基于表型性状和SNP标记构建桂花主要品种资源的分子身份证

摘要:【目的】筛选核心单核苷酸多态性(single nucleotide polymorphism,SNP)位点,建立基于KASP平台的桂花(Osmanthus fragrans)品种基因型快速检测方法,构建品种特异性分子身份证,为桂花品种鉴定、溯源、知识产权保护等提供理论基础。【方法】实地调查主流桂花栽培品种的重要表型特征。通过两轮严格筛选,从基因组SNP中保留一组能够完全鉴别测序品种的最优SNP标记,计算SNP 位点的多态信息含量(PIC)、期望杂合度(He)等信息。以‘日香桂’(‘Rixianggui’)基因组为参考,设计桂花特异性KASP引物并进行批量扩增,根据基因分型结果建立品种指纹图谱,评估核心SNP标记的品种鉴别力。结合品种表型信息码和品种分子指纹码构建桂花品种资源的分子身份证。【结果】筛选出14个能够完全鉴别测序品种的核心SNP位点。各位点PIC值的变化范围为0.246~0.375,平均值为0.335;He的变化范围为 0.288~0.500,平均值为0.431。针对核心位点设计的KASP引物基因分型准确。根据扩增结果构建DNA指纹图谱,可区分全部包括未测序品种在内的90个参试品种。对品种表型特征赋值,结合指纹码构建由34位数字组成的桂花品种资源分子身份证。【结论】确定了SNP1—SNP14共14个核心SNP位点,能够实现至少90个桂花品种的有效鉴别。结合品种群类型、表型特征和分子指纹码构建了90个桂花品种的唯一分子身份证,并生成对应的条形码和二维码。
关键词:桂花;品种鉴别;单核苷酸多态性;表型性状;分子身份证
中图分类号:S68"""""" 文献标志码:A开放科学(资源服务)标识码(OSID):
文章编号:1000-2006(2024)04-0012-13
Construction of molecular ID for Osmanthus fragrans cultivars based on phenotypic traits and single nucleotide polymorphisms (SNPs)
WANG Yihan1,2, LIU Jiaojiao1,2, JIN Peiquan1,2, LI Shuqing1,2, WEI Jianfen3, GUO Peng1,2, SHANG Fude1,2
(1. College of Life Sciences, Henan Agricultural University, Henan Engineering Research Center for Osmanthus Germplasm Innovation and Resource Utilization, Zhengzhou 450046, China; 2 National Osmanthus Germplasm Resource Repository, Hangzhou 310020, China)
Abstract: 【Objective】 This study aimed to select genomic core single-nucleotide polymorphism (SNP) loci to establish a rapid SNP genotyping method for cultivar identification based on the KASP platform, and to construct molecular IDs for Osmanthus fragrans cultivars. This will provide a theoretical basis for the identification, traceability, and intellectual property protection of O. fragrans cultivars. 【Methods】 Field surveys were carried to investigate the important phenotypic characteristics of O. fragrans cultivars. Through two rounds of rigorous screening, we retained a set of core SNP markers that could fully discriminate previously sequenced cultivars. The polymorphic information content (PIC) and expected heterozygosity (He) of each SNP loci were then analyzed. Using the genome sequences of ‘Rixianggui’ as a reference, we designed species-specific KASP primers and performed PCR amplifications. Based on the genotyping results, we constructed cultivar DNA fingerprints and evaluated the cultivar identification efficiency of the core SNP markers. The molecular IDs of O. fragrans cultivars were finally constructed by combining phenotypic information codes and molecular fingerprint codes. 【Results】 A total of 14 core SNP loci that could fully discriminate sequenced cultivars were retained from the genomic SNPs. The PIC values of these loci ranged from 0.246 to 0.375, with an average of 0.335, and the He indexes ranged from 0.288 to 0.500, with an average of 0.431. The KASP primers designed for the core SNP loci had accurate genotyping results, based on which we constructed DNA fingerprints that could distinguish all 90 tested cultivars, including those that had not been sequenced. A molecular ID of each cultivar, which composed of 34 digits, was formed finally. 【Conclusion】A total of 14 core SNP loci, SNP1 to SNP14, were identified, which could effectively discriminate at least 90 cultivars of O. fragrans. The unique molecular ID codes were constructed for the 90 cultivars using the DNA fingerprint codes, and the converted serial codes from cultivar group types and phenotypic characteristics. The bar codes and quick response (QR) codes were finally generated.
【Objective】 To analyze the phylogenetic relationships and evolutionary patterns of pollen morphology in Tsuga, we observed and compared the pollen characteristics of all 10 extant species of Tsuga along with pollen fossils. 【Methods】 Pollen morphology was examined using scanning electron microscopy (SEM), and hierarchical cluster analysis (HCA) was conducted using seven quantitative indicators and one qualitative indicator. 【Results】 Tsuga pollen typically exhibits characteristics of the N1P3C1 type, with a leptoma on the distal face. The equatorial length ranges from 43.74 to 95.30 μm, predominantly lacking sacci but occasionally with sparse saccate forms. The pollen surface is typically warty or sparsely micro-warty, occasionally with spines or without. According to the hierarchical cluster analysis, Tsuga could be divided into three categories: ①T. mertensiana and Nothotsuga longibracteata; ②T. caroliniana and T. canadensis; ③T. heterophylla, T. ulleungensis, T. diversifolia, T. forrestii, T. sieboldii, T. dumosa and T. chinensis. 【Conclusion】 Pollen morphology in Tsuga has evolved from saccate to non-saccate forms, and from lacking spines to possessing spines. These pollen characteristics are closely related to geographic distribution, indicating similarity among pollen from the same regions. The clustering analysis based on pollen morphology largely aligns with molecular phylogenetic trees, offering a method to distinguish extant species and fossils of Tsuga and providing valuable insights for phylogenetic studies of Tsuga.
【Objective】 This study"" selected core genomic single-nucleotide polymorphism (SNP) loci to" establish" a rapid SNP genotyping method on the KASP platform, and" to construct molecular IDs for Osmanthus fragrans cultivars. This study"" provides a theoretical foundation for identifying, tracing" and protecting the intellectual property of O. fragrans cultivars. 【Method】 Field surveys were conducted to investigate key phenotypic characteristics of O. fragrans cultivars. Following two rounds of rigorous screening, we identified a set of core SNP markers capable of completely distinguishing previously sequenced cultivars. Subsequently, we analyzed the polymorphic information content (PIC) and expected heterozygosity (He) of each SNP locus. Using the genome sequences of ‘Rixianggui’ as a reference, species-specific KASP primers were designed for PCR amplification. Based on the genotyping results, we constructed cultivar DNA fingerprints and assessed the efficiency of core SNP markers for cultivar identification. Molecular IDs for O. fragrans cultivars were established by integrating phenotypic information codes with molecular fingerprint codes. 【Result】 We retained a total of 14 core SNP loci from genomic SNPs that fully discriminated the sequenced cultivars. The PIC values of these loci ranged from 0.246 to 0.375, with an average of 0.335, and the He indices ranged from 0.288 to 0.500, averaging 0.431. The KASP primers designed for these core SNP loci produced accurate genotyping results, enabling us to construct DNA fingerprints capable of distinguishing all 90 tested cultivars, including those not previously sequenced. Each cultivar was assigned a molecular ID composed of 34 digits. 【Conclusion】 In conclusion, 14 core SNP loci (SNP1 to SNP14) were identified that effectively discriminate among at least 90 O. fragrans cultivars. Unique molecular ID codes were constructed using DNA fingerprint codes" along with serial codes derived from cultivar group types and phenotypic characteristics. Finally, barcode and quick response (QR) codes were generated for each cultivar.
Keywords:Osmanthus fragrans; cultivar identification; single nucleotide polymorphism (SNP); phenotype; molecular ID
桂花(Osmanthus fragrans)隶属木犀科(Oleaceae)木犀属(Osmanthus),为中国传统名花,在秦岭淮河流域以南、南岭以北的广大中亚热带和北亚热带地区广泛栽培[1]。经过两千多年的自然变异和人工繁育,桂花形成了丰富的种内变异和品种资源。根据花期、花色和叶色的不同,桂花可分为金桂(Luteus group)、银桂(Albus group)、丹桂(Aurantiacus group)、四季桂(Asiaticus group)和彩叶桂(Caiyegui group)五大品种群[2-4],已知品种有近300个[5]。近年来,随着野生资源的开发和杂交育种工作的系统深入,新优桂花品种不断涌现[6-7]。然而,在引种和品种资源交换过程中,“同物异名”和“同名异物”的现象时有发生。国家标准《植物新品种特异性、一致性和稳定性DUS测试指南 桂花》[8]的颁布为桂花品种鉴别和新品种测试提供了技术方法和标准。该标准确定了41个较稳定的特征(如花枝长度、叶腋内花芽数量、花色、花期等)作为DUS测试的性状特征。但由于表型性状极易受环境影响,国际植物新品种保护联盟在2013年提出了基于表型距离和分子距离联合筛选近似品种的模式[9]。但是目前在桂花品种认定中,除性状特异性外,仅要求其遗传稳定性与一致性,尚未在分子标记鉴定与分子条码上进行规范。
分子标记不依赖表型性状,稳定性强,是探索植物遗传变异、开展品种鉴别研究的重要工具。桂花现代栽培品种的遗传背景复杂,由于高质量基因组资源的缺乏,前期研究者主要采用传统标记进行品种鉴定、分子指纹图谱的构建和遗传连锁图谱的绘制,比如相关序列扩增多态性(sequence-related amplified polymorphism,SRAP)[10-11]、简单重复序列间扩增(inter-simple sequence repeat,ISSR)[12-13]、扩增片段长度多态性(amplified fragment length polymorphism,AFLP)[14-15]、微卫星(simple sequence repeats,SSR)[16-18]等。近年来, ‘日香桂’(‘Rixianggui’)[19]、‘柳叶金桂’(‘Liuye Jingui’)[20]、‘状元红’(‘Zhuangyuanhong’)和‘玉莲银丝’(‘Yulian Yinsi’)[21]4个桂花品种染色体水平的高质量基因组陆续公布,为挖掘和开发全基因组范围内的多态性分子标记(SSR、InDel、SNP等)提供了重要平台。与SSR和InDel相比,SNP标记更适合大量标记的高通量基因分型、数据库整合和数据共享,并且具有数量大、变异来源丰富、经济高效的优势[22-23],是BMT分子测试指南中构建DNA指纹数据库的推荐标记[24]。Chen等[20]基于重测序技术检测了桂花主流栽培品种基因组内的SNP标记,基于全基因组关联分析挖掘了与花色相关的SNP位点和候选基因。但仍亟须严格筛选全基因组的高多态性、非冗余的SNP位点,建立鉴定准确、检测高效的桂花品种分子鉴别技术。
新一代测序技术的发展推动了全基因组SNP标记的挖掘及其在观赏植物品种指纹图谱构建和品种鉴定上的应用,如月季(Rosa hybrida)[25]、鸢尾(Iris tectorum)[26]、波斯毛茛(Ranunculus asiaticus)[27]。研究者通过建立竞争性等位基因特异性PCR技术(competitive allele-specific PCR,KASP)[28-29]、SNP芯片技术(gene chip)[30]等可以进一步提高已有SNP的使用效率和分析通量。这些技术在作物、蔬菜等植物品种和种质资源的基因分型、功能基因克隆与鉴定等领域有广泛的应用[31-36],为桂花高效品种鉴别技术的研究提供了参考。
在前期研究[37]的基础上,本研究采用严格的过滤参数从桂花60个品种的全基因组重测序数据中筛选能够区分各品种的核心SNP位点,开发基于KASP平台的品种基因型快速检测方法,并建立品种特异性分子指纹图谱和分子身份证,以期为桂花品种鉴定、溯源、知识产权保护等提供理论参考。
1 材料与方法
1.1 试验材料和DNA的提取
试验所用的90个桂花品种,涵盖了4个桂花品种群,其中金桂30个、银桂28个、丹桂23个、四季桂9个,保存于浙江省杭州市余杭区的桂花国家级花卉种质资源库、河南省信阳市潢川金桂园和河南大学金明校区(附表1,nldxb.njfu.edu.cn,下同)。
于2019—2020年采集各品种的新鲜嫩叶,用硅胶充分干燥后置于4" ℃保存。采用改良的CTAB法[38]提取供试基因组DNA,采用1%(质量分数,下同)琼脂糖凝胶检测DNA的完整性,用NanoDrop 8000分光光度计(Thermo Fisher, DE, 美国)精确定量DNA浓度。提取的基因组DNA置于-20 ℃保存备用。
1.2 表型性状测定
桂花的花色、花香、花量和结实性状是品种分类的重要依据,也是产业上品种价值体现的核心品质特征[39-41]。本研究选取花色、花香、新梢顶芽和腋芽总数、着花密度和结实性共5个表型性状作为品种身份证中商品码的组成部分。参考《中国桂花品种图志》[2]《中国桂花》[3]和《桂花新品种DUS测试指南》[42] 制定调查标准,于2022—2023连续两年在样品采集地进行实地表型观测。调查时每个品种取3株重复,每株测定3次。
具体调查标准如下:
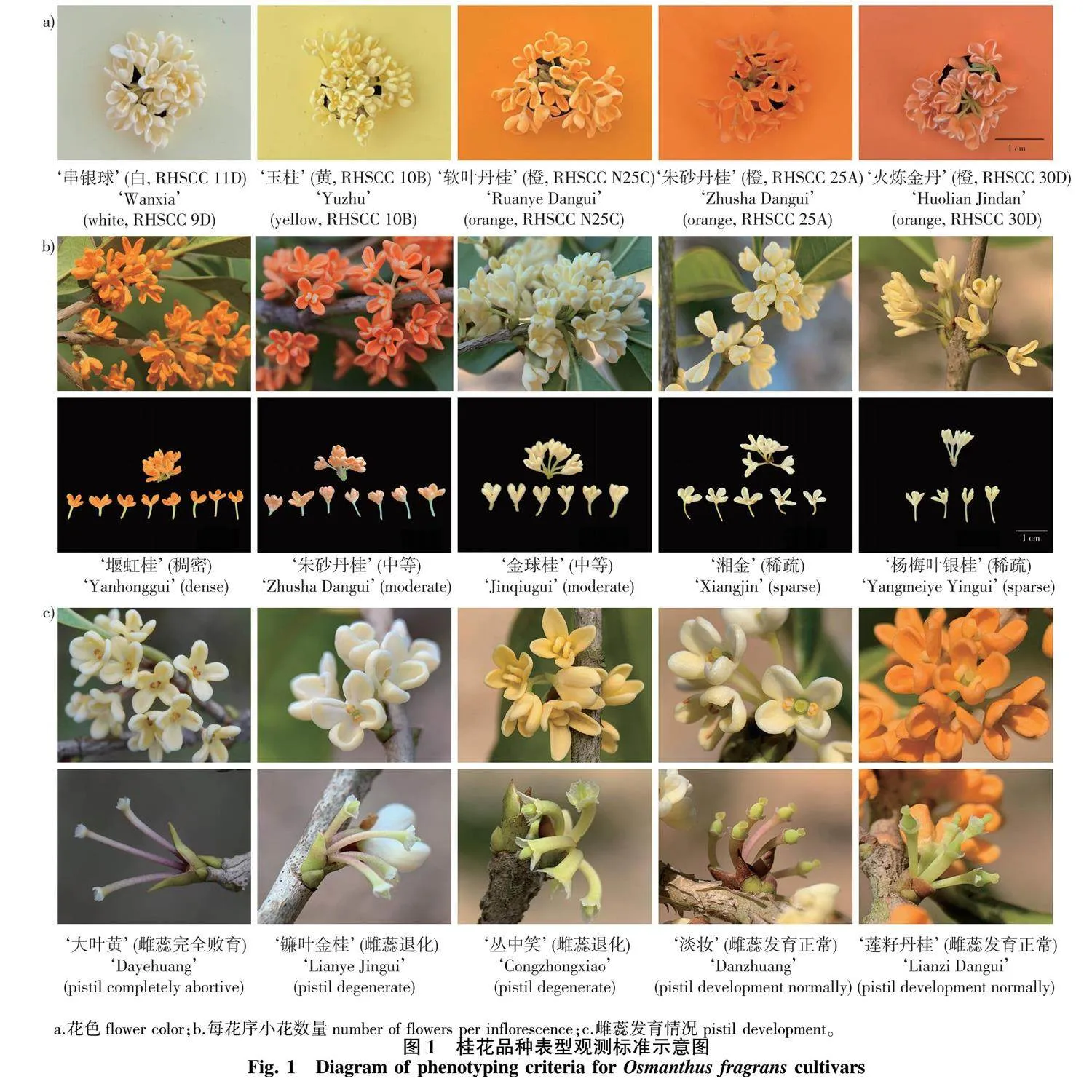
1)花色。在花初开放至全株1/3全开放期间观测,在晴天、自然光环境下,先把握全株整体的景观色彩,再细观向阳面树冠中上部花枝上的花瓣颜色,以英国皇家园艺学会(Royal Horticultural Society,RHS)出版的比色卡(RHS Color Chart)[43-45]为标准(图1a),进行“白”“黄”“橙”3色的定性描述。
2)花香。采用鼻闻定性法在品种头茬花的初花期首日进行花香鉴定,根据香气程度分为浓香、中香、微香、不香4种类型。
3)花量。采用新梢顶芽和腋芽总数、着花密度(即每花序小花数)两个指标反映[3]。新梢上的芽除了顶端少数是来年或当年秋季萌发的枝芽,绝大多数都是腋生的花芽。本研究于圆珠期随机选择9个1年生新生枝条,统计各枝条的顶芽和腋芽总数并计算平均值,根据结果将各品种划分为少花低产(<20枚/梢)、中花平产[[20,30)枚/梢]、多花丰产(≥30枚/梢)3个等级[3]。此外,于盛花期统计从新梢顶端向下第2对叶腋处花序的小花数量,判断各品种的着花密度为稀疏(<5朵/花序)、中等[[5,8)朵/花序或稠密(≥8朵/花序)][3](图1b)。
4)结实性。根据雌蕊发育状况和花后结实与否判断,是品种内的稳定性状。雌蕊发育状况常因品种而异,有3种情况[2-3, 46](图1c):一是雌蕊完全败育,属于雄花,常完全无雌蕊分化,或子房细长条形、柱头缺失;二是雌蕊退化,花内子房较狭长,呈长卵形,柱头短小,不能正常授粉受精或者果实发育异常;三是雌蕊和雄蕊均发育良好,为两性花,子房膨大呈圆球形或卵球形,能正常结实。雌蕊完全败育和雌蕊退化两种情况均记录为不结实品种。
1.3 品种鉴定核心SNP标记的筛选
前期李书情[37] 对60个桂花品种的重测序研究获得了1 599 694个高质量基因组SNP。本研究利用软件VCFtools V.0.1.13[47]对上述SNP进行严格过滤,过滤标准如下:最小等位基因频率(minor allele frequency,MAF)≥0.05、样本分型缺失率为0、多态性信息含量(polymorphism information content,PIC)为 0.2~0.5、独立遗传[连锁不平衡过滤参数为LD独立性过滤(indep) 与使用成对的LD计算方法(pairwise)窗口大小50、步长10、阈值(r2)0.2]、SNP位点前后50 bp无其他变异、SNP位点前后100 bp的侧翼序列在染色体上有唯一比对,SNP位于外显子区且尽量在染色体上均匀分布(密度按至少100 kb/SNP)。对于第1轮过滤后的SNP位点,采用李梓榕等[48]开发的R语言脚本,经过“初筛”和“精简”两步流程作进一步过滤,筛选出数量最少、能够完全区分60个桂花品种的5组SNP标记。通过后续引物设计和基因分型实验,保留一组KASP转化成功、分型准确的位点作为品种鉴定的核心SNP标记,并明确每个位点在各样本中的基因型。
1.4 KASP扩增与基因型检测
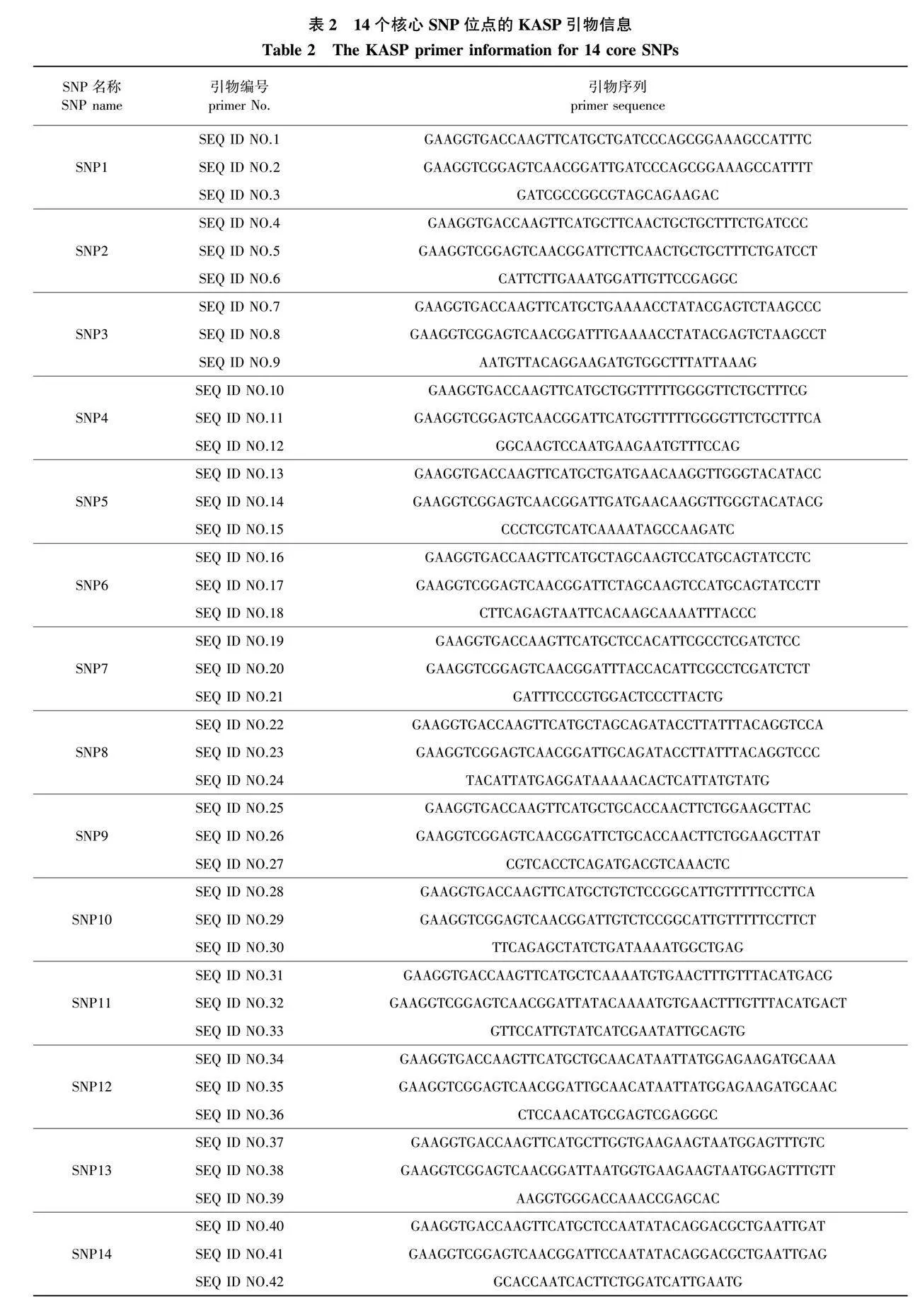
基于KASP平台,以‘日香桂’[19]基因组为参考,在每个核心SNP位点前后100 bp序列范围内设计引物。每个SNP位点分别设计3条引物:2条等位基因特异性的上(或下)游引物和一条下(或上)游通用引物。引物由北京擎科生物科技有限公司合成。KASP反应体系、反应程序和荧光信号判断方法按照Shen等[49]的方法进行。对于预实验中分型效果和准确率较好的核心标记组合,在60个测序品种的基础上进一步扩大样本(加入30个未测序品种),进行KASP 批量扩增,用于评估核心SNP标记的品种鉴别力,并获得90个受试桂花品种在核心SNP位点的基因型数据。利用PowerMarker V.3.25软件[50]计算各核心位点的次要等位基因频率(MAF)、多态信息含量(polymorphism information content,PIC)和期望杂合度(expected heterozygosity, He)。
1.5 分子指纹图谱和分子身份证的构建
通过KASP实验获得各品种在核心SNP位点的基因型数据后,将A、G、C、T依次转换为1、2、3、4,构建每个品种的分子指纹图谱,即分子指纹码。结合品种信息编码与分子指纹码,利用条码生成器和二维码生成器(http://qr-batch.com/)构建90个桂花品种的分子身份证。
2 结果与分析
2.1 表型性状多样性
2022—2023年连续2年对杭州市国家桂花种质资源库、信阳市潢川金桂园和河南大学金明校区的90个栽培桂花品种进行了花色、花香、花量和结实性的表型调查及图片拍摄。在调查的90个品种中,74个品种不结实,占比83.15%。浓香型桂花品种有23个,多集中在金桂和银桂品种群;四季桂品种群中,‘日香桂’和‘橙黄四季桂’(‘Chenghuang Sijigui’)花香较为浓郁,适合多季的香景营造。就花色而言,白色系桂花品种较少,代表的有银桂品种‘中华龙桂’(‘Zhonghua Longgui’)和‘串银球’(‘Chuanyinqiu’);金桂品种‘小花金桂’(‘Xiaohua Jingui’)和‘山茶金桂’(‘Shancha Jingui’)等;丹桂品种‘状元红’(‘Zhuangyuanhong’)和‘满条红’(‘Mantiaohong’)的花色最为鲜艳。桂花开花以当年新梢为主,各品种的花量与新梢花芽数和每花序小花数目有关[51]。综合考虑这两项发育指标,调查品种中的‘柳叶苏桂’(‘Liuye Sugui’)、‘玉玲珑’(‘Yulinglong’)和‘速生金桂’(‘Susheng Jingui’)在调查年份花芽量大(平均30~36/梢)、着花稠密(7~9朵/花序)且不结实,是适合用于采花的高产花量品种(附表1,nldxb.njfu.edu.cn)。
2.2 核心SNP的筛选和KASP标记转化
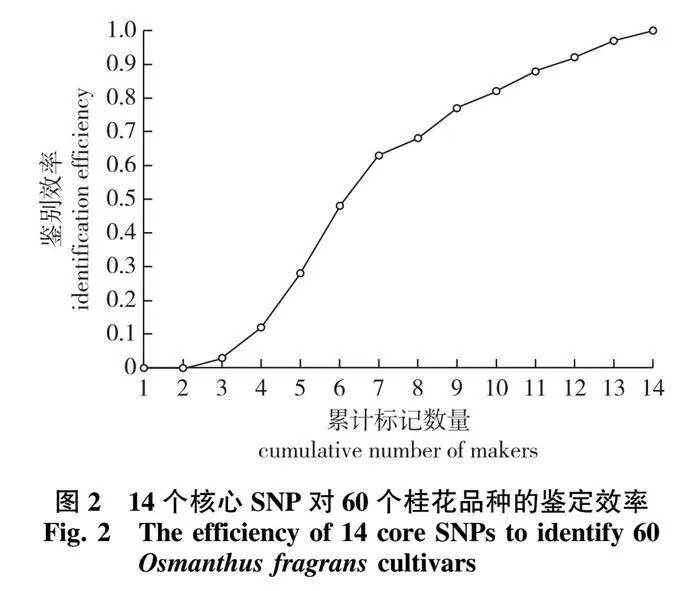
经过两轮的严格筛选和KASP预实验,最终保留一组由14个核心SNP 位点组成的最精简SNP标记组合,能完全区分60个桂花品种(图2)。
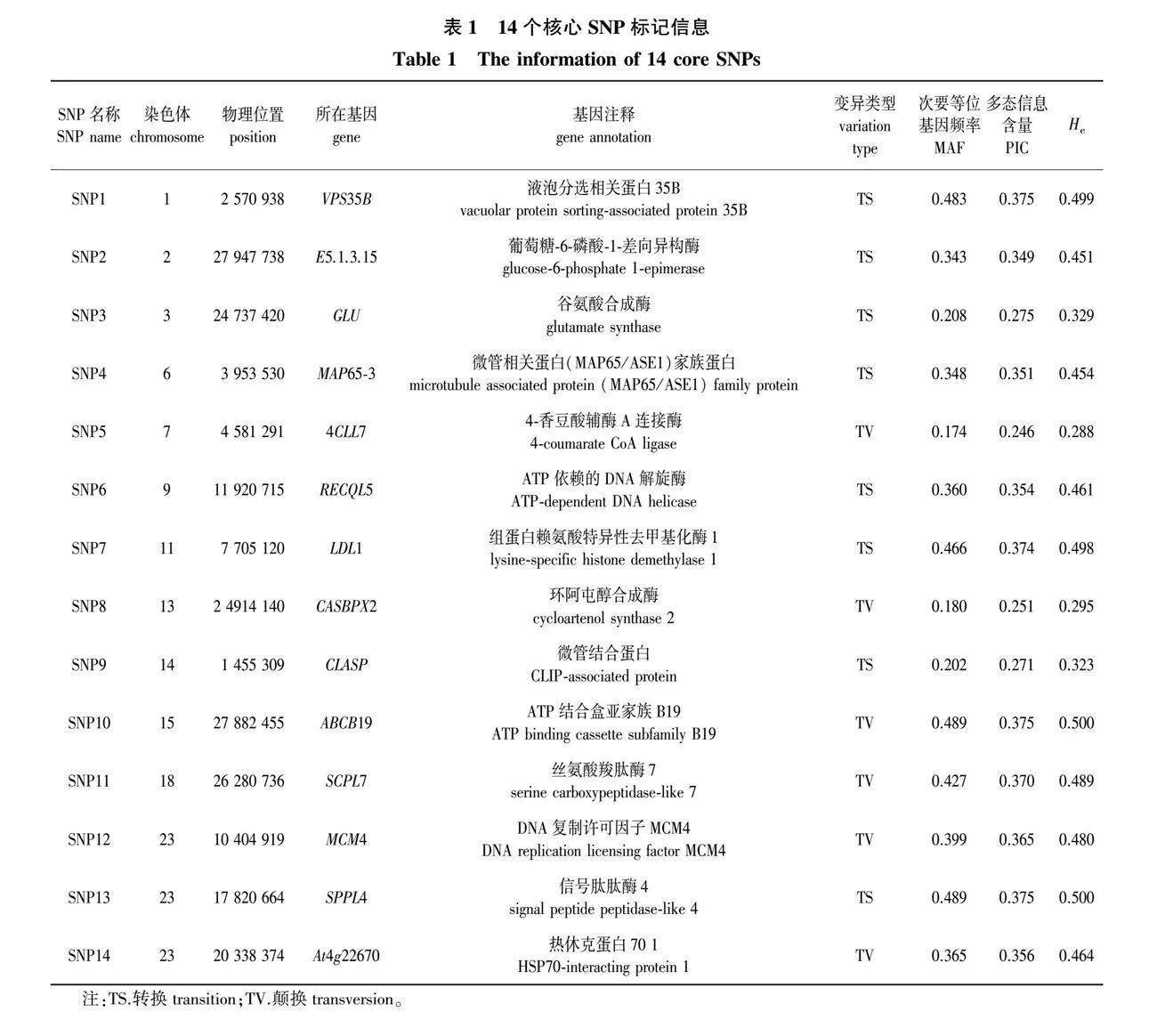
这14个SNP标记都位于基因编码区,包含4个非同义突变和10 个同义突变。6个SNP位点的变异类型是碱基颠换,8个SNP是碱基转换(表1)。经计算,14个SNP位点的多态性水平较高,PIC值的变异范围为0.246~0.375,平均值为0.335;He指数的变异范围为0.288~0.500,平均值为0.431(表1)。提取14个核心SNP位点前后各100 bp的侧翼序列,设计合成KASP引物,携带有FAM荧光标签的引物序列为GAAGGTGACCAAGTTCATGCT,携带有HEX荧光标签的引物序列为GAAGGTCGGAGTCAACGGATT(表2)。
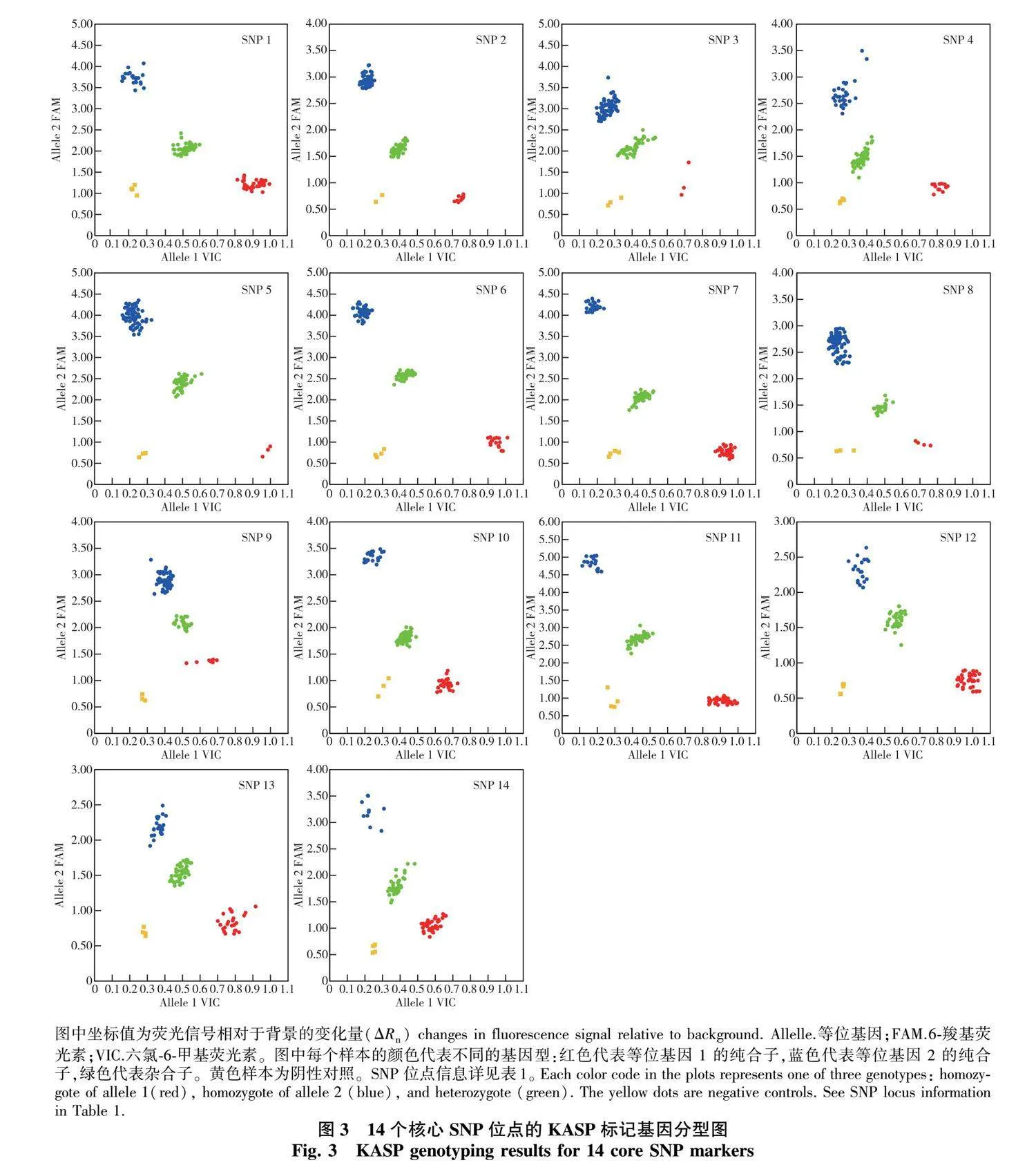
2.3 核心SNP鉴定能力检验和品种分子指纹图谱的构建
利用KASP引物对60个已测序品种和30个未测序品种进行批量扩增,通过荧光信号强弱结果判断各核心位点的基因型(图3),从而验证分型准确性。经对比,所有SNP位点的KASP分型结果与重测序结果一致,可用于后续桂花品种分子鉴别中基因型的快速检测。当供试品种数量增多至90个时,该SNP标记组合仍能鉴定所有品种。进一步将14个核心SNP位点在90个品种中的基因型数据转化为分子指纹,结果表明,多个品种间具有较高的遗传相似性(仅1个位点差异),如‘橙香丹桂’(‘Chengxiang Dangui’)和‘红桂’(‘Honggui’)、‘湘金’(‘Xiangjin’)和‘速生银桂’(‘Susheng Yingui’)、‘娇容’(‘Jiaorong’)和‘武夷丹桂’(‘Wuyi Dangui’)等。
图中坐标值为荧光信号相对于背景的变化量(ΔRn)" changes in fluorescence signal relative to background. Allelle.等位基因;FAM.6-羧基荧光素;VIC.六氯-6-甲基荧光素。
图中每个样本的颜色代表不同的基因型:红色代表等位基因 1 的纯合子,蓝色代表等位基因 2 的纯合子,绿色代表杂合子。黄色样本为阴性对照。SNP位点信息详见表1。 Each color code in the plots represents one of three genotypes: homozygote of allele 1(red), homozygote of allele 2 (blue), and heterozygote (green). The yellow dots are negative controls. See SNP locus information in Table 1.
2.4 桂花品种分子身份证的构建
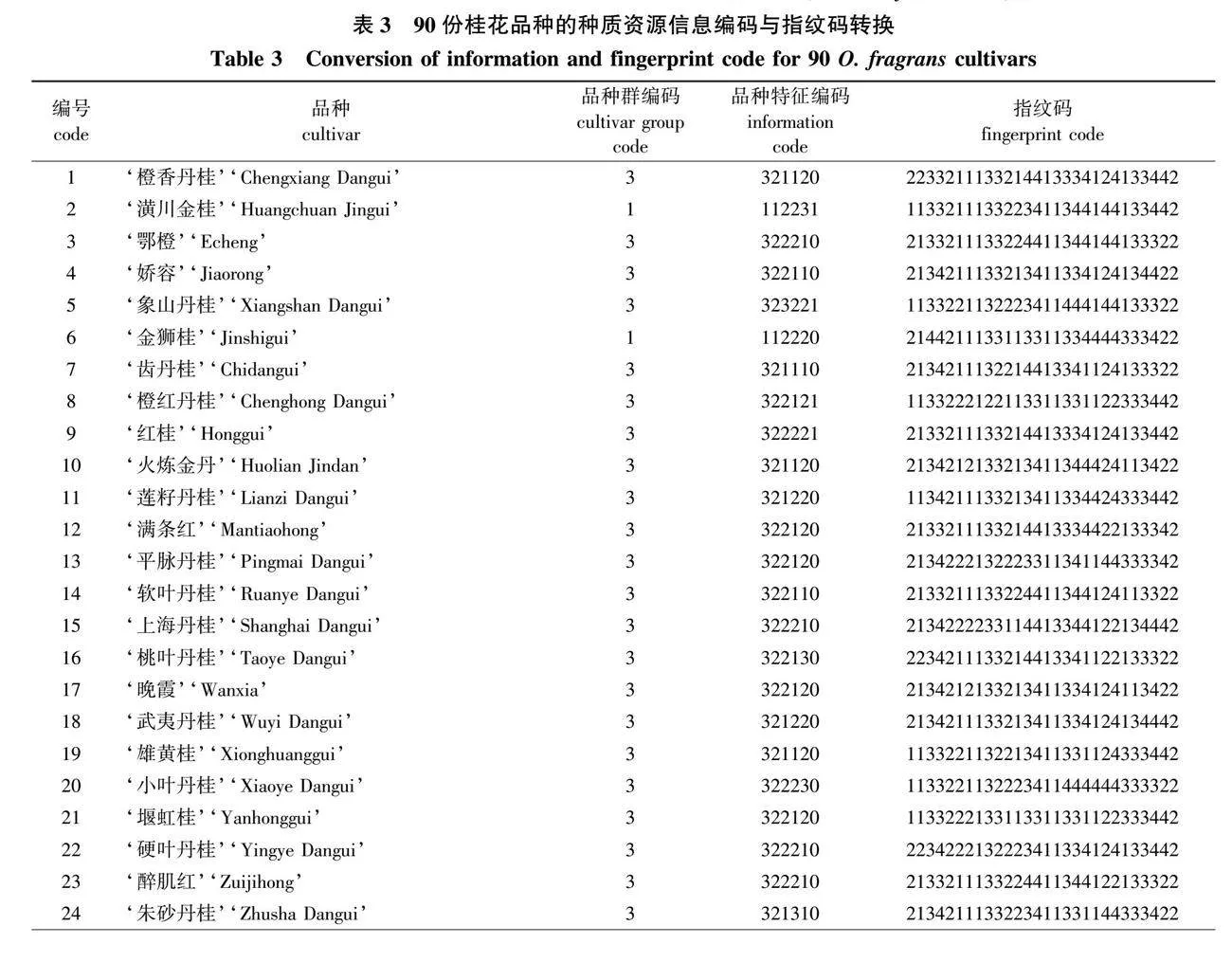
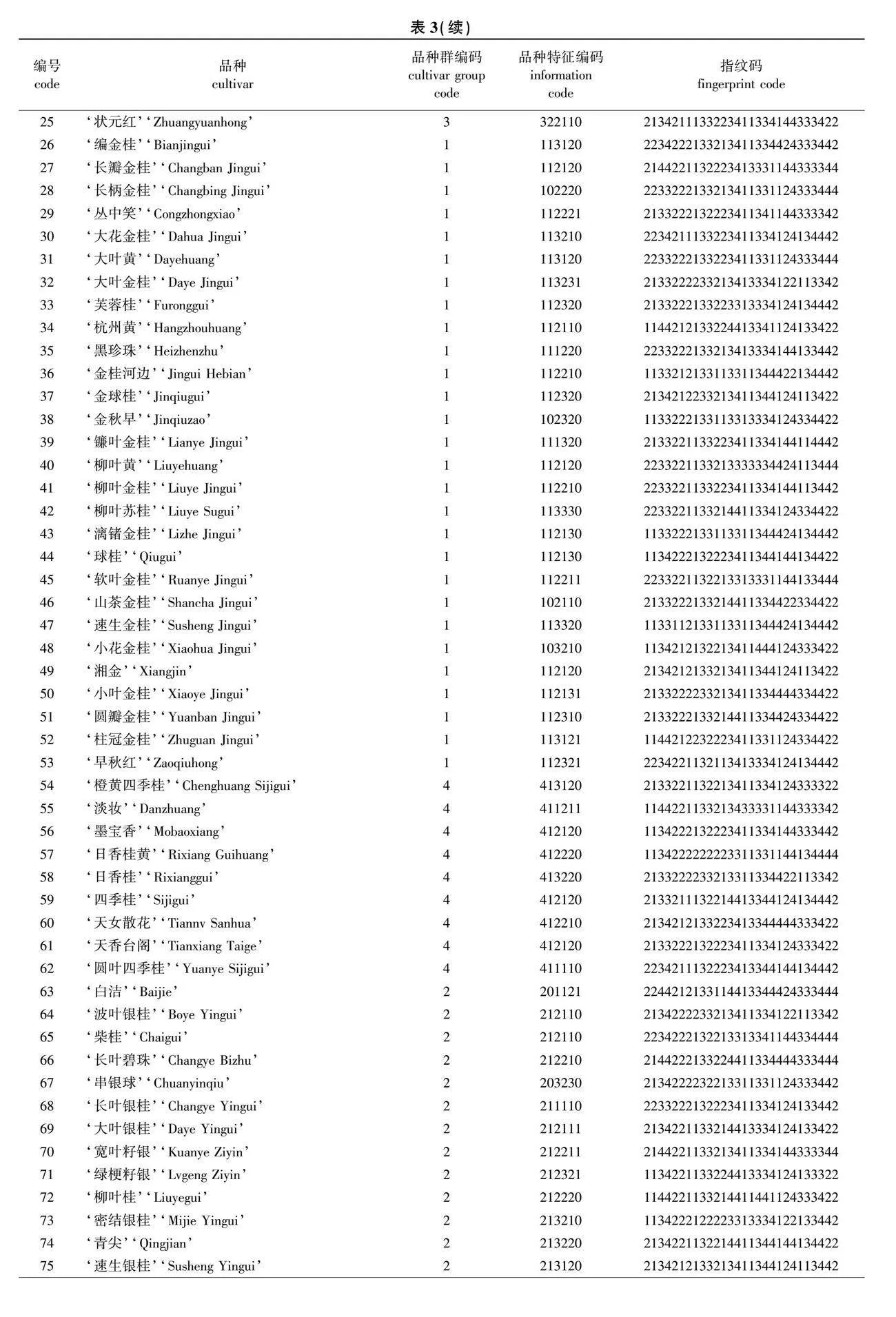
将14个核心SNP位点(SNP1—SNP14)在各品种的基因型数据转化为28位指纹码,与品种信息编码联合构建34位分子身份证码。
品种信息码为分子身份证的前6位,第1位数字为品种群类型编码,1~4分别代表金桂、银桂、丹桂和四季桂品种群;第2~6位数字为品种特征信息编码,通过对花色、花香、新梢顶芽和腋芽总数、每花序小花数量(着花密度)、结实性(雌蕊发育状况)5个品种表型特征赋值后生成。赋值方式如下:第2位“0、1、2”表示 “白、黄、橙”3种花色类型;第3位“0、1、2、3”表示 “不香、微香、中香、浓香”4种花香等级;第4位“1、2、3”依次表示新梢花芽数“少、中、多”;第5位“1、2、3”依次表示着花密度“稀疏、中等、稠密”;第6位“0、1”分别表示“不结实、结实”。以‘状元红’为例见图4)。
由图4可见,其分子身份证号码为3221102134211133223411334144333422,表示该品种属于丹桂品种群,花色为橙色,中香,新梢顶芽和腋芽总数少,着花密度稀疏,花后不结实,14个核心SNP标记的基因型依次为GA、CT、GA、AA、CG、
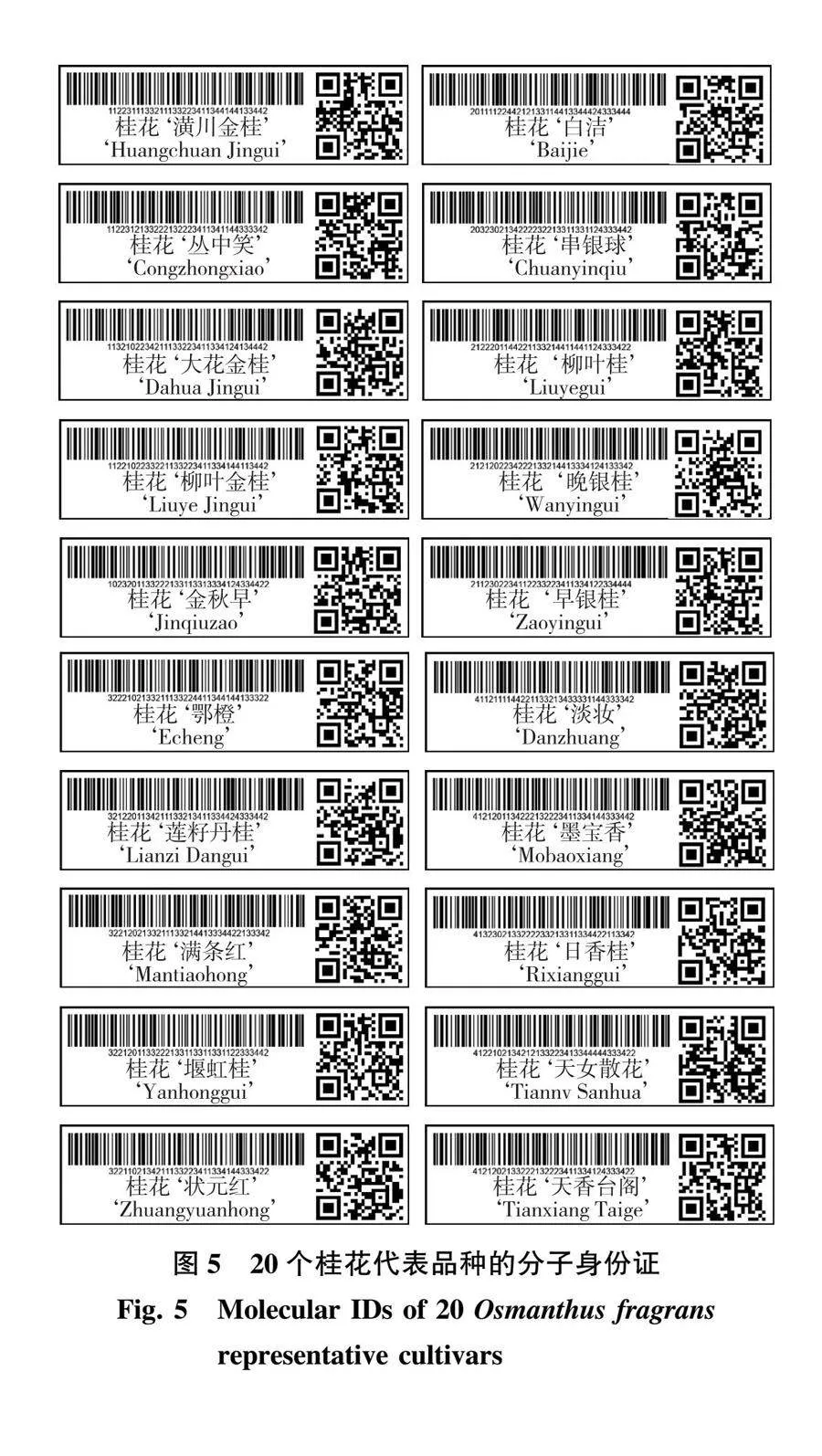
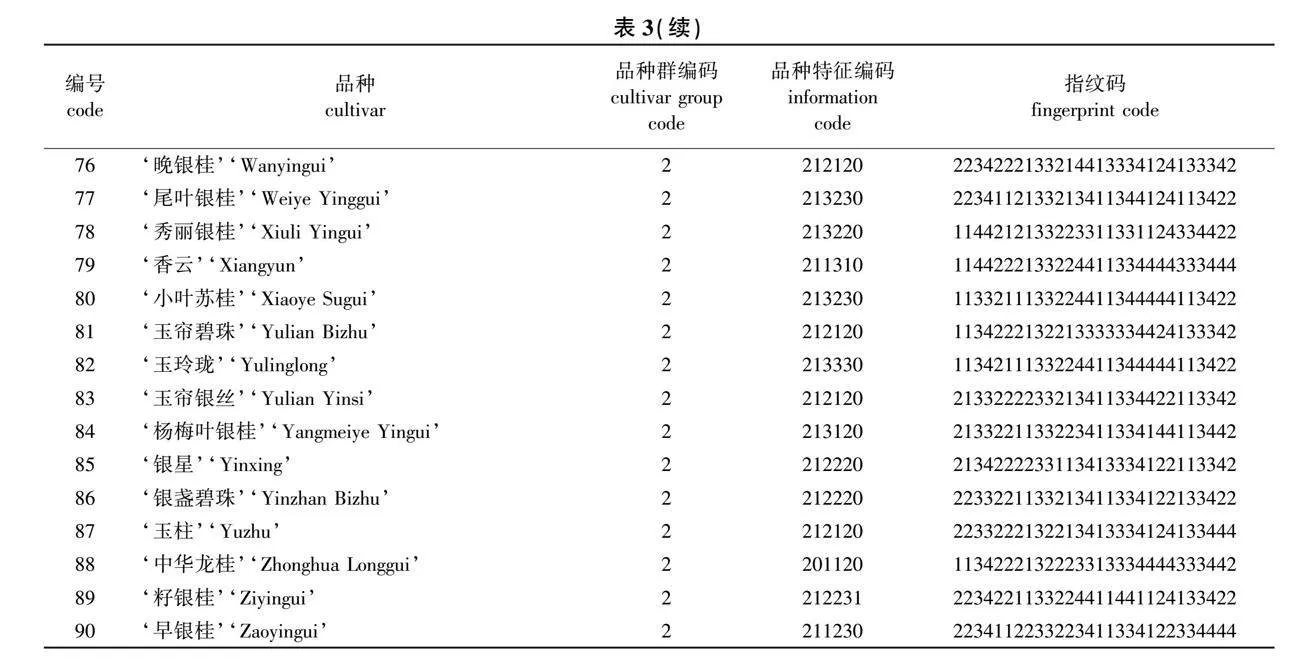
GG、CT、AA、CC、TA、TT、CC、CT、GG。按照此方法完成90份桂花品种分子身份证的构建(表3)。最终利用条码在线生成器分别制作出品种特异的条形码和二维码(图5),90个桂花品种的分子身份证详见附图1(nldxb.njfu.edu.cn)。
C-Red-Mean.树冠红色波段平均值 canopy red band mean;C-Blue-Mean.树冠蓝色波段平均值 canopy blue band mean;T-Red-Mean.树干红色波段平均值 trunk red band mean;T-Blue-Mean.树干蓝色波段平均值 trunk blue band mean;T-Green-Mean.树干绿色波段平均值 trunk green band mean;C-Red-Std.树冠红色波段标准差 canopy red band standard deviation;C-Green-Mean.树冠绿色波段平均值 canopy green band mean;T-Red-Std.树干红色波段标准差 trunk red band standard deviation;RL/H.冠幅树高比 canopy width to height ratio;C-Blue-Std.树冠蓝色波段标准差 canopy blue band standard deviation;T-Green-Std.树干绿色波段标准差 trunk green band standard deviation;T-Blue-Std.树干蓝色波段标准差 trunk blue band standard deviation;RA/H.树冠面积树高比 canopy area to height ratio;C-Green-Std.树冠绿色波段标准差 canopy green band standard deviation;RA/D.树冠面积胸径比 canopy area to diameter at breast height ratio;RV/D.树冠体积胸径比 canopy volume to diameter at breast height ratio;RV/H.树冠体积树高比 canopy volume to height ratio;RL/D.冠幅胸径比 canopy width to diameter at breast height ratio;0~10%、10%~20%、…、90%~100%表示分位点云数量占比0~10%、10%~20%、…、90%~100% represent quantile point cloud ratio。
3 讨 论
SSR和SNP分子标记是国际《UPOV-BMT测试指南》和中国《植物品种鉴定DNA指纹方法总则》(NY/T 2594—2016)中推荐的最佳分子标记[52]。SSR标记因通量有限、操作耗时、数据整合困难等问题,正逐渐被SNP标记取代[53]。基于第3代SNP标记的品种身份证构建在观赏物种中报道不多,仅在兰科(Orchidaceae)[54]、山茶(Camellia japonica)[55]等少数花卉中有应用,目前在桂花中尚无相关研究。桂花遗传背景复杂,基因组杂合度(1.02%~1.45%)和重复序列比例(49.35%~54.41%)较高[19-21],多态性位点的信息量大且解析难度较高。因此,以高质量基因组为参考,从海量的全基因组变异中发掘可靠且有“鉴别力”的共显性SNP标记是建立品种基因型快速检测体系的关键。
从降低检测成本的方面考虑,用于构建分子指纹图谱的位点选择原则是用尽可能少的SNP鉴定全部样本。常规的SNP筛选流程主要关注位点检出率和多态性指标[56-57]。前期桂花重测序研究[37]在121个品种中检测到SNP位点数约160万个,PIC值为0.2~0.5的SNP约有41万个。采用传统的高PIC值方法从基因组数据中筛选核心位点操作难度大,且常有精简优化的空间。为克服该问题,本研究选择近期开发的新方法[48],经过“初筛”和“精简”两步流程筛选能够鉴定全部60个供试品种的SNP标记组合。经计算,最终保留了14个(SNP1—SNP14)位于基因编码区、在染色体上均匀分布的SNP位点(PIC:0.246~0.375),并进行了KASP标记的转化。对比发现,这一组合的鉴别率明显高于高PIC值筛选的SNP组合(100% vs. 89%),并且当品种数量增多至90个时,仍能鉴定所有品种(1~13个位点差异)。在受试品种中,‘长叶碧珠’(‘Changye Bizhu’)和‘银盏碧珠’(‘Yinzhan Bizhu’)、‘雄黄桂’(‘Xionghuanggui’)和‘堰虹桂’(‘Yanhonggui’)之间的表型差异很小[2-3],加之环境影响,在引种栽培中容易混淆。利用本研究的14个核心KASP-SNP标记进行鉴定,两对近似品种分别有5个和9个位点的基因型差异,易于区分。理论上,14个标记区分目前已知的所有桂花品种存在可行性,但具体的实用性有待进一步检验。在实践中也可以重复分析多次,联合多套SNP组合提高鉴定方法的容错能力[48]。总之,本研究针对桂花核心SNP的KASP快速检测体系具有标记数量少、鉴别能力强、操作便捷、成本低的优势,检测结果可为今后桂花品种真实性鉴定和DUS测试标准的制定提供参考。
不同于单纯的分子指纹图谱,本研究将90个桂花品种的重要经济表型和14个SNP位点构建的分子指纹图谱信息结合起来,简化为标准化“身份”编码,再生成对应的条形码和二维码,可被设备快速扫码识别,便于产业上品种身份和品种应用特性的便捷化查询。同时,品种身份证的开发也可用于品种溯源和知识产权保护。例如,根据本研究的基因分型结果,国家地理标识产品‘潢川金桂’(‘Huangchuan Jingui’)在SNP2、SNP3、SNP7具有品种特异性的基因型组合(分别为C/C、G/A和C/T),可作为其在遗传上的明确标识特征,用于地理标志保护。但是,由于前期进行表型观测的种质资源圃中未引种彩叶桂品种,故本研究生成的品种身份证未涵盖该品种群。未来将收集市场主流的彩叶桂品种,如‘云田彩桂’(‘Yuntian Caigui’)、‘紫嫣公主’(‘Ziyan Gongzhu’)等,进一步检验SNP标记组合的品种鉴定力,并扩充桂花分子身份证库。
参考文献(reference):
[1]赵宏波,郝日明,胡绍庆.中国野生桂花的地理分布和种群特征[J].园艺学报,2015,42(9):1760-1770. ZHAO H B,HAO R M,HU S Q.Geographic distribution and population characteristics of Osmanthus fragrans[J].Acta Hortic Sin,2015,42(9):1760-1770.DOI: 10.16420/j.issn.0513-353x.2014-0939.
[2]向其柏,刘玉莲.中国桂花品种图志 [M].杭州:浙江科学技术出版社,2008. XIANG Q B,LIU Y L.An illustrated monograph of the sweet osmanthus cultivars in China[M].Hangzhou:Zhejiang Science amp; Technology Press,2008.
[3]杨康民.中国桂花[M].北京:中国林业出版社,2013. YANG K M.Chinese osmanthus[M].Beijing:China Forestry Publishing House,2013.
[4]向民,段一凡,向其柏.木犀属品种国际登录中心年报(1) 彩叶桂品种群的建立[J].南京林业大学学报(自然科学版),2014,38(1):2,187.XIANG M,DUAN Y F, XIANG Q B.International Cultivar Registration Center for Osmanthus,Nanjing Forestry University.Establishment of a new group-Osmanthus fragrans Colour Group[J].J Nanjing For Univ (Nat Sci Ed),2014,38(1):2,187.DOI: 10.3969/j.issn.1000-2006.2014.01.034.
[5]高丽,何岳球,孙志国,等.三个 “新发展” 下咸宁市中国桂花城传承创新发展研究[J].安徽农业科学,2023,51(12):256-259.GAO L,HE Y Q,SUN Z G,et al.Research on the inheritance,innovation and high-quality development of Chinese Osmanthus city in Xianning under three" “new developments”[J].J Anhui Agric Sci,2023,51(12):256-259.DOI: 10.3969/j.issn.0517-6611.2023.12.058.
[6]刘燕培,李书情,王长海,等. 桂花新品种‘潢川丹桂’[J]. 园艺学报, 2022, 49(S2): 229-230. LIU Y P, LI S Q, WANG C H, et al. A new Osmanthus fragrans cultivar ‘Huangchuan Dangui’ [J]. Hortic Plant J, 2022, 49(S2): 229-230 (in Chinese with English abstract). DOI:10.16420/j.issn.0513-353x.2022-0376.
[7]王良桂,潘多,丁卉芬,等.彩叶桂新品种‘南林彩云’[J].南京林业大学学报(自然科学版),2023,47(2):243-244.WANG L G,PAN D,DING H F,et al.Osmanthus fragrans ‘Nanlin Caiyun’:a new cultivar of Osmanthus[J].J Nanjing For Univ (Nat Sci Ed),2023,47(2):243-244.
[8]国家质量监督检验检疫总局,中国国家标准化管理委员会.植物新品种特异性、一致性、稳定性测试指 桂花:GB/T 24885—2010[S].北京:中国标准出版社,2011.General Administration of Quality Supervision,Inspection and Quarantine of the People’s Republic of China,Standardization Administration of the People’s Republic of China.Guidelines for the conduct of test for distinctness,uniformity and stability-Sweet osmanthus(Osmanthus fragrans L.):GB/T 24885-2010[S].Beijing:Standards Press of China,2011.
[9]UPOV.TGP/15. Guidance on the use of biochemical and molecular markers in the examination of distinctness,uniformity and stability(DUS). Geneva:UPOV, 2013.
[10]李梅.桂花种质资源遗传多样性研究及品种鉴定[D].南京:南京农业大学,2009.LI M.Study on genetic diversity and cultivar identification of Osmanthus fragrans germplasm[D].Nanjing:Nanjing Agricultural University,2009.
[11]邱帅,吴光洪,陈徐平,等.基于相关序列扩增多态性分子标记的桂花栽培品种演化分析[J].浙江大学学报(农业与生命科学版),2017,43(4):404-415.QIU S,WU G H,CHEN X P,et al.Evolution analysis of sweet Osmanthus(Osmanthus fragrans) cultivars based on sequence-related amplified polymorphism molecular marker[J].J Zhejiang Univ (Agric Life Sci),2017,43(4):404-415.DOI: 10.3785/j.issn.1008-9209.2016.08.241.
[12]乔中全,王晓明,李永欣,等.桂花优良品种‘珍珠彩桂’遗传多样性的ISSR分析及指纹图谱构建[J].湖南林业科技,2016,43(3):1-5.QIAO Z Q,WANG X M,LI Y X,et al.Genetic diversity and fingerprint construction of varieties of Osmanthus fragrans ‘Zhenzhu Caigui’ by ISSR markers[J].Hunan For Sci Technol,2016,43(3):1-5.DOI: 10.3969/j.issn.1003-5710.2016.03.001.
[13]罗仙英,桂敬飞,严治,等.贵州桂花种质资源遗传多样性的ISSR分析[J].种子,2017,36(6):62-66.LUO X Y,GUI J F,YAN Z,et al.Genetic diversity analyses of Osmanthus fragrans lour.in Guizhou with ISSR markers[J].Seed,2017,36(6):62-66.DOI: 10.16590/j.cnki.1001-4705.2017.06.062.
[14]韩远记.桂花品种资源遗传多样性的AFLP分析[D].开封:河南大学,2008.HAN Y J.Study on the genetic diversity of Osmanthus fragrans cultivars by AFLP markers[D].Kaifeng:Henan University,2008.
[15]YAN X Y,XIAO B L,HAN Y J,et al.AFLP analysis of genetic relationships and diversity of some Chinese Osmanthus fragrans cultivars[J].Life Sci J,2009,6(2):11-16.
[16]ZHANG Z R,FAN D M,GUO S Q,et al.Development of 29 microsatellite markers for Osmanthus fragrans (Oleaceae),a traditional fragrant flowering tree of China[J].Am J Bot,2011,98(12):e356-e359.DOI: 10.3732/ajb.1100241.
[17]DUAN Y F,WANG X R,XIANG Q B,et al.Genetic diversity of androdioecious Osmanthus fragrans (Oleaceae) cultivars using microsatellite markers[J].Appl Plant Sci,2013,1(6):apps.1200092.DOI: 10.3732/apps.1200092.
[18]李军,董彬,张超,等.桂花EST-SSR引物开发及在品种鉴定中的应用[J].浙江农林大学学报,2018,35(2):306-313.LI J,DONG B,ZHANG C,et al.EST-SSR primers and their application in cultivar identification of Osmanthus fragrans[J].J Zhejiang A F Univ,2018,35(2):306-313.DOI: 10.11833/j.issn.2095-0756.2018.02.015.
[19]YANG X L,YUE Y Z,LI H Y,et al.The chromosome-level quality genome provides insights into the evolution of the biosynthesis genes for aroma compounds of Osmanthus fragrans[J].Hortic Res,2018,5:72.DOI: 10.1038/s41438-018-0108-0.
[20]CHEN H G,ZENG X L,YANG J,et al.Whole-genome resequencing of Osmanthus fragrans provides insights into flower color evolution[J].Hortic Res,2021,8(1):98.DOI: 10.1038/s41438-021-00531-0.
[21]LI Y,ZHAO H,XIA H X,et al.Multi-omics analyses provide insights into the genomic basis of differentiation among four sweet osmanthus groups[J].Plant Physiol,2024:kiae280.DOI: 10.1093/plphys/kiae280.
[22]魏中艳,李慧慧,李骏,等.应用SNP精准鉴定大豆种质及构建可扫描身份证[J].作物学报,2018,44(3):315-323.WEI Z Y,LI H H,LI J,et al.Accurate identification of varieties by nucleotide polymorphisms and establishment of scannable variety IDs for soybean germplasm[J].Acta Agron Sin,2018,44(3):315-323.DOI: 10.3724/SP.J.1006.2018.000315.
[23]DU H S,YANG J J,CHEN B,et al.Target sequencing reveals genetic diversity,population structure,core-SNP markers,and fruit shape-associated loci in pepper varieties[J].BMC Plant Biol,2019,19(1):578.DOI: 10.1186/s12870-019-2122-2.
[24]BUTTON P.The international union for the protection of new varieties of plants (upov) recommendations on variety denominations[J].Acta Hortic,2008(799):191-200.DOI: 10.17660/actahortic.2008.799.27.
[25]HEO M S,HAN K,KWON J K,et al.Development of SNP markers using genotyping-by-sequencing for cultivar identification in rose (Rosa hybrida)[J].Hortic Environ Biotechnol,2017,58(3):292-302.DOI: 10.1007/s13580-017-0268-0.
[26]贾清香.基于RAD测序的野鸢尾和射干的SSR及SNP特征分析[D].沈阳:沈阳农业大学,2018.JIA Q X.Characteristics of SSR and SNP in I. dichotoma and I. domestica using RAD sequencing[D].Shenyang:Shenyang Agricultural University,2018.
[27]MARTINA M,ACQUADRO A,PORTIS E,et al.Diversity analyses in two ornamental and large-genome Ranunculaceae species based on a low-cost Klenow NGS-based protocol[J].Front Plant Sci,2023,14:1187205.DOI: 10.3389/fpls.2023.1187205.
[28]SEMAGN K,BABU R,HEARNE S,et al.Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR (KASP):Overview of the technology and its application in crop improvement[J].Mol Breed,2014,33(1):1-14.DOI: 10.1007/s11032-013-9917-x.
[29]HE C L,HOLME J,ANTHONY J.SNP genotyping: the KASP assay[M]. New York:Humana Press,2014:75-86. DOI: 10.1007/978-1-4939-0446-4_7.
[30]GANAL M W,WIESEKE R,LUERSSEN H,et al.High-throughput SNP profiling of genetic resources in crop plants using genotyping arrays[M]//Genom Plant Genet Resour.Dordrecht:Springer,2014:113-130. DOI: 10.1007/978-94-007-7572-5_6.
[31]DO KIM K,KANG Y N,KIM C.Application of genomic big data in plant breeding:past,present,and future[J].Plants,2020,9(11):1454.DOI: 10.3390/plants9111454.
[32]NGUYENen, TOAN" K, HA S T T, et al. Analysis of Chrysanthemum genetic diversity by genotyping-by-sequencing[J]. Hortic Environ Biotechnol,2020,61: 903-913. DOI:10.1007/s13580-020-00274-2.
[33]WANG F Q, FAN X C, ZHANG Y, et al.Establishment and application of an SNP molecular identification system for grape cultivars[J].J Integr Agric,2022,21(4):1044-1057.DOI: 10.1016/S2095-3119(21)63654-7.
[34]LI Z Y,YU H L,LI X,et al.Kompetitive allele-specific PCR (KASP) genotyping and heterotic group classification of 244 inbred lines in cabbage (Brassica oleracea L.var.capitata)[J].Euphytica,2020,216(7):106.DOI: 10.1007/s10681-020-02640-8.
[35]RASHEED A,WEN W E,GAO F M,et al.Development and validation of KASP assays for genes underpinning key economic traits in bread wheat[J].Theor Appl Genet,2016,129(10):1843-1860.DOI: 10.1007/s00122-016-2743-x.
[36]GREWAL S,HUBBART-EDWARDS S,YANG C Y,et al.Rapid identification of homozygosity and site of wild relative introgressions in wheat through chromosome-specific KASP genotyping assays[J].Plant Biotechnol J,2020,18(3):743-755.DOI: 10.1111/pbi.13241.
[37]李书情.基于全基因组SNP的桂花品种精准鉴定体系的构建及应用[D].郑州:河南农业大学,2023.LI S Q.Construction and application of accurate identification system of Osmanthus fragrans varieties based on whole-genome SNP[D].Zhengzhou:Henan Agricultural University,2023.DOI: 10.27117/d.cnki.ghenu.2023.000780.
[38]POREBSKI S,BAILEY L G,BAUM B R.Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components[J].Plant Mol Biol Report,1997,15(1):8-15.DOI: 10.1007/BF02772108.
[39]CAI X,MAI R Z,ZOU J J,et al.Analysis of aroma-active compounds in three sweet Osmanthus (Osmanthus fragrans) cultivars by GC-olfactometry and GC-MS[J].J Zhejiang Univ Sci B,2014,15(7):638-648.DOI: 10.1631/jzus.B1400058.
[40]FU J X,HOU D,ZHANG C,et al.The emission of the floral scent of four Osmanthus fragrans cultivars in response to different temperatures[J].Molecules,2017,22(3):430.DOI: 10.3390/molecules22030430.
[41]HAN Y J,LI L X,DONG M F,et al.cDNA cloning of the phytoene synthase (PSY) and expression analysis of PSY and carotenoid cleavage dioxygenase genes in Osmanthus fragrans[J].Biologia,2013,68(2):258-263.DOI: 10.2478/s11756-013-0002-z.
[42]路飞.桂花新品种DUS测试指南的研制[D].南京:南京林业大学,2009.LU F.Study on the design of DUS testing guideline for new varieties of Osmanthus fragrans[D].Nanjing:Nanjing Forestry University,2009.
[43]VOSS D H,HALE W N.A comparison of the three editions of the royal horticultural society colour chart[J].Hortscience,1998,33:13-17.DOI: 10.21273/HORTSCI.33.1.13.
[44]臧德奎,向其柏.中国桂花品种分类研究[J].中国园林,2004,20(11):40-49.ZANG D K,XIANG Q B.Studies on the cultivar classification of Chinese sweet Osmanthus[J].J Chin Landsc Archit,2004,20(11):40-49.DOI: 10.3969/j.issn.1000-6664.2004.11.011.
[45]尚富德,陈仲芳,刘玉莲,等.桂花品种资源调查方法研究[J].河南大学学报(自然科学版),2003,33(1):9-13.SHANG F D,CHEN Z F,LIU Y L,et al.Study on the investigation of cultivar resources of Osmanthus fragrans[J].J Henan Univ (Nat Sci),2003,33(1):9-13.DOI: 10.15991/j.cnki.411100.2003.01.003.
[46]GLEISER G,VERD M.Repeated evolution of dioecy from androdioecy in Acer[J].New Phytol,2005,165(2):633-640.DOI: 10.1111/j.1469-8137.2004.01242.x.
[47]DANECEK P,AUTON A,ABECASIS G,et al.The variant call format and VCFtools[J].Bioinformatics,2011,27(15):2156-2158.DOI: 10.1093/bioinformatics/btr330.
[48]李梓榕,袁雄,陈叶,等.基于全基因组SNP高效鉴定水稻种质资源并构建指纹图谱[J].分子植物育种,2020,18(18):6050-6057.LI Z R,YUAN X,CHEN Y,et al.Effective identification for varieties by genome-wide SNPs and establishment of fingerprint for rice germplasm[J].Mol Plant Breed,2020,18(18):6050-6057.DOI: 10.13271/j.mpb.018.006050.
[49]SHEN Y S,WANG J S,SHAW R K,et al.Development of GBTS and KASP panels for genetic diversity,population structure,and fingerprinting of a large collection of broccoli (Brassica oleracea L.var.italica) in China[J].Front Plant Sci,2021,12:655254.DOI: 10.3389/fpls.2021.655254.
[50]LIU K J,MUSE S V.PowerMarker:An integrated analysis environment for genetic marker analysis[J].Bioinformatics,2005,21(9):2128-2129.DOI: 10.1093/bioinformatics/bti282.
[51]杨康民,朱文江.桂花[M].上海:上海科学技术出版社,1999.YANG K M,ZHU W J.Osmanthus fragrans[M].Shanghai:Shanghai Scientific amp; Technical Publishers,1999 (in Chinese).
[52]任海龙,许东林,张晶,等.菜薹KASP-SNP指纹图谱构建及品种鉴定[J].园艺学报,2023,50(2):307-318.REN H L,XU D L,ZHANG J,et al.Establishment of SNP fingerprinting and identification of Chinese flowering cabbage varieties based on KASP genotyping[J].Acta Hortic Sin,2023,50(2):307-318.DOI: 10.16420/j.issn.0513-353x.2021-1046.
[53]王富强,樊秀彩,张颖,等.SNP分子标记在作物品种鉴定中的应用和展望[J].植物遗传资源学报,2020,21(5):1308-1320.WANG F Q,FAN X C,ZHANG Y,et al.Application and prospect of SNP molecular markers in crop variety identification[J].J Plant Genet Resour,2020,21(5):1308-1320.DOI: 10.13430/j.cnki.jpgr.20200309002.
[54]LI H L,XIAO W J,TONG T,et al.The specific DNA barcodes based on chloroplast genes for species identification of Orchidaceae plants[J].Sci Rep,2021,11(1):1424.DOI: 10.1038/s41598-021-81087-w.
[55]樊晓静,于文涛,蔡春平,等.利用SNP标记构建茶树品种资源分子身份证[J].中国农业科学,2021,54(8):1751-1772.FAN X J,YU W T,CAI C P,et al.Construction of molecular ID for tea cultivars by using of single-nucleotide polymorphism(SNP) markers[J].Sci Agric Sin,2021,54(8):1751-1772.DOI: 10.3864/j.issn.0578-1752.2021.08.014.
[56]赵仁欣,李森业,郭瑞星,等.利用SNP芯片构建我国冬油菜参试品种DNA指纹图谱[J].作物学报,2018,44(7):956-965.ZHAO R X,LI S Y,GUO R X,et al.Construction of DNA fingerprinting for Brassica napus varieties based on SNP chip[J].Acta Agron Sin,2018,44(7):956-965.DOI: 10.3724/SP.J.1006.2018.00956.
[57]朱国忠,张芳,付洁,等.适于陆地棉品种身份鉴定的SNP核心位点筛选与评价[J].作物学报,2018,44(11):1631-1639.ZHU G Z,ZHANG F,FU J,et al.Genome-wide screening and evaluation of SNP core loci for identification of upland cotton varieties[J].Acta Agron Sin,2018,44(11):1631-1639.DOI: 10.3724/SP.J.1006.2018.01631.9.
(责任编辑 吴祝华)
收稿日期Received:2024-05-15 """修回日期Accepted:2024-06-07
基金项目:国家自然科学基金项目(32371911);河南省重大公益专项(201300110900)。
第一作者:王一涵(yihanwang@vip.163.com),副教授。
*通信作者:尚富德(shangfude@henau.edu.cn),教授。
引文格式:王一涵,刘姣姣,金沛权,等.
基于表型性状和SNP标记构建桂花主要品种资源的分子身份证[J]. 南京林业大学学报(自然科学版),2024,48(4):12-24.
WANG Y H, LIU J J, JIN P Q, et al.
Construction of molecular ID for Osmanthus fragrans cultivars based on phenotypic traits and single nucleotide polymorphisms (SNPs)[J]. Journal of Nanjing Forestry University (Natural Sciences Edition),2024,48(4):12-24.
DOI:10.12302/j.issn.1000-2006.202405026.

