A novel Effective Panicle Number per Plant 4 haplotype enhances grain yield by coordinating panicle number and grain number in rice
Yun Wang, Xiaoqian Wang, Laiyuan Zhai, Sundus Zafar, Congong Shen, Shuangbing Zhu,Kai Chen, Yun Wang*, Jianlong Xua,,e,*
a State Key Laboratory of Crop Gene Resources and Breeding, Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100081, China
b State Key Laboratory of Crop Biology, College of Agronomy, Shandong Agricultural University, Tai’an 271018, Shandong, China
c Shenzhen Branch,Guangdong Laboratory for Lingnan Modern Agriculture,Agricultural Genomics Institute at Shenzhen,Chinese Academy of Agricultural Sciences,Shenzhen 518120,Guangdong, China
d Rice Research Institute, Shenyang Agricultural University, Shenyang 110866, Liaoning, China
e National Nanfan Research Institute (Sanya), Chinese Academy of Agricultural Sciences, Sanya 572024, Hainan, China
Keywords:Rice Effective panicle number per plant Grain number per panicle Haplotype Grain yield potential
ABSTRACT Increasing effective panicle number per plant (EPN) is one approach to increase yield potential in rice.However, molecular mechanisms underlying EPN remain unclear.In this study, we integrated mapbased cloning and genome-wide association analysis to identify the EPN4 gene, which is allelic to NARROW LEAF1 (NAL1).Overexpression lines containing the Teqing allele (TQ) of EPN4 had significantly increased EPN.NIL-EPN4TQ in japonica (geng) cultivar Lemont (LT) exhibited significantly improved EPN but decreased grain number and flag leaf size relative to LT.Haplotype analysis indicated that accessions with EPN4-1 had medium EPN, medium grain number, and medium grain weight, but had the highest grain yield among seven haplotypes, indicating that EPN4-1 is an elite haplotype of EPN4 for positive coordination of the three components of grain yield.Furthermore, accessions carrying the combination of EPN4-1 and haplotype GNP1-6 of GNP1 for grain number per panicle showed higher grain yield than those with other allele combinations.Therefore, pyramiding of EPN4-1 and GNP1-6 could be a preferred approach to obtain high yield potential in breeding.
1.Introduction
Rice (Oryza sativa L.) is a staple food, feeding more than half of the global population[1].High rice productivity will be essential in meeting food requirements as the human population grows, coupled with the severe constraints of a changing environment and declining land and water resources [2].Rice grain yield is determined by three key component traits:EPN(effective panicle number per plant), GNP (grain number per panicle), and grain weight.However, these three traits are inversely correlated [3].GNP and EPN are the major contributors to grain yield compared to grain weight [4].The trade-off between EPN and GNP largely limits rice yield improvement, as there is a negative correlation between them [5].Rice production has remained stagnant since the significant improvements from the Green Revolution and introduction of hybrid rice last several decades [6].Therefore, further increasing panicle and grain number and balancing the sourcesink relationship to break the rice yield plateau is a huge challenge to rice breeders worldwide.
Several QTL were identified for the three yield component traits in the last few years using bi-parental cross analysis and genomewide association studies (GWAS) [7–11].So far, researchers have paid particular attention to grain weight and grain number,resulting in the isolation of many major QTL using map-based cloning,including qSW5 [12], GS3 [13], and GW2 [14] for grain size and weight, and Gn1a [15], DEP1 [16], IPA1 [17], GNP1 [18], and SPIKE[19] for grain number.Several studies focused on understanding the genetic regulation of tiller number by molecular analysis of mutants, such as moc1 [20], ostb1 [21], prog1 [22,23], tad1 [24],and moc3 [25].However, elite grain number alleles, such as IPA1[17], SPIKE [19], GNP1 [26], showed trade-off effects between EPN and GNP.Song et al.[5] solved the trade-off between GNP and EPN by editing a 54-base pair cis-regulatory region of IPA1 through CRISPR/Cas9, thus significantly increasing yield per plant.Thus, a search for superior haplotypes of genes affecting grain number or panicle number that better balance the relationship between GNP and EPN may be another effective strategy to overcome trade-off effects.
Here, a major QTL, EPN4 (LOC_Os04g52479), which significantly increased EPN in rice, was isolated using an integrated strategy combining genome-wide association analysis and map-based cloning.In previous studies, LOC_Os04g52479 (NAL1) has pleiotropic effects in regulation of several traits affecting leaf size[6,27],photosynthesis[28,29],chlorophyll content[30],spikelet number[19],vascular bundle phloem area in the panicle neck[31],root volume[32], and adventitious root development [33] in different genetic backgrounds.NAL1 degraded a corepressor factor OsTPR2,promoting histone acetylation levels and expression of the genes in hormone signaling pathways, causing pleiotropic physiological effects [34].In this study, we identified EPN4TQthat enhanced EPN from the high-yielding xian rice variety Teqing (TQ).p35S::EPN4TQtransgenic lines were used to validate the function of EPN4.Near-isogenic lines containing EPN4TQexhibited improved panicle number but decreased grain number per panicle and smaller flag leaf size.Our studies provide new insight into understanding the relationship between panicle number and other yieldrelated traits (leaf size and panicle size).We found an elite haplotype of EPN4-1 for EPN4,which achieved an optimal balance among the three yield components,EPN,GNP,and grain weight.This combination led to the highest yield among seven tested haplotypes.
2.Materials and methods
2.1.Materials and measurements of EPN
Six hundred and fifty-two accessions from the 3000 Rice Genomes Project(RGP)[35]were used for association mapping,including 303 xian, 250 geng, 5 basmati/sadri, 81 aus/boro, and 13 admixture (adm) accessions (Table S1).The association panel was grown at Jingzhou (30.3°N, 112.2°E) in Hubei province in the summer of 2016 and 2017.Each accession in three replications was sown as a two-row plot with ten plants per row using 17 cm × 25 cm spacing.Eight mid-row plants were investigated for EPN at maturity.
Reciprocal introgression lines(ILs)previously generated using a geng cultivar (Lemont, LT) and a xian cultivar (Teqing, TQ) for QTL mapping by linkage analysis.These included 201 ILs in LT background and 252 ILs in TQ background [36] planted in two-row plots as above in two replications in winter 2015(at Sanya in Hainan province,18.3°N,109.3°E),and summer 2016(Beijing,40.2°N,116.2°E).EPN was measured as described above.
Zhonghua 11(ZH11,geng variety)was used in transgenic experiments.Wild-type ZH11 and transgenic lines were planted in a randomized block design at 17 cm and 25 cm hill and row spacing,respectively, with three replications in Sanya in winter 2021.EPN measurements at maturity were as described above.
2.2.Genome-wide association study and haplotype analysis of QTL affecting EPN
GWAS was conducted using a mixed-model analysis, principal component analysis (PCA) + kinship (K), with EMMA eXpedited(EMMAX) software [37].The Rice SNP-Seek Database (http://www.oryzasnp.org/) retrieved 3K RGP 4.8mio SNP data [38].Plink software was used to filter SNPs with missing rates exceeding 20%and minor allele frequency(MAF)<5%[39].EMMAX software built on the batch normalization (BN) method based on high-quality SNPs was used to calculate the K matrix.Best linear unbiased predictors(BLUPs)were used for the association analysis to reduce the effects of environmental changes [40].GEC software was used to measure effective number of independent markers (N) [41], and suggestive P-value thresholds of association (1/N) were 1.23E-06,3.28E-06, and 1.9E-06 for the whole, geng, and xian populations,respectively.
Haplotype and gene-based association analysis was performed to identify candidate genes for the QTL (qEPN4) for EPN.The linkage disequilibrium (LD) block was defined by significant trait-associated SNPs at the candidate gene region of qEPN4.The software ‘‘Haploview v4.2” was used to analyze the LD block[42].An LD block was generated when the above 95% confidence boundaries of D’ value surpassed 0.98 and the lower boundaries surpassed 0.70 [43].All available SNPs for the genes in the qEPN4 region were retrieved from 3 K RGP 18mio SNP data set in the Rice SNP-Seek Database [38].The remaining SNPs, after filtering(using the same screening criteria as the GWAS above),were used to conduct gene-based association analyses through a mixed linear model(MLM)using the PCA and K applications in GWAS.The suggestive Pvalue threshold was 4.65E-04.All candidate genes with significant SNPs detected by gene-based association analysis were used for haplotype analysis.Haplotype analysis was conducted using all non-synonymous SNPs within the gene coding sequence(CDS).
2.3.QTL mapping by linkage analysis and fine-mapping of qEPN4
IciMapping V4.1 software was used to perform QTL mapping[44].QTL identification was conducted using 154 high-quality SSR markers.Threshold values logarithm of the odds (LOD) > 3.0 were used to declare the presence of a QTL.
For map-based cloning of qEPN4, an IL named GG276 (BC3F6),containing chromosome region RM317–RM348 from TQ and 87.3% of LT genetic background was selected from 201 LT-ILs,and backcrossed twice to LT using marker-assisted selection(MAS).Self-pollination of BC5F1heterozygous plants produced heterozygote near-isogenic lines (NILs, BC5F2), comprising almost all LT genetic background, except the RM317–RM348 region from TQ (Fig.S1).Self-pollination was conducted to obtain segregating NIL-BC (BC5F3, BC5F4,and BC5F5) populations for fine mapping of qEPN4 and selection of NIL-EPN4TQ(Fig.S1).Primers designed fine mapping are listed in Table S2.
2.4.Vector construction and plant transformation
The CDS (1.7-kb) of EPN4 from TQ was amplified using genespecific primers(SmaI-EPN4-F/SalI-EPN4-R)to construct an overexpression vector (Table S2).The amplified DNA was digested by restriction enzyme (SmaI/SalI) and inserted into a binary vector(pCAMBIA2300) with the constitutive CaMV35S promoter.The overexpression plasmid was inserted into Agrobacterium tumefaciens strain EHA105 and transformed into ZH11.
2.5.Phenotypic evaluation of NIL-EPN4TQ and identification of EPN4 haplotypes
To evaluate the effects of EPN4 on EPN and other yield-related traits, we developed the reciprocal NILs, NIL-EPN4TQ, and NILEPN4LT, differing in ~50.3 kb region of EPN4 derived from LT in TQ genetic background [6].The NILs and their corresponding isogenic controls(LT and TQ)were grown in three different provinces,i.e., Jiangxi (Pingxiang, 27.6°N, 113.9°E), Hubei (Jingzhou, 30.3°N,112.2°E), and Guangdong (Jiangmen, 21.3°N, 111.6°E) in summer,2020 in randomized plot design with three replications.The area of each plot was 13.3 m2, with a single plant transplanted per hill at 25 d after sowing and a spacing of 17 cm between hills and 20 cm between rows.At the full heading stage, flag leaf width(FLW, cm) and flag leaf length (FLL, cm) were measured on the main stems of eight plants in each line.Flag leaf area (FLA, cm2)was calculated by FLL × FLW × 0.75.At maturity, the whole plot was harvested for grain yield measurement based on 14%moisture content after air-drying.Eight plants were harvested and dried in an oven at 70°C for five days for trait analysis,including EPN,grain number per panicle (GNP), primary branch number (PBN), secondary branch number (SBN), filled grain number per panicle(FGN), thousand-grain weight (TGW, g), panicle length (PL, cm),grain length (GL, mm), and grain width (GW, mm).
To identify elite haplotypes of EPN4 and GNP1 for GNP, haplotype analysis was executed using all SNPs in the EPN4 and GNP1(from the promoter to 3′UTR) searched from the 3 K RGP 18mio SNP data set in the Rice SNP-Seek Database [38].Yield and yieldrelated traits in accessions carrying major haplotypes (> 20 accessions) were investigated at Jingzhou in Hubei province (30.3°N,112.2°E) in summer 2016 and 2017.FLL and FLW were measured for each plant at the heading stage.FLA was calculated by FLL×FLW×0.75.Eight uniform plants were harvested at maturity and dried in an oven at 70 °C for five days to measure EPN, GNP,TGW, and grain yield per plant (GY, g).
2.6.RNA isolation and qRT-PCR
Total RNA was extracted from flag leaves at full heading using an RNA Pure Plant Kit (TIANGEN, China).cDNA was synthesized using M-MLV Reverse Transcriptase, and qRT–PCR analyses were performed using the SYBR Premix Ex Taq kit (TIANGEN) on a Step One System (Applied Biosystems).A 7500 Real-Time PCR System(Applied Biosystems) was used to conduct qRT-PCR.The quantification method (2-ΔΔCT) was used and variation in expression was estimated using three biological replications.PCR conditions consisted of an initial denaturation step at 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 56 °C for 30 s.The rice OsActin2 gene was used as the internal reference.
2.7.Statistical analysis
Phenotypic differences betweenthe haplotypesof EPN4(containing more than 20 accessions)and haplotype combinations of EPN4 and GNP1(containing more than 15 accessions)were examined by a one-way ANOVA or two-tailed Student’s t-tests.Duncan’s multiple range tests were used to determine the significance of differences(P<0.05).All the analyses were performed in R software[45].
3.Results
3.1.Phenotypic variation of EPN in a natural population
The 652 rice accessions from 58 countries exhibited a distinctive population structure.Among the 652 accessions,303 were categorized into indica (xian) subpopulation, including 79 (xian-1A),85 (xian-1B), 13 (xian-2), 1 (xian-3), and 125 (xian-adm), and 250 were categorized into japonica (geng) subpopulation, including 151 (geng-tmp), 61 (geng-trp), 35 (geng-adm), and 3 (geng-sbtrp)(Fig.1A; Table S1).EPN showed significant phenotypic changes in the whole population ranging from 2.8 to 24.0 (Fig.1A).There was also a significant difference in EPN between two different subpopulations (xian and geng) (Fig.1A).
3.2.Identification of QTL for EPN by GWAS
We identified single-nucleotide polymorphisms (SNPs) related to EPN by conducting an MLM analysis in a GWAS using the whole,xian and geng populations.16 significant SNPs were identified in the whole population, whereas 6 and 337 SNPs were identified in xian and geng subpopulations, respectively (Fig.1B).SNPs in the estimated LD block were denoted as a single QTL [46].5, 1, and 2 QTL affecting EPN were detected in the whole, xian and geng populations, respectively (Fig.1C; Table S3).QTL qEPN4.2 located on chromosome (chr) 4 was present in both the whole and geng populations (Fig.1C; Table S3).
qEPN4.2 was located in the 30.74–31.33 Mb region (Fig.1D);2,152 SNPs(from promoter to 3′UTR)found in 79 genes were used for association analysis, and 10 significant SNPs associated with EPN were located in seven genes (LOC_Os04g51990, LOC_Os04g52130, LOC_Os04g52280, LOC_Os04g52479, LOC_Os04g52500,LOC_Os04g52520, and LOC_Os04g52614) (Fig.1E).There were differences in EPN among different haplotypes at four genes,
LOC_Os04g52280 (FC1), LOC_Os04g52479 (NAL1), LOC_Os04g52520,and LOC_Os04g52614 in the whole population (Table S4).Among them, NAL1 (LOC_Os04g52479) plays an integral part in leaf morphogenesis by regulating auxin transport and contributing to grain number per panicle and GY in rice[6,19,28,30].Three haplotypes of LOC_Os04g52479 were identified (Fig.1F) based on three nonsynonymous SNPs and where haplotype 1 (Hap 1) and Hap 3 genotypes produced more EPN than Hap 2 genotypes (Fig.1G).Thus,NAL1 was considered the most likely candidate gene for qEPN4.2.
3.3.Map-based cloning of qEPN4
Five and three QTL for EPN were identified in TQ-ILs and LT-ILs,respectively, in two environments (Table S5).Among them, the major QTL (qEPN4) was consistently detected in the RM317–RM348 region on chr 4 in both TQ-and LT-ILs across different environments, and was responsible for the high genetic variation in EPN (Table S5).The TQ allele was associated with higher EPN(Table S5).
To further validate and clone qEPN4, we conducted segregation from BC5F2to BC5F5populations by continuous self-pollination using the recombinant heterozygote NILs(BC5F2)for fine mapping of qEPN4 and construction of NIL-EPN4TQ(Fig.2A).In total, 6000 BC5F3plants derived from eight BC5F2heterozygous plants were used for fine mapping of qEPN4 in the RM317–RM348 interval.Ten recombinants were identified from three genotypes using four new makers under the target region (RM317–RM348).Multiple comparisons of BC5F4lines (homozygous recombinant) for EPN with the non-recombinant controls delimited qEPN4 in a 143.4 kb region between WY25 and RM3534 (Fig.2B).We identified four recombinants and two genotypes in the target area through fine mapping of 8,500 BC5F4plants with three new markers between WY25 and RM3534.We finally narrowed qEPN4 down to a 29.2 kb region flanked by cleaved amplified polymorphic sequence(CAPS) makers FL42 and FL98, using the linkage map by progeny testing of BC5F5homozygous recombinant plants(Fig.2B).According to the Rice Genome Annotation Project, there were two annotated genes, LOC_Os04g52460 (retrotransposon protein), and
LOC_Os04g52479 (NAL1/SPIKE/GPS/LSCHL4/SS1/LVPA4) in this region.We concluded that LOC_Os04g52479, tentatively termed EPN4,was the target gene associated with effective panicle number.
3.4.Allelic sequence comparison of EPN4 in Teqing and Lemont
Based on the predicted data, we sequenced the genomic sequence of EPN4 from the promoter (1.5 kb upstream of ATG) to the 3′UTR in TQ and LT.There were three single-base substitutions between TQ and LT, located in the promoter region, 3′UTR, and CDS, respectively (Fig.2C).A single-base substitution in the third exon of the coding region caused a change from arginine (R) in TQ to histidine(H)in LT(Fig.2C).Based on this amino acid substitution, a significant change in the predicted 3-D structure was observed in the trypsin-like serine and cysteine protease domain of the NAL1 protein between the LT and TQ [6].In addition, insertion of a retrotransposon(5895 bp)was observed at the junction site of the first and second exons (Fig.2C).
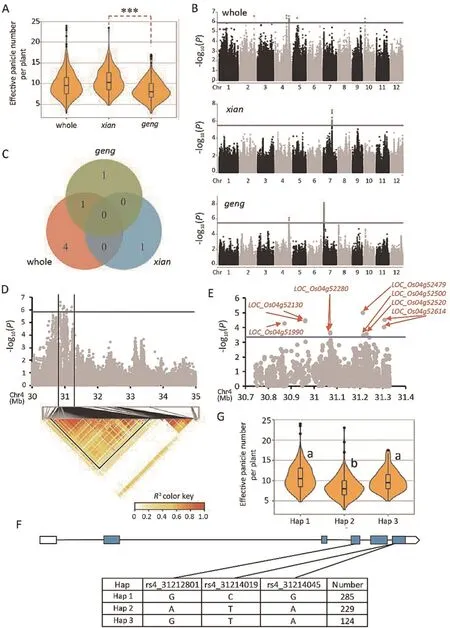
Fig.1.Candidate genes identified in chromosome (chr) 4 via genome-wide association analysis study (GWAS).(A) Box plots of effective panicle number per plant(EPN).***,P<0.001 by Student’s t-tests.(B)Manhattan plots of GWAS for the EPN in the whole, xian, and geng populations.Horizontal lines represent the significance threshold.The significant P-value thresholds of association(Bonferroni correction method) are 1.23E-06, 3.28E-06, and 1.9E-06 for the whole, xian, and geng populations, respectively.(C) Venn diagram showing unique and shared QTL mapped by GWAS among the whole, xian, and geng populations.(D) Local Manhattan plot (top) and LD heatmap (bottom) surrounding the peak on chr 4.Vertical solid lines indicate the candidate region for the peak.(E) Gene-based association analysis of targeted genes within the LD heatmap.The horizontal line represents a significance threshold.The significant P-value thresholds of association(Bonferroni correction method) is 4.65E-04.(F) Exon–intron structure of LOC_Os04g52479 and DNA polymorphisms in the gene.(G) Boxplots for EPN based on the haplotypes (Hap) of LOC_Os04g52479 in the whole population.Different letters on boxplots(a and b)indicate significant differences at a P<0.05 for multiple comparisons by Duncan’s multiple range test.
3.5.Effects of EPN4 overexpression on panicle number
To test this prediction, we constructed a binary vector containing the CDS sequence of EPN4 driven by a CaMV35S promoter,which we used to transform a geng rice (O.sativa L.) variety ZH11, whose EPN4 CDS matches that of LT.EPN4 showed 14.6-and 58.7-fold higher expression than ZH11 in flag leaves (Fig.3A)and the p35S::EPN4TQ-1 and p35S::EPN4TQ-2 lines were 11.3% and 22.5%, respectively, higher than ZH11 (Fig.3B, C).These outcomes confirmed that LOC_Os04g52479 is the same gene as EPN4,and that the EPN4TQallele increased effective panicle number in accordance with its expression.
3.6.NIL- EPN4TQ increases EPN but decreases FLW and GNP
The heading date for NIL-EPN4TQwas 1–2 d earlier than LT in three environments.The width, length, and area of flag leaves in NIL-EPN4TQwere considerably smaller than those in LT (Table 1).Compared with LT, NIL-EPN4TQexhibited a significant increase in EPN (+35.7%) but significant decreases in FGN (-16.4%), GNP(-14.1%), PBN (-13.8%), and SBN (-22.9%), a slight decrease in TGW(-5.5%)and GL(-2.3%),and no effect on PL and GW.Finally,the NIL-EPN4TQproduced higher (3.1%–4.0%) grain yield than LT across the three environments (Table 1).
The heading date for NIL-EPN4LTwas 1–2 d later than TQ across three environments.GNP for NIL- EPN4LTwas an average 16.0%higher than that of TQwith much higher(17.5%)secondary branch number, and more (18.9%) FGN.No significant difference was detected in EPN and TGW.Finally, NIL-EPN4LTproduced 4.7%–8.6% higher grain yield than TQ (Table 1).
3.7.Haplotype analysis of EPN4 in rice
Twenty-four SNPs with MAF > 5% were identified in the EPN4 genomic region among 652 accessions.Of these polymorphisms,6 SNPs were in the promoter region of about 2 kb upstream of the start codon (Fig.4A).Two synonymous SNPs, and one and two nonsynonymous SNPs were identified in the first, third and fifth exons, respectively.The nonsynonymous SNP(rs4_31212801,G/A)in the third exon resulted in the replacement of an arginine (R) by a histidine (H) at the 233rd residue.Among the other 13 SNPs,9 and 4 were present in introns and the untranslated region (3′UTR), respectively (Fig.4A).
We identified 13 haplotypes(EPN4-1 to EPN4-13)based on SNPs in 652 diverse accessions and four haplotypes (WR1 to WR4) in eight wild accessions.Haplotypes EPN4-10 and EPN4-11 from cultivated rice were the same as WR1 and WR2,respectively(Fig.4A,B), suggesting that EPN4-10 and EPN4-11 were the original haplotypes.Differences in yield and yield-related traits were analyzed using seven major haplotypes(EPN4-1 to EPN4-7),each comprising more than 20 rice accessions (Fig.4C–H).Among them, aus accessions carrying EPN4-7 had the most EPN (Fig.4C), suggesting that EPN4-7 was an elite haplotype associated with effective panicle number.Germplasms with the EPN4-6 haplotype (represented by LT) exhibited the most GNP, widest FLW, and largest FLA among all seven haplotypes (Fig.4D–F), indicating that EPN4-6 was an elite haplotype related to flag leaf size and grain number.Interestingly, the grain yields of accessions carrying the two elite haplotypes (EPN4-7 and EPN4-6) were not the highest among the seven haplotypes (Fig.4H).Indeed, accessions carrying EPN4-6 had the lowest yields due to the lowest EPN(Fig.4C,H).Accessions with EPN4-1 and EPN4-5 had medium EPN and GNP, but the final grain yields were substantially higher than those of the other five haplotypes (Fig.4C, D, H).Among them, the accessions with EPN4-1 exhibited significantly higher TGW than those with EPN4-5(Fig.4G),resulting in slightly higher but no significant difference in final grain yield (Fig.4H), and indicating that EPN4-1 was an elite haplotype of EPN4 for positive coordination among the three components of grain yield.
Association analysis of EPN4 showed that SNP rs4_31214479(A/G) in the 3′UTR was significantly associated with the GNP and FLW(Table S6).A SNP in the third exon causing an amino acid change from arginine (R) in TQ to histidine (H) in LT was the only variation between EPN4-3 and EPN4-4,and the cultivars containing EPN4-3 had considerably higher GNP and wider FLW than those containing EPN4-4 (Fig.4D, E).Furthermore, a SNP (rs4_31212801, G/A) was significantly associated with FLW, GNP, and EPN (Table S6).Only one nucleotide variation in rs4_31212801 was detected between EPN4-2 and EPN4-4 (Fig.4A), and the same nucleotide variation was identified between TQ and LT.Cultivars with EPN4-2 (H-type) had significantly more GNP and wider FLW but less EPN than those cultivars with EPN4-4(R-type).A comparison of accessions containing R-type and H-type showed that R-type accessions exhibited significantly more EPN but less GNP and narrower FLW than H-type accessions(Fig.S2),which supporting previous reports [46].Based on these results, we speculated that the SNP rs4_31212801 was also the functional SNP for EPN4.However, the favorable allele of EPN4 associated with EPN was G,whereas those associated with GNP and FLW of NAL1 were A.The different base sequences of the functional SNP might contribute to the pleiotropic effect of NAL1.Interestingly,all eight wild rice germplasms detected in this study were R-type (Fig.4A), suggesting that the R-type was the ancestral form.Accessions carrying the R-type haplotype (EPN4-1, EPN4-3, EPN4-4, EPN4-5, and EPN4-7) had higher EPN than accessions with the H-type haplotype(EPN4-2 and EPN4-6).R-type haplotypes EPN4-10 and EPN4-11 were present in six wild accessions(Fig.4B);these haplotypes with higher EPN having R-type might be derived from an O.rufipogon accession with higher panicle number.H-type accessions with higher EPN might be derived from R-type accessions.

Fig.3.Transgenic analysis for EPN4 through overexpression in rice.(A)Relative expression levels of EPN4 in flag leaves of two EPN4TQ overexpression lines and Zhonghua 11(ZH11).The values are means ± SD (n = 5).(B) Gross morphology of two independent pS35::EPN4TQ overexpression lines and ZH11 (Scale bar, 10 cm).(C) Effective panicle number per plant in two independent pS35::EPN4TQ overexpression lines and ZH11.The values are means ± SD (n = 20).
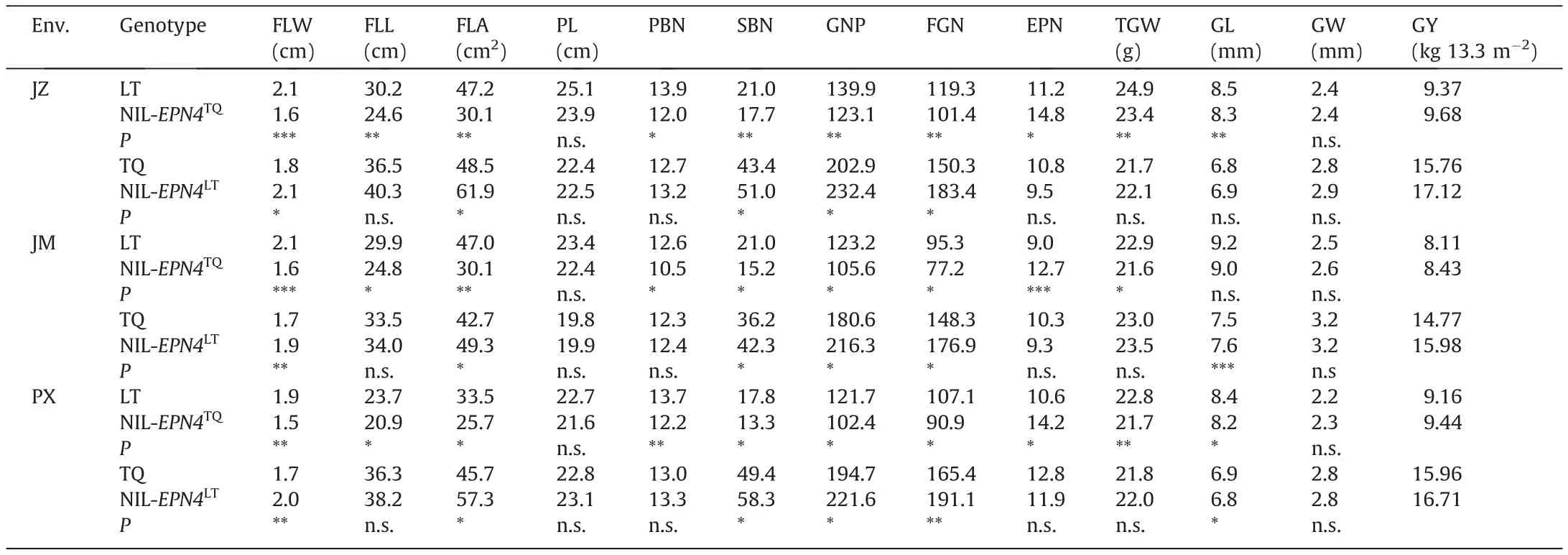
Table 1 Agronomic traits for NILs across three environments.
3.8.Effects of EPN4 and GNP1 haplotype combinations on grain yield and related traits
In our previous study,specific haplotype combinations of GNP1 and NAL1, both of which affect GNP, significantly enhanced grain yield in geng rice cultivars [47].To dissect the effects of combinations of EPN4 and GNP1 haplotypes, we conducted a haplotype analysis of GNP1 in 652 accessions.There were 33 SNP variations in the GNP1 genomic region, and a total of 12 haplotypes (GNP1-1 to GNP1-12) were detected (Fig.S3A).As in the EPN4 haplotype analysis, phenotypic differences in GNP and GY were analyzed using six major haplotypes (GNP1-1 to GNP1-6) comprising more than 20 rice accessions.The results demonstrated that accessions carrying GNP1-6 had significantly higher GNP than accessions carrying the other five haplotypes (Fig.S3B) and produced the highest mean grain yield of 19.3 g per plant (Fig.S3C).Thus GNP1-6 was an elite haplotype of GNP1 for grain number.
Based on the major haplotypes of EPN4 and GNP1 in germplasms,differences in EPN,GNP,and GY phenotypes were analyzed using 11 two-gene combinations (above 15 accessions) (Table 2).Among them, accessions carrying EPN4-1/GNP1-6 showed the highest mean GNP of 219.9 and produced the highest grain yield of 21.4 g per plant (Table 2).A comparison of yield and yieldrelated traits of accessions carrying EPN4-1 and GNP1-6 with those of accessions carrying only EPN4-1(Fig.5)showed that the average GNP of germplasms having EPN4-1 and GNP1-6 was significantly increased by 26.9% relative to germplasms carrying only EPN4-1(Fig.5A).However, there were no significant differences in EPN,TGW, FLW, and FLA between the combined EPN4-1/GNP1-6 and EPN4-1 haplotypes (Fig.5B–E).Finally, cultivars carrying the EPN4-1/GNP1-6 combination significantly increased grain yield by 13.9% on average compared with cultivars having only EPN4-1(Fig.5F).These results indicated that the EPN4-1/GNP1-6 combination had potential to increase grain yield.
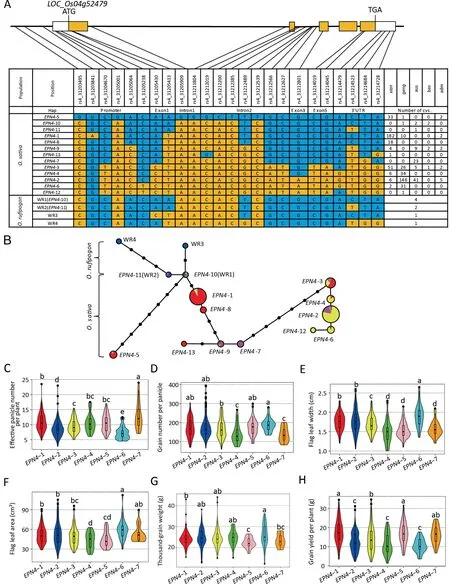
Fig.4.Haplotype analysis of EPN4.(A) Gene structure of EPN4.Five exons are indicated as light orange boxes, 3′UTR and 5′UTR are shown as white boxes, and single nucleotide polymorphism(SNP)positions are connected to the haplotype table by lines(SNP frequency>5%).Thirteen haplotypes,classified by SNP variations,were detected in the collection (indicated as either blue or light orange).The number and subpopulation identity of varieties carrying each haplotype are displayed in the right columns.Eight wild rice (O.rufipogon) varieties of are indicated by WR1–4.(B) A minimum-spanning tree for EPN4.A circle represents each haplotype group, and the circle size represents the number of accessions within each haplotype, as in Fig.4A.Blue, red, yellow, pink, green, and cyan represent O.rufipogon, xian, geng, aus, bas, admix,respectively.(C–H)Effective panicle number per plant(C),grain number per panicle(D),flag leaf width(E),flag leaf area(F),thousand-grain weight(G),and grain yield per plant (H) are compared among accessions (more than 20 accessions) carrying EPN4-1 to EPN4-7.Letters on violin plots (a, b, c, d, and e) are ranked by Duncan’s tests at P < 0.05.
4.Discussion
4.1.EPN4 from xian cultivar Teqing plays a positive role in EPN
Plant morphology can be improved to acquire high yield; for example,semi-dwarf varieties with lodging-resistance were developed to sustain high yield potential in the‘‘Green Revolution”[48].Thus, panicle number is a key plant morphological trait to acquire high grain yield in rice.The plant with tillering ability (primary,secondary, and tertiary tillers) can produce effective panicle numbers.To understand the regulation of tillering, various mutants,such as moc1 [20], ostb1 [21], prog1 [22,23], tad1 [24], and moc3[25], were molecularly analyzed.QTL analyses showed that variation in tiller number in segregating populations tended to be quantitative rather than qualitative indicating different genetic regulatory mechanisms.Therefore, cloning QTL for tiller number,especially EPN, is essential for understanding and utilizing such variation.However, little research has been reported on cloning genes relevant to EPN in rice.
This study used map-based cloning and genome-wide association analysis to isolate a major QTL, EPN4, that significantly increased EPN.We validated the function of EPN4 by haplotype analysis, genetic transformation, and NIL comparison.Introgression of the EPN4 allele from a high-yielding xian variety TQ into the geng rice cultivar LT significantly increased EPN by 32.1%–41.1% in the three environments (Table 1).Overexpression lines with EPN4 from TQ had significantly increased EPN(Fig.3),demonstrating that EPN4, which is identical to previously named NAL1,affected panicle number.Yano et al.[46]identified NAL1 associated with the EPN trait using a panel of 176 geng rice cultivars developed in breeding programs conducted in Japan.They found that varieties with haplotype B (belonging to R-type) from geng varieties produced more EPN than varieties those containing haplotype A (belonging to H-type).A comparison of the two studies showed that although both EPN4 and NAL1 affected the EPN trait, their respective favorable alleles came from xian and geng rice cultivars,respectively.In this study,the plants overexpressing EPN4 from TQ(carrying haplotype R-type) showed significantly increased EPN compared with the ZH11 background (carrying haplotype Htype).In contrast, lines transformed with haplotype B (belonging to R-type)showed no apparent difference compared to the controls[46].Based on these results,we proposed that superior NAL1 superior alleles from xian and geng rice varieties might differ in how they regulate EPN trait.
4.2.Effects of EPN4 alleles on plant type and panicle type
Grain yield per plant in rice is mainly determined by traits EPN,GNP, and grain weight.Grain weight is less significant than GNP and EPN in determining grain yield [4].Genetically, TGW showed no association with EPN, and a negative correlation was found between TGW and GNP,mainly due to genetic linkage rather than pleiotropy.However, panicle development and tillering share the same regulatory mechanisms and other botanical developments like branching and apical growth[49].EPN was significantly negatively correlated with GNP and FLW [7,11,50].Previous research showed that NILs containing grain number genes, such as IPA1[17], SPIKE [19], and GNP1 [26], showed significantly enhanced GNP but considerably reduced EPN.The same trend was found in NIL-EPN4LTin this research.
NIL-EPN4LTcontaining the EPN4(NAL1)allele from LT increased the GNP by 13.8%–19.8% but increased actual plot yield by 4.7%–8.6% compared with TQ in three environments (Table 1).The limited increase in grain yield of NIL-EPN4LTmainly resulted from the decrease in EPN (-12% to -7%) (Table 1).Conversely, NILEPN4TQwith the EPN4 allele from TQ produced more EPN (32.1%–41.1%) but 12.0%–15.9% fewer GNP than its isogenic control (LT).Finally,the NIL-EPN4TQonly produced 3.1%–4.0%higher grain yield than LT across the three environments(Table 1).Wu et al.[51]also reported that large culm cultivars exhibited larger panicles, larger flag leaf size, more grains per panicle, and lower tiller numbers.Accessions with EPN4 R-type haplotypes produced more but smaller panicles, and narrower flag leaves.In contrast, H-type accessions had less but larger panicles and wider flag leaves (Fig.S2).In other words,the reduction of panicle number in H-type cultivars might provide more assimilate to growing organs(e.g.,flag leaves),or the initiation and pre-dimensioning of larger organs (e.g., grain number) at the meristem level, thus decreasing tiller bud outgrowth and reducing tiller and panicle numbers [52].Thus, variation in the EPN4 gene could better clarify the relationship between organ size(leaf size and panicle size)and panicle number.
4.3.Implications of EPN4 in rice breeding of high yield potential
The ‘yield component compensation’ is thought to be largely responsible for failure of breeding to enhance yield potential by selecting yield components in major cereals [53].The trade-off between EPN and GNP is the main limiting factor in rice.Although the accessions carrying EPN4-7 had the highest EPN among the seven haplotypes in this study, the yield was not as high as expected due to lower GNP (Fig.4C, D, H).On the contrary, germplasms with EPN4-6 had the highest GNP but lowest EPN,resulting in the lowest grain yield among the seven haplotypes (Fig.4C, D,H).Therefore, selection for extremely high EPN or extremely high GNP is not conducive to obtaining the highest yield.The balance between the panicle and grain numbers seems more essential than emphasizing a single trait like the panicle number or gain number.In this study, the average grain yield of accessions carrying haplotypes EPN4-1 or EPN4-5 were substantially higher than accessions with five other haplotypes (Fig.4H), mainly due to the fact that accessions carrying the two haplotypes were able to better balance the relationship between EPN and GNP.However,in rice breeding,there are successful examples of cultivars with larger panicles or panicle numbers that are adapted to specific ecological regions.For instance, IRRI-released rice cultivars with higher panicle numbers are more productive in the dry season with more solar radiation.In contrast, cultivars with large panicles are more productive in the wet season with less solar radiation [54].Zhou developed a three-line intersubspecific F1hybrid (between xian and geng)with high panicle number and well adapted to areas like Sichuan province with less solar radiation,high humidity,and high temperature[55].Large panicles are often associated with reduced percentage of grain filling.lower panicle number, longer growth period,increased plant height[56],and greater proneness to lodging resistance.Rice breeding practice indicates that panicle number-type cultivar usually has broader adaptation than large panicle-type cultivar.It is therefore advisable to improve grain yield potential by enlarging sink capacity by increasing spikelet number per panicle based on medium panicle numbers [57].As indicated in this study, the accessions carrying a combination of the elite haplotypes EPN4-1 and GNP1-6 could significantly increase final grain yield via increasing GNP compared to those carrying only EPN4-1 (Fig.5A, F).
Previous reports indicated that introgression of genes associated with grain number, such as DEP1 [16], SPIKE [19], and GNP1[18], significantly enhanced GNP but considerably reduced grain weight in rice.Due to the significant reduction in grain weight,the yield potential of these grain number genes was limited.Thus,attention should be paid to coordinating the three main rice yield components to develop high-yield rice varieties.Although accessions with EPN4-1 or EPN4-5 had similar GNP and EPN, the latterdisplayed had significantly reduced TGW by 7.5%, and GY was significantly reduced by 4.7% (P > 0.05).We concluded that EPN4-1 was the preferred elite haplotype of EPN4 for positive coordination of the three components of grain yield and should be used as a potential genetic resource for increasing rice yield.Two xian accessions carrying with haplotype EPN4-1, B011 (with mean GNP of 193.2, mean EPN of 12, mean TGW of 22.3 g, and mean GY of 29.9 g/plant) and IRIS_313-7911 (with mean GNP of 193.3, mean EPN of 15,mean TGW of 27.3 g,and mean GY of 32.4 g/plant)were identified as elite resources in the accession panel.The favorable haplotypes from B011 and IRIS_313-7911 could be introgressed into geng and xian cultivars by MAS to develop high-yielding rice varieties with balanced panicle number and grain number.
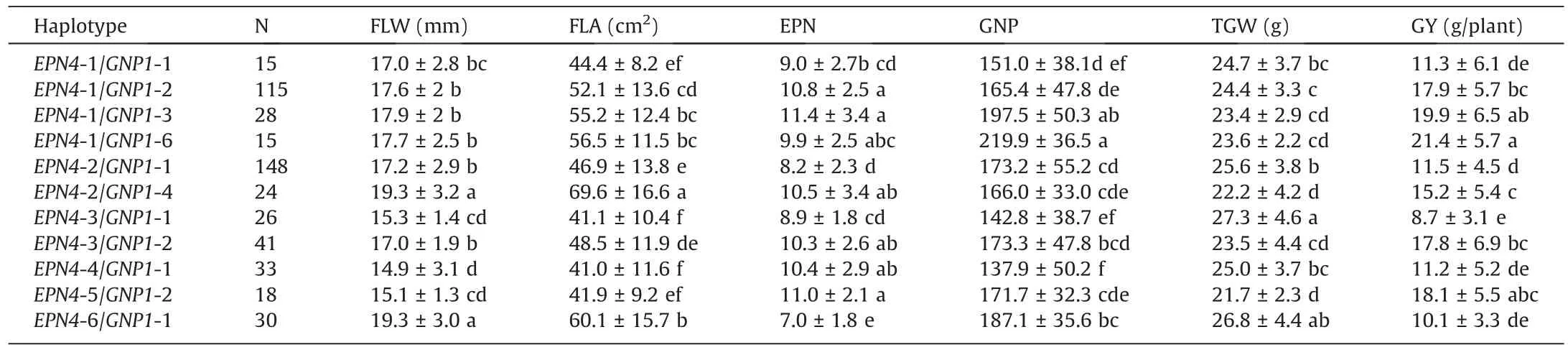
Table 2 Comparison of grain yield and their related traits among genotype combinations at EPN4 and GNP1.
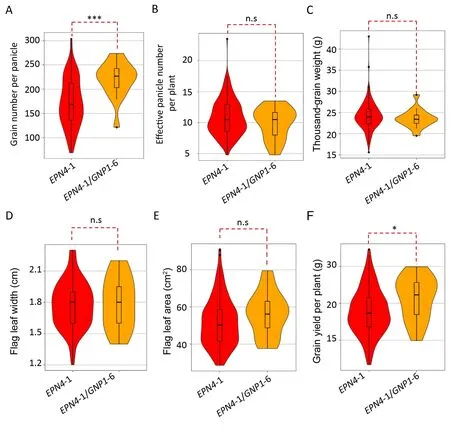
Fig.5.Comparison of yield and yield-related traits between accessions carrying an EPN4-1/GNP1-6 combination and accessions carrying the EPN4-1 haplotype.
5.Conclusions
The EPN4 gene for effective panicle number (EPN) was genetically dissected by GWAS and map-based cloning and then validated by transformation and near-isogenic lines.The elite haplotype EPN4-1 conferred medium EPN, medium grain number,and medium grain weight, indicating a balanced coordination of EPN, GNP, and grain weight, in determination of the highest grain yield among seven haplotypes.Among six major haplotypes for grain number per panicle(GNP1-1 to GNP1-6), GNP1-6 had significantly more GNP than the other five haplotypes and produced the highest grain yield.Accessions carrying the combination of EPN4-1 and GNP1-6 showed the highest grain yield, indicating a positive coordination of panicle number, grain number, and grain weight.Our results suggest that increased grain yield can be achieved by positively coordinating three yield component traits as indicated in the combination of EPN4-1 for EPN and GNP1-6 for GNP via a marker-assisted pyramiding approach.
CRediT authorship contribution statement
Yun Wang:Investigation, Writing – original draft.Xiaoqian Wang:Investigation, Writing – original draft.Laiyuan Zhai:Data curation, Formal analysis.Sundus Zafar:Writing – review & editing.Congcong Shen:Investigation.Shuangbing Zhu:Investigation.Kai Chen:Investigation.Jianlong Xu:Funding acquisition,Project administration, Conceptualization, Writing – review &editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Dr.Xianjin Qiu for management of materials at Jingzhou.This work was funded by the National Key Research and Development Program of China (2023YFF1000404), the Shenzhen Basic Research and Development Key Program of China(JCYJ20200109150713553), and Hainan Key Research and Development in Modern Agriculture of China (ZDYF2021Y128).
Appendix A.Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2023.11.003.
- The Crop Journal的其它文章
- Flowering-time regulation by the circadian clock: From Arabidopsis to crops
- Global characterization of OsPIP aquaporins reveals that the H2O2 transporter OsPIP2;6 increases resistance to rice blast
- Drought-triggered repression of miR166 promotes drought tolerance in soybean
- The OsBSK1-2-MAPK module regulates blast resistance in rice
- Natural variation of an autophagy-family gene among rice subspecies affects grain size and weight
- Rice gene OsUGT75A regulates seedling emergence under deep-sowing conditions

