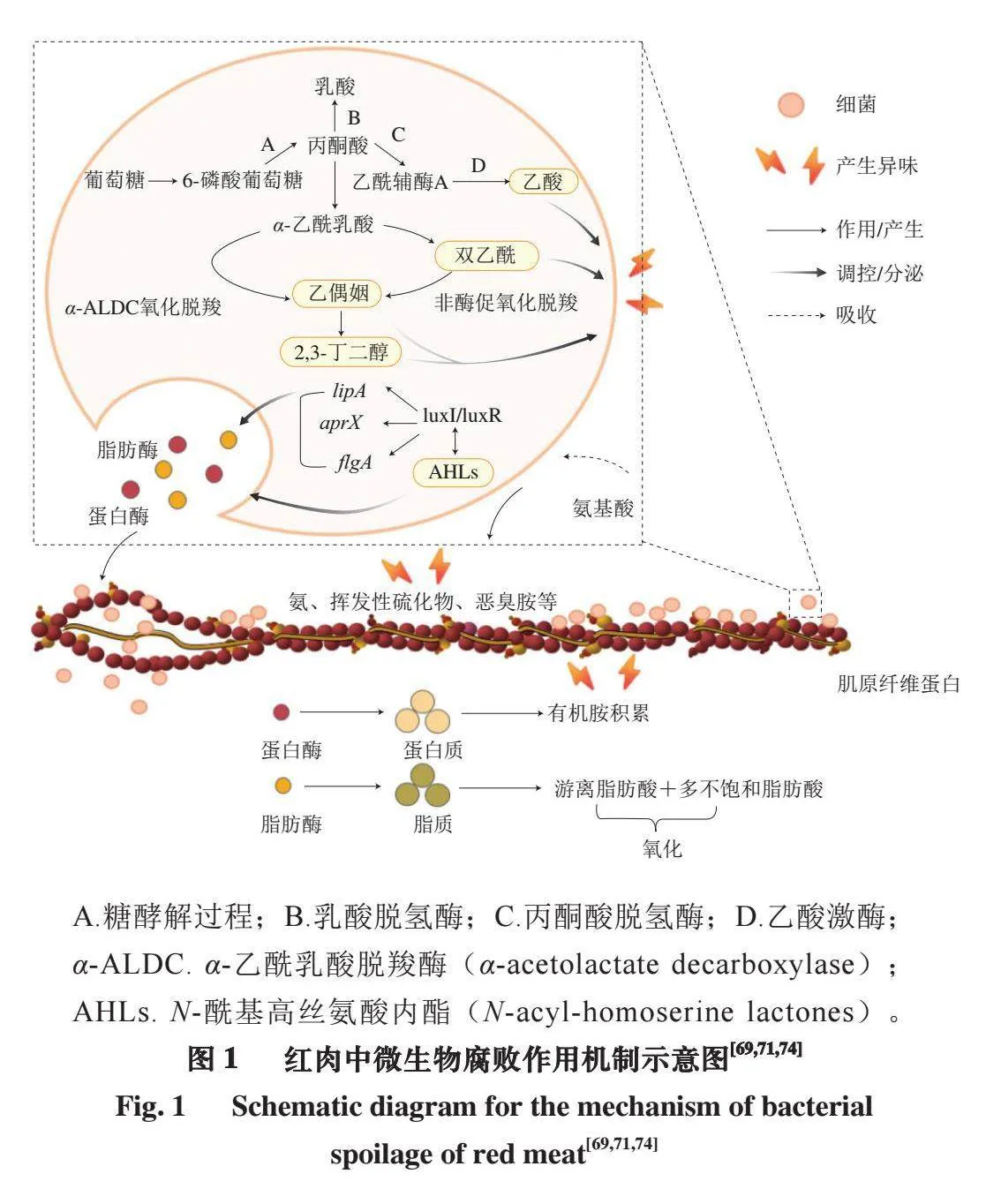
肉类腐败通常与细菌的代谢活动和酶活性有关,细菌可以利用底物通过丙酮酸代谢生成与腐败相关的化合物,这些化合物往往会导致肉类产生不正常或难闻的气味。同时细菌在信号分子的刺激和相关基因的调控下向胞外分泌酶,一方面造成细胞外物质积累,另一方面参与蛋白降解、脂肪分解等,进而影响肉的结构和品质。因此,一些学者从细菌代谢、信号分子产生、细菌胞外酶分泌(脂肪酶、蛋白酶等)和蛋白降解几个方面对肉类腐败的内在机制进行分析[69-74]。图1从细菌代谢、胞外酶分泌和蛋白降解3 个方面对红肉中微生物的腐败作用机制进行总结。
3.1 细菌代谢在红肉腐败中的作用
细菌代谢活动影响腐败类型、速率,而代谢途径丰富程度决定食品腐败状态[75]。在鲜肉中,细菌群落主要通过磷酸戊糖途径获取能量,丙酮酸代谢是第二大碳水化合物代谢途径,排名第三的是糖酵解途径,糖酵解相关基因在整个肉类贮藏期间均呈现高表达[76]。肉中的葡萄糖是多数微生物参与代谢活动首先利用的物质,有氧包装条件下的优势腐败菌假单胞菌可以充分利用肉中的碳源和能源,优先分解葡萄糖尤其是D-葡萄糖[4],但也有例外,如莓实假单胞菌NMC25,其在肉中主要以氨基酸和短肽作为碳源和氮源[77]。Kolbeck等[78]推测无氧条件下的假单胞菌可能通过Entner-Doudoroff途径进行葡萄糖代谢,通过形成乙醇进行烟酰胺腺嘌呤二核苷酸(nicotinamide adenine dinucleotide,NAD+)回收,从而实现长期存活。研究[79-80]表明,真空包装和CO气调包装中的优势菌——乳酸菌主要通过碳水化合物代谢途径维持生长活动,碳水化合物吸收和代谢对乳酸菌生长和将肉中的糖转化为乳酸极为重要。乳酸菌偏好利用葡萄糖,同时也能代谢常见的己糖、戊糖和核糖[81]。Bell[82]研究发现,葡萄糖初始浓度较高的肉出现腐败现象(黏液、异味)比葡萄糖浓度较低的肉需要更大的细菌数量。肉中葡萄糖浓度的限制促使细菌代谢由糖分解向氨基酸降解转换。Rood等[83]报道,添加0.5%~10%葡萄糖可以延长真空包装冷藏羊肉的货架期。随着贮藏时间延长,肉中葡萄糖水平不断下降,在缺乏糖的情况下,丙酮酸的葡萄糖异生较为活跃。Zareian等[84]研究表明,丙酮酸代谢活动由丙酮酸甲酸裂合酶、丙酮酸脱氢酶、乳酸脱氢酶、乙酸激酶参与,与腐败相关化合物2,3-丁二醇、乙偶姻、乙酸等的生成密切相关。一旦肉中葡萄糖耗尽,细菌会继续代谢次级产物,如游离氨基酸和乳酸[85]。
碳水化合物被消耗殆尽后,肉中参与氨基酸代谢的途径丰度显著增加[62]。例如,有氧条件下莓实假单胞菌和热杀索丝菌的协同作用由组氨酸和核苷酸代谢途径支持,可促进冷藏猪肉发生腐败[86]。含硫氨基酸的分解导致硫化氢、二甲基硫化物和二甲基二硫化物等恶臭味含硫挥发性化合物形成,释放难闻气味[87]。
3.2 细菌分泌信号分子在红肉腐败中的作用
细菌分泌的信号分子中,AHLs可通过刺激细菌的群体感应(quorum sensing,QS)系统调控蛋白酶和脂肪酶相关基因表达。AHLs由luxI蛋白负责产生,当AHLs浓度达到一定阈值时,受体luxR与内源性AHLs及相邻细胞产生的外源性AHLs相结合,形成受体-AHL复合物,触发细菌特定基因flgA、aprX、lipA的表达,发挥调节胞外蛋白酶和脂肪酶分泌、生物膜形成等生理功能,从而促进肉贮藏期间蛋白、脂肪、糖的分解,导致总挥发性盐基氮含量增加,提高细菌腐败活性[71,88]。与肉类腐败密切相关的荧光假单胞菌主要采用luxI/R型QS系统,AHL依赖性QS由luxI同源物合成,并通过luxR型转录调控因子检测。信号通过细胞膜自由扩散,当细胞密度较高时,luxR结合其同源自诱导剂,luxR自诱导剂复合物结合目标基因启动子并激活生物膜形成、运动和酶的产生等途径,导致肉类腐败[89]。莓实假单胞菌经常被报道为腐败肉中的优势菌,但Ferrocino等[90]研究发现,从新鲜和腐败肉中分离的72 株莓实假单胞菌均不能产生AHLs,由此猜测莓实假单胞菌缺少与AHLs合成相关的基因,Quintieri等[89]验证了这一假设。常见的腐败菌哈夫尼菌和沙雷氏菌均具有较强的AHLs生成能力,AHLs介导的QS系统在细菌在不同表面的运动和生物膜形成中起关键作用[71,91]。抑制细菌的QS系统已经成为一种新型防腐策略。目前,已有多种天然和合成化合物被证实具有群体感应抑制剂(quorum sensing inhibitory,QSI)活性[92]。Zhang Ying等[93]研究表明,乙醛能够与荧光假单胞菌中EcbI和PcoI蛋白相互作用,降低特殊AHLs(N-辛酰基-L-高丝氨酸内酯)浓度,从而抑制假单胞菌生物膜形成、降低细胞外酶活性。Gopu等[94]证明孜然、茴香和胡椒可以抑制腌制牛肉中AHLs产生,从而降低微生物负荷并延迟腐败。酚类化合物可能通过与AHLs信号分子竞争性结合QS受体,抑制QS系统[95]。例如,姜黄素可以抑制铜绿假单胞菌、大肠杆菌、沙雷氏菌的几种表型;白藜芦醇能够抑制细菌生物膜形成;没食子酸能够引起细菌细胞膜破裂和孔隙形成等不可逆变化;辣椒素具有抑制微生物生长能力[95-98]。
3.3 胞外酶分泌及肌肉蛋白降解在红肉腐败中的作用
肉类富含蛋白质,这对维持肌肉骨架至关重要。腐败微生物通过分泌胞外蛋白酶,水解肌肉中的蛋白质(如肌原纤维蛋白、胶原蛋白等),产生微生物生长所必需的氨基酸,从而诱导和加速肉类腐败[72]。目前已经鉴定和表征了多种细菌胞外蛋白酶,包括金属蛋白酶、丝氨酸蛋白酶和胶原蛋白酶。例如,Wang Guangyu等[88]发现,金属蛋白酶AprA是莓实假单胞菌产生的唯一胞外蛋白酶,在8 ℃下可以降解肉肌原纤维蛋白。Jia Shiliang等[99]研究证明,嗜冷假单胞菌和腐败希瓦氏菌分泌的丝氨酸蛋白酶是肉类腐败过程中参与蛋白质降解的主要蛋白酶。有报道称在4~15 ℃下蜂房哈夫尼菌中编码丝氨酸蛋白酶的serp基因具有高表达[100]。假单胞菌、沙雷氏菌和气单胞菌的一些菌株还表现出强胶原蛋白酶活性,可促进带皮肉类产品腐败[101]。根据肽链中蛋白酶活性位点的位置,细菌分泌的蛋白酶可初步分为内肽酶和外肽酶[72]。M4家族金属肽酶大多数为内肽酶,当其与蛋白质相结合时,酶上的金属离子可能通过增强水分子的亲核性实现肽键攻击,促进蛋白质降解[72-73]。氨肽酶是一种催化肽和蛋白质N端氨基酸残基水解的外肽酶[102]。Tan Chunming等[103]已经证明腐败菌可以通过分泌氨肽酶破坏蛋白质结构,促进贮藏后期肌肉结构分解和蛋白质降解。肉贮藏期间的优势腐败菌普遍具有较高的蛋白水解潜力,Scatamburlo等[104]研究发现,假单胞菌在7、25、35 ℃下均表现出蛋白水解活性,Alves等[105]观察到一些假单胞菌还可以产生冷活性蛋白酶和脂肪酶,即使在冷藏条件下酶仍具有高活性和较高表达量。假单胞菌具有较强的肌原纤维蛋白和肌基质蛋白降解能力,易导致肉类外表面形成黏液和肉质软化[106]。腐败菌分泌的脂肪酶可以将肉中的脂肪和脂质分解成分子质量更小的成分,如脂肪酸和甘油,产生酸败和难闻的味道[107]。酶源性的脂肪分解产生的游离脂肪酸还是挥发性物质的主要前体。Ercolini等[108]研究发现,莓实假单胞菌分泌的一些脂肪酶可以更特异性地水解肉类脂肪,且大多在4 ℃有脂解作用,一小部分在20 ℃活性更强。从肉和肉制品中分离出的芽孢杆菌、热杀索丝菌和沙雷氏菌均可以分泌脂肪酶,加速肉类腐败[109-111]。
4 结 语
细菌主导的红肉腐败非常复杂,不仅受到多种外界因素的影响,还受到内在多个生化系统的调控。目前,肉类的保鲜多涉及温度、包装方面,通过调节贮藏环境条件抑制微生物的生长繁殖。未来,通过影响细菌腐败机制来抑制腐败现象的产生可能成为新的研究热点。因此,为控制肉类腐败、延长货架期,必须全面深入了解细菌的腐败机制。虽然本文已经揭示了细菌引起的肉类腐败的基本原理,但不同肉类腐败过程中优势菌作用机制的差异还所知甚少。肉类腐败过程中各种主要细菌作用机制之间的特定相互作用(如促进、抑制和干预)还有待进一步研究,各种代谢产物对菌群的反馈效应还有待进一步探索。
参考文献:
[1] HU G W, MU X Z, XU M, et al. Potentials of GHG emission reductions from cold chain systems: case studies of China and the United States[J]. Journal of Cleaner Production, 2019, 239: 118053. DOI:10.1016/j.jclepro.2019.118053.
[2] KAUR M, WILLIAMS M, BISSETT A, et al. Effect of abattoir, livestock species and storage temperature on bacterial community dynamics and sensory properties of vacuum packaged red meat[J]. Food Microbiology, 2021, 94: 103648. DOI:10.1016/j.fm.2020.103648.
[3] MORSHDY A E M, MEHREZ S M, THARWAT A E, et al. A review on the microbial surface contaminants of the animal carcasses[J]. Journal of Advanced Veterinary Research, 2023, 13(6): 1248-1251.
[4] DOULGERAKI A I, ERCOLINI D, VILLANI F, et al. Spoilage microbiota associated to the storage of raw meat in different conditions[J]. International Journal of Food Microbiology, 2012, 157(2): 130-141. DOI:10.1016/j.ijfoodmicro.2012.05.020.
[5] United States Department of Agriculture (USDA). Livestock and poultry: world markets and trade[EB/OL]. (2023-10-12) [2024-06-03]. https://www.fas.usda.gov/data/livestock-and-poultry-world-markets-and-trade.
[6] 韩吉娜, 张佳, 罗欣, 等. 肉牛屠宰过程中的减菌技术研究进展[J]. 食品科学, 2019, 40(15): 330-337. DOI:10.7506/spkx1002-6630-20180625-465.
[7] 田盼. 冷鲜牛肉生产过程的HACCP体系建立与减菌措施[D]. 石河子: 石河子大学, 2015. DOI:10.7666/d.D717838.
[8] MARTÍNEZ B, CELDA M F, MILLÁN M E, et al. Assessment of the microbiological conditions of red-meat carcasses from bacterial counts recovered by sampling via excision or swabbing with cotton wool[J]. International Journal of Food Science amp; Technology, 2009, 44(4): 770-776. DOI:10.1111/j.1365-2621.2008.01895.x.
[9] KANG S S, RAVENSDALE J, COOREY R, et al. A comparison of 16S rRNA profiles through slaughter in Australian export beef abattoirs[J]. Frontiers in Microbiology, 2019, 10: 2747. DOI:10.3389/fmicb.2019.02747.
[10] VAN BA H, SEO H W, PIL-NAM S, et al. The effects of pre-and post-slaughter spray application with organic acids on microbial population reductions on beef carcasses[J]. Meat Science, 2018, 137: 16-23. DOI:10.1016/j.meatsci.2017.11.006.
[11] MOHAMED H A, VAN KLINK E G, ELHASSAN S M. Damage caused by spoilage bacteria to the structure of cattle hides and sheep skins[J]. International Journal of Animal Health and Livestock Production Research, 2016, 2(1): 39-56.
[12] SUI L C, YI Z K, XIAO X N, et al. Investigation of microbial communities across swine slaughter stages and disinfection efficacy assessment in a pig slaughterhouse[J]. LWT-Food Science and Technology, 2023, 187: 115334. DOI:10.1016/j.lwt.2023.115334.
[13] TERGNEY A, BOLTON D. Validation studies on an online monitoring system for reducing faecal and microbial contamination on beef carcasses[J]. Food Control, 2006, 17(5): 378-382. DOI:10.1016/j.foodcont.2005.01.004.
[14] HAUGE S J, NAFSTAD O, SKJERVE E, et al. Effects of shearing and fleece cleanliness on microbiological contamination of lamb carcasses[J]. International Journal of Food Microbiology, 2011, 150(2/3): 178-183. DOI:10.1016/j.ijfoodmicro.2011.07.038.
[15] ZDOLEC N, KOTSIRI A, HOUF K, et al. Systematic review and meta-analysis of the efficacy of interventions applied during primary processing to reduce microbial contamination on pig carcasses[J]. Foods, 2022, 11(14): 2110. DOI:10.1111/dme.13509.
[16] ZWIRZITZ B, WETZELS S U, DIXON E D, et al. The sources and transmission routes of microbial populations throughout a meat processing facility[J]. npj Biofilms Microbomes, 2020, 6(1): 12. DOI:10.1038/s41522-020-0136-z.
[17] WHEATLEY P, GIOTIS E S, MCKEVITT A I. Effects of slaughtering operations on carcass contamination in an Irish pork production plant[J]. Irish Veterinary Journal, 2014, 67(1): 1. DOI:10.1186/2046-0481-67-1.
[18] LABAN S, MASHALY M, ALY A, et al. Evaluation of different hygienic practices applied in slaughterhouses and its effect on beef quality[J]. Advances in Animal and Veterinary Sciences, 2021, 9(3): 429-437.
[19] WANG H, HE A N, YANG X Q. Dynamics of microflora on conveyor belts in a beef fabrication facility during sanitation[J]. Food Control, 2018, 85: 42-47. DOI:10.1016/j.foodcont.2017.09.017.
[20] LAVILLA LERMA L, BENOMAR N, CASADO MUNOZ M D C,
et al. Antibiotic multiresistance analysis of mesophilic and psychrotrophic Pseudomonas spp. isolated from goat and lamb slaughterhouse surfaces throughout the meat production process[J]. Applied and Environmental Microbiology, 2014, 80(21): 6792-6806. DOI:10.1128/AEM.01998-14.
[21] SHEDLEUR-BOURGUIGNON F, DUCHEMIN T, THÉRIAULT W P, et al. Distinct microbiotas are associated with different production lines in the cutting room of a swine slaughterhouse[J]. Microorganisms, 2023, 11(1): 133. DOI:10.3390/microorganisms11010133.
[22] NYCHAS G J E, SKANDAMIS P N, TASSOU C C, et al. Meat spoilage during distribution[J]. Meat Science, 2008, 78(1/2): 77-89. DOI:10.1016/j.meatsci.2007.06.020.
[23] HAN J N, DONG P C, HOLMAN B W, et al. Processing interventions for enhanced microbiological safety of beef carcasses and beef products: a review[J]. Critical Reviews in Food Science and Nutrition, 2024, 64(8): 2105-2129. DOI:10.1080/10408398.2022.2121258.
[24] CASTILLO A, HARDIN M D, ACUFF G R, et al. Reduction of microbial contaminants on carcasses[M]//JUNEJA V K, SOFOS J N. Control of foodborne microorganisms. Boca Raton: CRC Press, 2001: 351-381. DOI:10.1201/b16945-13.
[25] DAN S D, MIHAIU M, REGET O, et al. Pathogens contamination level reduction on beef using organic acids decontamination methods[J]. Bulletin of University of Agricultural Sciences and Veterinary Medicine CLUJ-NAPOCA. Veterinary Medicine, 2017, 74(2): 52. DOI:10.15835/buasvmcn-vm:0052.
[26] HAN J N, LUO X, ZHANG Y M, et al. Effects of spraying lactic acid and peroxyacetic acid on the bacterial decontamination and bacterial composition of beef carcasses[J]. Meat Science, 2020, 164: 108104. DOI:10.1016/j.meatsci.2020.108104.
[27] STANBOROUGH T, FEGAN N, POWELL S M, et al. Genomic and metabolic characterization of spoilage-associated Pseudomonas species[J]. International Journal of Food Microbiology, 2018, 268: 61-72. DOI:10.1016/j.ijfoodmicro.2018.01.005.
[28] WICKRAMASINGHE N N, RAVENSDALE J, COOREY R, et al. The predominance of psychrotrophic pseudomonads on aerobically stored chilled red meat[J]. Comprehensive Reviews in Food Science and Food Safety, 2019, 18(5): 1622-1635. DOI:10.1111/1541-4337.12483.
[29] ZHOU C, WANG J J, LI R, et al. High-throughput sequencing analysis of the bacterial community for assessing the differences in extraction methods of bacteria separation from chilled pork[J]. LWT-Food Science and Technology, 2020, 134: 110213. DOI:10.1016/j.lwt.2020.110213.
[30] MANSUR A R, SONG E J, CHO Y S, et al. Comparative evaluation of spoilage-related bacterial diversity and metabolite profiles in chilled beef stored under air and vacuum packaging[J]. Food Microbiology, 2019, 77: 166-172. DOI:10.1016/j.fm.2018.09.006.
[31] RUSSO F, ERCOLINI D, MAURIELLO G, et al. Behaviour of Brochothrix thermosphacta in presence of other meat spoilage microbial groups[J]. Food Microbiology, 2006, 23(8): 797-802. DOI:10.1016/j.fm.2006.02.004.
[32] PELLISSERY A J, VINAYAMOHAN P G, AMALARADJOU M A R, et al.
Spoilage bacteria and meat quality[M]//BISWAS A K, MANDAL P K.
Meat quality analysis. Amsterdam: Academic Press, 2020: 307-334. DOI:10.1016/B978-0-12-819233-7.00017-3.
[33] JOHANSSON P, JÄÄSKELÄINEN E, NIEMINEN T, et al. Microbiomes in the context of refrigerated raw meat spoilage[J]. Meat and Muscle Biology, 2020, 4(2): 10369. DOI:10.22175/mmb.10369.
[34] XU M M, KAUR M, PILLIDGE C J, et al. Effect of protective cultures on spoilage bacteria and the quality of vacuum-packaged lamb meat[J]. Food Bioscience, 2022, 50: 102148. DOI:10.1016/j.fbio.2022.102148.
[35] YANG J, YANG X Y, LIANG R R, et al. The response of bacterial communities to carbon dioxide in high-oxygen modified atmosphere packaged beef steaks during chilled storage[J]. Food Research International, 2022, 151: 110872. DOI:10.1016/j.foodres.2021.110872.
[36] DJORDJEVIC J, BOSKOVIC M, DOKMANOVIC M, et al. Vacuum and modified atmosphere packaging effect on Enterobacteriaceae behaviour in minced meat[J]. Journal of Food Processing and Preservation, 2017, 41(2): e12837. DOI:10.1111/jfpp.12837.
[37] NIEMINEN T T, DALGAARD P, BJÖRKROTH J. Volatile organic compounds and Photobacterium phosphoreum associated with spoilage of modified-atmosphere-packaged raw pork[J]. International Journal of Food Microbiology, 2016, 218: 86-95. DOI:10.1016/j.ijfoodmicro.2015.11.003.
[38] LI N, ZHANG Y X, WU Q P, et al. High-throughput sequencing analysis of bacterial community composition and quality characteristics in refrigerated pork during storage[J]. Food Microbiology, 2019, 83: 86-94. DOI:10.1016/j.fm.2019.04.013.
[39] YANG X Y, LUO X, ZHANG Y M, et al. Effects of microbiota dynamics on the color stability of chilled beef steaks stored in high oxygen and carbon monoxide packaging[J]. Food Research International, 2020, 134: 109215. DOI:10.1016/j.foodres.2020.109215.
[40] ODEYEMI O A, ALEGBELEYE O O, STRATEVA M, et al. Understanding spoilage microbial community and spoilage mechanisms in foods of animal origin[J]. Comprehensive Reviews in Food Science and Food Safety, 2020, 19(2): 311-331. DOI:10.1111/1541-4337.12526.
[41] ERCOLINI D, FERROCINO I, NASI A, et al. Monitoring of microbial metabolites and bacterial diversity in beef stored under different packaging conditions[J]. Applied and Environmental Microbiology, 2011, 77(20): 7372-7381. DOI:10.1128/AEM.05521-11.
[42] 牟广磊. 不同包装方式对冷却牛肉品质及微生物影响的研究[D].
泰安: 山东农业大学, 2015. DOI:10.7666/d.D01212179.
[43] 岑璐伽, 唐善虎, 郝小倩, 等. 冷却牦牛肉贮藏过程中优势菌的PCR-变性梯度凝胶电泳分析[J]. 肉类研究, 2012, 25(1): 36-40. DOI:10.3969/j.issn.1001-8123.2012.01.010.
[44] CHEN X, ZHANG Y M, YANG X Y, et al. Shelf-life and microbial community dynamics of super-chilled beef imported from Australia to China[J]. Food Research International, 2019, 120: 784-792. DOI:10.1016/j.foodres.2018.11.039.
[45] ROVIRA P, BRUGNINI G, RODRIGUEZ J, et al. Microbiological changes during long-storage of beef meat under different temperature and vacuum-packaging conditions[J]. Foods, 2023, 12(4): 694. DOI:10.3390/foods12040694.
[46] YANG J, WEI W, HOLMAN B W, et al. Effects of low-energy electron beam irradiation on the shelf-life and quality of vacuum-packaged beef steaks during chilled storage[J]. Meat Science, 2022, 193: 108932. DOI:10.1016/j.meatsci.2022.108932.
[47] CHEN X, ZHU L X, LIANG R R, et al. Shelf-life and bacterial community dynamics of vacuum packaged beef during long-term super-chilled storage sourced from two Chinese abattoirs[J]. Food Research International, 2020, 130: 108937. DOI:10.1016/j.foodres.2019.108937.
[48] REID R, FANNING S, WHYTE P, et al. Comparison of hot versus cold boning of beef carcasses on bacterial growth and the risk of blown pack spoilage[J]. Meat Science, 2017, 125: 46-52. DOI:10.1016/j.meatsci.2016.11.012.
[49] ERCOLINI D, RUSSO F, TORRIERI E, et al. Changes in the spoilage-related microbiota of beef during refrigerated storage under different packaging conditions[J]. Applied and Environmental Microbiology, 2006, 72(7): 4663-4671. DOI:10.1128/AEM.00468-06.
[50] JÄÄSKELÄINEN E, HULTMAN J, PARSHINTSEV J, et al. Development of spoilage bacterial community and volatile compounds in chilled beef under vacuum or high oxygen atmospheres[J]. International Journal of Food Microbiology, 2016, 223: 25-32. DOI:10.1016/j.ijfoodmicro.2016.01.022.
[51] 杨鸿博, 杨啸吟, 张一敏, 等. 包装方式对牛排贮藏期间微生物数量和演替的影响[J]. 食品科学, 2021, 42(13): 166-173. DOI:10.7506/spkx1002-6630-20200519-217.
[52] SÄDE E, PENTTINEN K, BJÖRKROTH J, et al. Exploring lot-to-lot variation in spoilage bacterial communities on commercial modified atmosphere packaged beef[J]. Food Microbiology, 2017, 62: 147-152. DOI:10.1016/j.fm.2016.10.004.
[53] SEQUINO G, COBO-DIAZ J F, VALENTINO V, et al. Microbiome mapping in beef processing reveals safety-relevant variations in microbial diversity and genomic features[J]. Food Research International, 2024, 186: 114318. DOI:10.1016/j.foodres.2024.114318.
[54] WANG X H, DENG Y H, SUN J S, et al. Unraveling characterizations of bacterial community and spoilage profiles shift in chilled pork during refrigerated storage[J]. Food Science and Technology, 2021, 42: e80321. DOI:10.1590/FST.80321.
[55] JIANG Y, GAO F, XU X L, et al. Changes in the composition of the bacterial flora on tray-packaged pork during chilled storage analyzed by PCR-DGGE and real-time PCR[J]. Journal of Food Science, 2011, 76(1): M27-M33. DOI:10.1111/j.1750-3841.2010.01879.x.
[56] ZHAO F, ZHOU G H, YE K P, et al. Microbial changes in vacuum-packed chilled pork during storage[J]. Meat Science, 2015, 100: 145-149. DOI:10.1016/j.meatsci.2014.10.004.
[57] JIANG Y, GAO F, XU X L, et al. Changes in the bacterial communities of vacuum-packaged pork during chilled storage analyzed by PCR-DGGE[J]. Meat Science, 2010, 86(4): 889-895. DOI:10.1016/j.meatsci.2010.05.021.
[58] 王真真. 不同包装冷却猪肉的腐败特性研究[D]. 郑州: 河南农业大学, 2012. DOI:10.7666/d.y2157052.
[59] 江芸. 托盘和真空包装冷却猪肉冷藏过程中菌相变化规律研究[D]. 南京: 南京农业大学, 2010. DOI:10.7666/d.Y1986600.
[60] BASSEY A P, CHEN Y F, ZHU Z S, et al. Assessment of quality characteristics and bacterial community of modified atmosphere packaged chilled pork loins using 16S rRNA amplicon sequencing analysis[J]. Food Research International, 2021, 145: 110412. DOI:10.1016/j.foodres.2021.110412.
[61] WANG T J, GUO H Y, ZHANG H, et al. Dynamics of bacterial communities of lamb meat packaged in air and vacuum pouch during chilled storage[J]. Food Science of Animal Resources, 2019, 39(2): 209-221. DOI:10.5851/kosfa.2019.e16.
[62] LIANG C, ZHANG D Q, ZHENG X C, et al. Effects of different storage temperatures on the physicochemical properties and bacterial community structure of fresh lamb meat[J]. Food Science of Animal Resources, 2021, 41(3): 509-526. DOI:10.5851/kosfa.2021.e15.
[63] WEN X Y, LIANG C, ZHANG D Q, et al. Effects of hot or cold boning on the freshness and bacterial community changes of lamb cuts during chilled storage[J]. LWT-Food Science and Technology, 2022, 170: 114063. DOI:10.1016/j.lwt.2022.114063.
[64] WEN X Y, ZHANG D Q, LI X, et al. Dynamic changes of bacteria and screening of potential spoilage markers of lamb in aerobic and vacuum packaging[J]. Food Microbiology, 2022, 104: 103996. DOI:10.1016/j.fm.2022.103996.
[65] WANG T J, ZHAO L, SUN Y N, et al. Changes in the microbiota of lamb packaged in a vacuum and in modified atmospheres during chilled storage analysed by high-throughput sequencing[J]. Meat Science, 2016, 121: 253-260. DOI:10.1016/j.meatsci.2016.06.021.
[66] KAUR M, SHANG H S, TAMPLIN M, et al. Culture-dependent and culture-independent assessment of spoilage community growth on VP lamb meat from packaging to past end of shelf-life[J]. Food Microbiology, 2017, 68: 71-80. DOI:10.1016/j.fm.2017.06.015.
[67] CARRIZOSA E, BENITO M J, RUIZ-MOYANO S, et al. Bacterial communities of fresh goat meat packaged in modified atmosphere[J]. Food Microbiology, 2017, 65: 57-63. DOI:10.1016/j.fm.2017.01.023.
[68] RUBIO B, VIEIRA C, MARTÍNEZ B. Effect of post mortem temperatures and modified atmospheres packaging on shelf life of suckling lamb meat[J]. LWT-Food Science and Technology, 2016, 69: 563-569. DOI:10.1016/j.lwt.2016.02.008.
[69] SHAO L T, CHEN S S, WANG H D, et al. Advances in understanding the predominance, phenotypes, and mechanisms of bacteria related to meat spoilage[J]. Trends in Food Science amp; Technology, 2021, 118: 822-832. DOI:10.1016/j.tifs.2021.11.007.
[70] BRUHN J B, CHRISTENSEN A B, FLODGAARD L R, et al. Presence of acylated homoserine lactones (AHLs) and AHL-producing bacteria in meat and potential role of AHL in spoilage of meat[J]. Applied and Environmental Microbiology, 2004, 70(7): 4293-4302. DOI:10.1128/AEM.70.7.4293-4302.2004.
[71] MACHADO I, SILVA L R, GIAOURIS E D, et al. Quorum sensing in food spoilage and natural-based strategies for its inhibition[J]. Food Research International, 2020, 127: 108754. DOI:10.1016/j.foodres.2019.108754.
[72] SHAO L T, DONG Y, CHEN S S, et al. Revealing extracellular protein profile and excavating spoilage-related proteases of Aeromonas salmonicida based on multi-omics investigation[J]. International Journal of Biological Macromolecules, 2024, 265: 130916. DOI:10.1016/j.ijbiomac.2024.130916.
[73] ADEKOYA O A, SYLTE I. The thermolysin family (M4) of enzymes: therapeutic and biotechnological potential[J]. Chemical Biology amp; Drug Design, 2009, 73(1): 7-16. DOI:10.1111/j.1747-0285.2008.00757.x.
[74] DAI J Y, FANG L M, WU Y, et al. Effects of exogenous AHLs on the spoilage characteristics of Pseudomonas koreensis PS1[J]. Journal of Food Science, 2022, 87(2): 819-832. DOI:10.1111/1750-3841.16038.
[75] DAVE D, GHALY A E. Meat spoilage mechanisms and preservation techniques: a critical review[J]. American Journal of Agricultural and Biological Sciences, 2011, 6(4): 486-510. DOI:10.3844/ajabssp.2011.486.510.
[76] HULTMAN J, JOHANSSON P, BJÖRKROTH J. Longitudinal metatranscriptomic analysis of a meat spoilage microbiome detects abundant continued fermentation and environmental stress responses during shelf life and beyond[J]. Applied and Environmental Microbiology, 2020, 86(24): e01575-20. DOI:10.1128/AEM.01575-20.
[77] WANG G Y, MA F, CHEN X J, et al. Transcriptome analysis of the global response of Pseudomonas fragi NMC25 to modified atmosphere packaging stress[J]. Frontiers in Microbiology, 2018, 9: 1277. DOI:10.3389/fmicb.2018.01277.
[78] KOLBECK S, ABELE M, HILGARTH M, et al. Comparative proteomics reveals the anaerobic lifestyle of meat-spoiling Pseudomonas species[J]. Frontiers in Microbiology, 2021, 12: 664061. DOI:10.3389/fmicb.2021.664061.
[79] PENNACCHIA C, ERCOLINI D, VILLANI F. Spoilage-related microbiota associated with chilled beef stored in air or vacuum pack[J]. Food Microbiology, 2011, 28(1): 84-93. DOI:10.1016/j.fm.2010.08.010.
[80] YANG X Y, ZHU L X, ZHANG Y M, et al. Microbial community dynamics analysis by high-throughput sequencing in chilled beef longissimus steaks packaged under modified atmospheres[J]. Meat Science, 2018, 141: 94-102. DOI:10.1016/j.meatsci.2018.03.010.
[81] 焦晶凯. 乳酸菌代谢研究进展[J]. 乳业科学与技术, 2020, 43(2): 49-55. DOI:10.15922/j.cnki.jdst.2020.02.009.
[82] BELL R G. Meat packaging: protection, preservation and presentation[M]//HUI Y H, NIP W K, ROGERS R W, et al. Meat science and applications. Boca Raton: CRC Press, 2001: 479-506. DOI:10.1201/9780203908082.ch19.
[83] ROOD L, BOWMAN J P, ROSS T, et al. The effects of glucose on microbial spoilage of vacuum-packed lamb[J]. Meat Science, 2022, 188: 108781. DOI:10.1016/j.meatsci.2022.108781.
[84] ZAREIAN M, BOHNER N, LOOS H M, et al. Evaluation of volatile organic compound release in modified atmosphere-packaged minced raw pork in relation to shelf-life[J]. Food Packaging Shelf Life, 2018, 18: 51-61. DOI:10.1016/j.fpsl.2018.08.001.
[85] AJAYKUMAR V J, MANDAL P K. Modern concept and detection of spoilage in meat and meat products[M]//BISWAS A K, MANDAL P K.
Meat quality analysis. Amsterdam: Academic Press, 2020: 335-349. DOI:10.1016/B978-0-12-819233-7.00018-5.
[86] ZHOU Z L, REN F Q, HUANG Q L, et al. Characterization and interactions of spoilage of Pseudomonas fragi C6 and Brochothrix thermosphacta S5 in chilled pork based on LC-MS/MS and screening of potential spoilage biomarkers[J]. Food Chemistry, 2024, 444: 138562. DOI:10.1016/j.foodchem.2024.138562.
[87] BEKHIT A A, HOLMAN B, GITERU S G, et al. Total volatile basic nitrogen (TVB-N) and its role in meat spoilage: a review[J]. Trends in Food Science amp; Technology, 2021, 109: 280-302. DOI:10.1016/j.tifs.2021.01.006.
[88] WANG G Y, LI Q, TANG W Q, et al. AprD is important for extracellular proteolytic activity, physicochemical properties and spoilage potential in meat-borne Pseudomonas fragi[J]. Food Control, 2021, 124: 107868. DOI:10.1016/j.foodcont.2021.107868.
[89] QUINTIERI L, CAPUTO L, BRASCA M, et al. Recent advances in the mechanisms and regulation of QS in dairy spoilage by Pseudomonas spp.[J]. Foods, 2021, 10(12): 3088. DOI:10.3390/foods10123088.
[90] FERROCINO I, ERCOLINI D, VILLANI F, et al. Pseudomonas fragi strains isolated from meat do not produce N-acyl homoserine lactones as signal molecules[J]. Journal of Food Protection, 2009, 72(12): 2597-2601. DOI:10.4315/0362-028x-72.12.2597.
[91] ZHU Y L, HOU H M, ZHANG G L, et al. AHLs regulate biofilm formation and swimming motility of Hafnia alvei H4[J]. Frontiers in Microbiology, 2019, 10: 1330. DOI:10.3389/fmicb.2019.01330.
[92] CHEN X H, YU F H, LI Y Q, et al. The inhibitory activity of p-coumaric acid on quorum sensing and itsenhancement effect on meat preservation[J]. CyTA-Journal of Food, 2020, 18(1): 61-67. DOI:10.1080/19476337.2019.1701558.
[93] ZHANG Y, YU H, XIE Y F, et al. Inhibitory effects of hexanal on acylated homoserine lactones (AHLs) production to disrupt biofilm formation and enzymes activity in Erwinia carotovora and Pseudomonas fluorescens[J]. Journal of Food Science and Technology, 2023, 60(1): 372-381. DOI:10.1007/s13197-022-05624-9.
[94] GOPU V, SHETTY P H. Regulation of acylated homoserine lactones (AHLs) in beef by spice marination[J]. Journal of food science and technology, 2016, 53: 2686-2694. DOI:10.1007/s13197-016-2240-x.
[95] SANTOS C, LIMA E, FRANCO BDGDM P U. Exploring phenolic compounds as quorum sensing inhibitors in foodborne bacteria[J]. Frontiers in Microbiology, 2021, 12: 735931. DOI:10.3389/fmicb.2021.735931.
[96] DING T, LI T T, WANG Z, et al. Curcumin liposomes interfere with quorum sensing system of Aeromonas sobria and in silico analysis[J]. Scientific Reports, 2017, 7(1): 8612. DOI:10.1038/s41598-017-08986-9.
[97] DUARTE A, ALVES A C, FERREIRA S, et al. Resveratrol inclusion complexes: antibacterial and anti-biofilm activity against Campylobacter spp. and Arcobacter butzleri[J]. Food Research International, 2015, 77: 244-250. DOI:10.1016/j.foodres.2015.05.047.
[98] SORRENTINO E, SUCCI M, TIPALDI L, et al. Antimicrobial activity of gallic acid against food-related Pseudomonas strains and its use as biocontrol tool to improve the shelf life of fresh black truffles[J]. International Journal of Food Microbiology, 2018, 266: 183-189. DOI:10.1016/j.ijfoodmicro.2017.11.026.
[99] JIA S L, JIA Z F, AN J, et al. Insights into the fish protein degradation induced by the fish-borne spoiler Pseudomonas psychrophila and Shewanella putrefaciens: from whole genome sequencing to quality changes[J]. International Journal of Food Microbiology, 2024, 416: 110675. DOI:10.1016/j.ijfoodmicro.2024.110675.
[100] ZHU Y L, SANG X, LI X, et al. Effect of quorum sensing and quorum sensing inhibitors on the expression of serine protease gene in Hafnia alvei H4[J]. Applied Microbiology and Biotechnology, 2020, 104: 7457-7465. DOI:10.1007/s00253-020-10730-9.
[101] WANG H D, SHAO L T, ZHANG J H, et al. Insight into the spoilage heterogeneity of meat-borne bacteria isolates with high-producing collagenase[J]. Food Science and Human Wellness, 2024, 13(3): 1402-1409. DOI:10.26599/FSHW.2022.9250118.
[102] NANDAN A, NAMPOOTHIRI K M. Molecular advances in microbial aminopeptidases[J]. Bioresource Technology, 2017, 245: 1757-1765. DOI:10.1016/j.biortech.2017.05.103.
[103] TAN C M, HU J R, GAO B Y, et al. Effects of the interaction between Aeromonas sobria and Macrococcus caseolyticus on protein degradation of refrigerated sturgeon fillets: novel perspective on fish spoilage[J]. LWT-Food Science and Technology, 2023, 183: 114908. DOI:10.1016/j.lwt.2023.114908.
[104] SCATAMBURLO T M, YAMAZI A K, CAVICCHIOLI V Q, et al.
Spoilage potential of Pseudomonas species isolated from goat milk[J]. Journal of Dairy Science, 2015, 98(2): 759-764. DOI:10.3168/jds.2014-8747.
[105] ALVES M P, SALGADO R L, ELLER M R, et al. Temperature modulates the production and activity of a metalloprotease from Pseudomonas fluorescens 07A in milk[J]. Journal of Dairy Science, 2018, 101(2): 992-999. DOI:10.3168/jds.2017-13238.
[106] KATIYO W, DE KOCK H L, COOREY R, et al. Sensory implications of chicken meat spoilage in relation to microbial and physicochemical characteristics during refrigerated storage[J]. LWT-Food Science and Technology, 2020, 128: 109468. DOI:10.1016/j.lwt.2020.109468.
[107] ABRIL A G, CALO-MATA P, VILLA T G, et al. Comprehensive shotgun proteomic characterization and virulence factors of seafood spoilage bacteria[J]. Food Chemistry, 2024, 448: 139045. DOI:10.1016/j.foodchem.2024.139045.
[108] ERCOLINI D, CASABURI A, NASI A, et al. Different molecular types of Pseudomonas fragi have the same overall behaviour as meat spoilers[J]. International Journal of Food Microbiology, 2010, 142(1/2): 120-131. DOI:10.1016/j.ijfoodmicro.2010.06.012.
[109] ÖZDEMIR F, ARSLAN S. Molecular characterization and toxin profiles of Bacillus spp. isolated from retail fish and ground beef[J]. Journal of Food Science, 2019, 84(3): 548-556. DOI:10.1111/1750-3841.14445.
[110] NOWAK A, PIOTROWSKA M. Biochemical activities of Brochothrix thermosphacta[J]. Meat Science, 2012, 90(2): 410-413. DOI:10.1016/j.meatsci.2011.08.008.
[111] FOUGY L, COEURET G, CHAMPOMIER-VERGÈS M C, et al. Draft genome sequence of Serratia proteamaculans MFPA44A14-05, a model organism for the study of meat and seafood spoilage[J]. Microbiology Resource Announcements, 2017, 5(23): e00491-17. DOI:10.1128/genomeA.00491-17.

