Drought-triggered repression of miR166 promotes drought tolerance in soybean
Chen Zho, Jingjing M, Chen Yn, Yu Jing, Yohu Zhng, Yudn Lu, Ye Zhng, Suxin Yng,Xinzhong Feng,*, Jun Yn,*
a School of Life Sciences, East China Normal University, Shanghai 200241, China
b Key Laboratory of Soybean Molecular Design Breeding, Northeast Institute of Geography and Agroecology, Chinese Academy of Sciences, Changchun 130102, Jilin, China
Keywords:Soybean Drought stress miRNA ABA signaling
ABSTRACT Drought stress limits agricultural productivity worldwide.Identifying and characterizing genetic components of drought stress-tolerance networks may improve crop resistance to drought stress.We show that the regulatory module formed by miR166 and its target gene,ATHB14-LIKE,functions in the regulation of drought tolerance in soybean (Glycine max).Drought stress represses the accumulation of miR166, leading to upregulation of its target genes.Optimal knockdown of miR166 in the stable transgenic line GmSTTM166 conferred drought tolerance without affecting yield.Expression of ABA signaling pathway genes was regulated by the miR166-mediated regulatory pathway, and ATHB14-LIKE directly activates some of these genes.There is a feedback regulation between ATHB14-LIKE and MIR166 genes, and ATHB14-LIKE inhibits MIR166 expression.These findings reveal that drought-triggered regulation of the miR166-mediated regulatory pathway increases plants drought resistance, providing new insights into drought stress regulatory network in soybean.
1.Introduction
Drought stress reduces plant growth and crop yield [1].As sessile organisms, plants have evolved mechanisms to adapt to such environmental stress.The abscisic acid (ABA) signaling pathway mediates plant response to drought stress.In ABA signaling, ABA is perceived by PYRABACTIN RESISTANCE1 (PYR1)/PYR1-likes(PYL)/REGULATORY COMPONENT OF ABA RECEPTORs (RCARs)receptors [2,3].This leads to the recruitment of PROTEIN PHOSPHATASE 2C (PP2C) [4] and the formation of a PYR/RCAR-PP2C complex, which in turn releases the repression by PP2C of the activity of SNF1-RELATED PROTEIN KINASE2 (SnRK2) kinases[5–7].The activated SnRK2s can phosphorylate a series of downstream target substrates to trigger the activation of ABA response.Multiple physiological responses can be regulated by SnRK2s via phosphorylation of targets including ion channels, transcription factors, and transporters [8,9].
The highly conserved microRNA166 (miR166) is present in all land plants.microRNAs (miRNAs) are endogenous small noncoding RNA molecules that function in various biological processes in both plants and animals.In plants, miRNAs negatively regulate the expression of their target genes mainly at the posttranscriptional level.In Arabidopsis, miR166 target genes encode Class III HD-ZIP transcription factors and include PHABULOSA(PHB), PHAVOLUTA (PHV), REVOLUTA (REV), ATHB-8, and ATHB-15[10–12].In soybean, ATHB14-LIKE, which is homologous to Arabidopsis PHB, has been validated as the direct target of miR166[13], and is upregulated in miR166 knockdown lines compared to the wild type[14].miR166 is a regulator of multiple developmental processes including organ morphogenesis and pattern formation [15–19].miR166 functions in abiotic stress responses, such as drought tolerance and cadmium resistance [20–23].The miR166-mediated regulatory module is also integrated into hormone networks that include auxin,cytokinin,ABA,and gibberellin[14,20,24,25].
In this study, we showed that the accumulation of mature miR166 is dramatically repressed by drought stress in soybean,which in turn derepresses the expression of miR166 target genes such as ATHB14-LIKE.Knockdown of miR166 to optimal levels in the stable transgenic line GmSTTM166 confers drought tolerance without affecting the yield.Moreover, we found that that the miR166-mediated regulatory pathway regulates the expression of ABA signaling pathway genes.Furthermore, we showed that ATHB14-LIKE negatively regulate the expression of MIR166 genes in soybean.Collectively, these findings reveal that the miR166-mediated regulatory pathway is involved in the regulation of drought tolerance in soybean.
2.Materials and methods
2.1.Plant materials and stress treatments
The soybean cultivar Dongnong 50(DN50)was used as the wild type (WT) control.Soybean plants were grown in a controlledclimate chamber under a 16 h light and 8 h dark photoperiod with 60% relative humidity at 25°C.For comparison of gene expression before and after drought stress treatment, the roots of 10-dayseedlings were washed gently with water and the plants were placed on filter paper to induce drought stress.For drought sensitivity tests, 20-day-old seedlings were grown in separate pots in a 2:1 soil vermiculite mixture with watering every 3 d, subjected to drought by withholding water, and photographed for phenotype evaluation.
2.2.Development of stable soybean transgenic plants and hairy roots
Stable transgenic soybean plants were generated with DN50 as previously described [14,26].Transgenic soybean plants were selected by PAT/bar test paper based on the manufacturer’s instructions (PAT/bar EPSPS LFD Strips, Youlong Biotech, China).To screen GmSTTM166 knockdown lines, RT-qPCR was used to measure the expression levels of STTM structure using primers STTM166-F and STTM166-R.For lines with higher expression of STTM, RT-qPCR was used to measure the abundance of mature miR166.GmSTTM166-2 and GmSTTM166-3 lines, in which miR166 abundance but not yield was reduced were selected for further study.Agrobacterium rhizogenes-mediated transformation was used to produce transgenic soybean hairy roots, as described previously [27].
2.3.Physiological experiments
For measurement of drought-mediated stomatal closure, second trifoliate leaves were cut from 2-week-old plants and exposed to air for 1 h.The mesophyll cells were removed and the guard cells were photographed under a microscope.IMAGEJ software was used to determine the aperture of the stomata[28].For measuring the water loss rate of the detached leaves, second trifoliate leaves were cut from 2-week-old plants, placed on a piece of weighing paper under light in the climate chamber, and photographed and weighed every 0.5 h during 2.5 h periods.They were exposed to drought for 7 d, and 0.2 g of second trifoliate leaves was used to measure superoxide dismutase(SOD),peroxidase(POD),and catalase (CAT) activity and malondialdehyde (MDA), chlorophyll, and free proline content as previously described [29–31].
2.4.RT-qPCR analysis and small RNA gel blot analysis
RT-qPCR measurement of gene expression and of miRNA abundance was performed as previously described [14].The primer sequences are listed in Table S1.Small RNA gel blotting was performed as previously described [14].
2.5.RNA sequencing and processing
Leaves of 3-week-old WT and GmSTTM166 transgenic plants were collected and total RNA was isolated using Trizol reagent(Invitrogen; Thermo Fisher Scientific Inc.) according to the manufacturer’s protocol.RNA sequencing was performed by Oebiotech Company (Shanghai, China).Clean data were mapped to the soybean genome (https://phytozome.jgi.doe.gov/) with TopHat 2.0 software (https://tophat.cbcb.umd.edu/) and analyzed with Cufflinks software (https://cufflinks.cbcb.umd.edu/).
2.6.Plasmid construction
For an miR166 overexpression construct,the miR166 precursor gene MIR166c was amplified and inserted into the vector directly following the cauliflower mosaic virus 35S promoter sequence.The 35S:ATHB14-LIKE-GFP and GST-ATHB14-LIKE plasmid were described previously[14].For dual-luciferase assay,the 2.5-kb promoter regions of genes interested were cloned into pGreen0800II vectors containing Luciferase (LUC) sequence to generate reporter vectors.
2.7.Chromatin immunoprecipitation (ChIP)
ChIP was performed as described previously[32].ATHB14-LIKEGFP transgenic hairy roots were collected and fixated in 1%formaldehyde.Soluble chromatin was subjected to ChIP with anti-GFP (Abcam) or anti-IgG (Abcam) antibodies.Precipitated DNA was purified and used for ChIP–quantitative polymerase chain reaction (ChIP-qPCR).
2.8.Electrophoretic mobility shift assay (EMSA)
The oligonucleotides used in EMSA were synthesized (Shenggong Company, Shanghai, China) and the sense sequence was 3′end-labeled.The recombinant protein GST-ATHB14-LIKE was expressed in Escherichia coli BL21 (DE3).Gel mobility shift (gel shift) assay employed a Light Shift Chemiluminescent EMSA Kit according to the manufacturer’s directions (Thermo).
2.9.Dual-luciferase assay in Nicotiana benthamiana leaves
The reporter construct and the plasmid containing ATHB14-LIKE-GFP were transformed into Agrobacterium tumefaciens strain GV3101 and expressed transiently in N.benthamiana leaves.This assay was performed as previously described [33].
3.Results
3.1.Drought repressed the accumulation of mature miR166 in soybean
Under drought treatment, miR166 accumulation was strongly repressed(Fig.1A,B)and transcript levels of all tested miR166 target genes including ATHB14-LIKE were upregulated(Fig.1C).These observations suggest that drought stress inhibits the expression of miR166, releasing the repression of its target genes.miR166 is derived from more than 20 MIR166 genes in soybean.We next compared their transcript levels before and after drought treatment.The expression of highly abundant MIR166 genes MIR166c and MIR166d was significantly reduced by drought treatment(Fig.1D).This finding suggests that the reduced abundance of mature miR166 in response to drought treatment may be caused by the decreased expression of its precursors.

Fig.1.Expression of soybean miR166, miR166 target genes, MIR166 genes.(A) RT-qPCR measurement of relative abundance of mature miR166 before and after drought treatment.(B)Abundance of mature miR166 in wild-type(WT)plants and T3 GmSTTM166 transformants under drought treatment using RNA gel blotting.U6 served as the internal control.(C)RT-qPCR measurement of miR166 target gene expression under drought treatment.(D)RT-qPCR measurement of MIR166 gene expression under drought treatments.The experiment was conducted with three independent biological replicates and means were compared by Student’s t-test(*,P<0.05;**,P<0.01;***,P<0.001).Bars indicate means ± SD.
3.2.Knockdown of miR166 in soybean conferred drought tolerance
The repression of miR166 by drought stress in soybean led us to investigate whether reducing the abundance of miR166 would increase soybean drought tolerance.Using short tandem target mimic (STTM) technology, we generated and selected two independent GmSTTM166 stable transgenic lines,in which the expression of miR166 was decreased to optimal level without affecting yield (Figs.S1, S2).Both wild-type and GmSTTM166 plants grew well under normal conditions.Under drought conditions, wildtype plants displayed a severely damaged phenotype, whereas the GmSTTM166 plants were healthier and appeared to be less affected by drought stress (Fig.2A, B).
To better understand the function of soybean miR166 for drought tolerance, we compared leaves of wild type and GmSTTM166 plants with respect to water loss rate, a parameter that is correlated with plant drought tolerance.Water loss was slower from GmSTTM166 than from wild-type plants (Fig.2C, D).Under normal conditions, the stomatal apertures of the wild type and GmSTTM166 did not differ (Fig.2E, F).Under drought stress,the stomatal aperture was narrower in GmSTTM166 (Fig.2E, F).The reduced water loss rate in leaves of GmSTTM166 plants could be a consequence of narrower stomatal aperture under drought conditions.
Next, we compared the effects of drought stress on selected physiological parameters in wild type and GmSTTM166 plants.Under normal conditions, there were no significant differences in contents of proline, chlorophyll, and malondialdehyde (MDA)between the wild type and GmSTTM166 (Fig.3A–C).Under drought stress conditions, proline and chlorophyll contents were higher in GmSTTM166 than wild type, but the concentration of MDA was lower in GmSTTM166 than in the wild type (Fig.3A–C).Activities of peroxidase (POD), superoxide dismutase (SOD),and catalase (CAT) did not differ between the wild type and GmSTTM166 under normal conditions (Fig.3D–F), but the activities of all three of these enzymes were higher in GmSTTM166 under drought stress (Fig.3D–F).
3.3.GmSTTM166 displayed altered ABA response
Without ABA treatment, there was no discernible difference in seedling growth between WT and GmSTTM166 (Fig.4A, B).However, when cultivated in ABA solution, seedlings of the GmSTTM166 plants were much shorter than those of WT, indicating that the GmSTTM166 plants are sensitive to ABA in terms of seedling growth(Fig.4A,B).Expression of the ABA-response genes GmRD29A and GmABI5 did not differ between WT and GmSTTM166 plants without ABA treatment.However,after ABA treatment,their expression levels were much higher in GmSTTM166 than those of the WT control (Fig.4C, D).Taken together, our findings suggest that knockdown of miR166 altered soybean plants response to ABA.
3.4.The miR166-mediated regulatory pathway affected the expression of PYLs in soybean
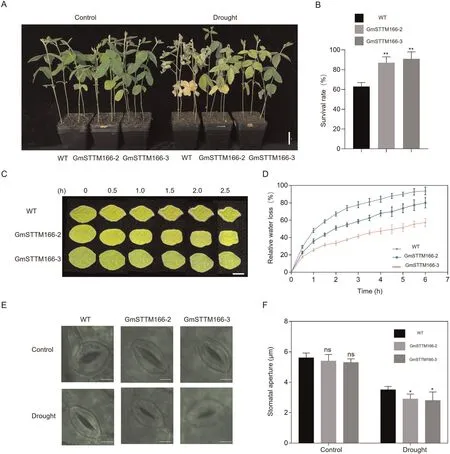
Fig.2.Knockdown of miR166 increased drought tolerance in soybean plants.(A) Phenotypes of WT and GmSTTM166 transgenic plants under drought stress.For drought treatment, 3-week-old GmSTTM166-2, GmSTTM166-3 and wild-type plants were grown in the greenhouse under optimal irrigation, and then exposed to drought by withholding water for 7 d.Drought tolerance was measured four times in more than 20 plants of each line per measurement.Representative images are shown.Scale bar,3 cm.(B) Comparison of the survival rates of WT and GmSTTM166 transgenic plants under drought stress.The experiment was conducted by three independent biological replicates(20 plants per replicate)and mean rates were compared with Student’s t-test(**,P<0.01).Bars indicate means±SD.(C)Water loss phenotypes of detached leaves in WT and GmSTTM166 transgenic plants.Three biological replicates were performed and six individual leaves were used in each replicate.Representative images are shown.Scale bar,2 cm.(D)Relative water loss in WT and GmSTTM166 transgenic plants.The second trifoliate leaves of 2-week-old plants were clipped off and weighed every 0.5 h.Values are mean± SD(n= 3).(E)Drought-promoted stomatal closure phenotype of WT and GmSTTM166 transgenic soybean plants.Leaves were detached from plants and exposed to air for 1 h before being photographed.Scale bars, 20 μm.(F) Comparison of stomatal apertures in WT and GmSTTM166 transgenic plants.The experiment was conducted in three independent biological replicates(20 stomata from one plant per replicate)and means were compared by Student’s t-test(*,P<0.05;ns,no significance).Bars indicate means ± SD.

Fig.3.Comparison of physiological parameters between WT and GmSTTM166.(A) Proline content, (B) chlorophyll content, (C) MDA content in soybean leaves of WT and GmSTTM166 transgenic plants under control and drought treatments.(D)POD activity,(E)SOD activity,(F)CAT activity of soybean leaves in WT and GmSTTM166 transgenic plants under control and drought treatments.Means were compared by Student’s t-test(*,P<0.05;**,P<0.01;***,P<0.001;ns,no significance).Bars indicate means±SD.
The leaf transcriptome profiles of GmSTTM166 and wild-type plants revealed many differentially expressed genes(DEGs)among which were GmPYLs.By RT-qPCR, four GmPYL genes were more highly expressed in GmSTTM166 than in WT plants (Fig.5A).To further confirm this, we generated miR166 overexpression(miR166-OE) transgenic hairy soybean roots by overexpressing MIR166c and analyzed the expression of these GmPYL genes in miR166-OE lines.Compared with empty vector (EV) control lines,the transcript levels of GmPYL genes were significantly reduced relative to empty vector (EV) control lines (Fig.5B).We also generated miR166 target gene overexpression transgenic hairy soybean roots, 35S:ATHB14-LIKE-GFP, and 35S:ATHB14-LIKE G202G-GFP, which is a miR166 cleavage resistant version of ATHB14-LIKE.We compared the expression of these GmPYL genes between EV control lines and 35S:ATHB14-LIKE-GFP or 35S:ATHB14-LIKE G202G-GFP transgenic hairy soybean roots.As expected, Transcript levels of GmPYL genes were upregulated in 35S:ATHB14-LIKE-GFP or 35S:ATHB14-LIKE G202G-GFP transgenic hairy soybean roots (Fig.5C, D).These indicate that the miR166 mediated regulatory pathway regulate the expression of GmPYL genes.
3.5.ATHB14-LIKE directly regulated the expression of PYL genes in soybean
The promoter of Glyma.17G229900 (GmPYL2) contained cisacting elements of ATHB14-LIKE recognition motif (Fig.6A).A chromatin immunoprecipitation (ChIP) qPCR assay was thus performed.The ‘‘IgG” samples were immunoprecipitated with IgG antibody and served as a negative control.Compared with IgG,enrichment of the ChIP signal was detected in 35S:ATHB14-LIKEGFP transgenic hairy soybean roots using GFP antibody,suggesting that ATHB14-LIKE is associated with the Glyma.17G229900(GmPYL2) promoter region (Fig.6A).Electrophoretic mobility shift assay (EMSA) assay further demonstrated that ATHB14-LIKE directly bound to Glyma.17G229900 (GmPYL2) promoter (Fig.6B).ATHB14–LIKE activated the promoter activities of Glyma.17G229900 (GmPYL2) in dual-luciferase assays, suggesting that ATHB14-LIKE could activate the transcription of Glyma.17G229900(GmPYL2) (Fig.6C, D).Taken together, these results suggest that ATHB14-LIKE promotes Glyma.17G229900 (GmPYL2) expression by directly binding to its promoter.
3.6.ATHB14-LIKE negatively regulated the expression of MIR166c and MIR166d
Under drought stress,expression of soybean miR166 was inhibited and the miR166 target gene ATHB14-LIKE was upregulated(Fig.1A–C).These observations suggest that drought stress inhibits miR166 expression and released the repression of its target genes in soybean.Drought treatment reduced the expression of the highly abundant MIR166 genes MIR166c and MIR166d (Fig.1D).This finding suggested that the reduced abundance of mature miR166 in response to drought treatment might be caused by the decreased expression of its precursors.These findings led us to examine whether there is a feedback regulation between
ATHB14-LIKE and MIR166.In 35S:ATHB14-LIKE-GFP transgenic hairy roots, the expression of MIR166c and MIR166d was lower than in the EV control (Figs.7A, S3A).The promoters of MIR166c and MIR166d harbored ATHB14-LIKE binding motifs (Figs.7B, S3B).ATHB14-LIKE was enriched in the promoter regions of MIR166c and MIR166d (Figs.7B, S3B).ATHB14-LIKE bound directly to the promoters of MIR166c and MIR166d(Figs.7C,S3C).Overexpression of ATHB14-LIKE inhibited the promoter activity of MIR166c and MIR166d in N.benthamiana leaves (Figs.7D, S3D).These results indicate that there is a feedback regulation between ATHB14-LIKE and MIR166c and MIR166d in soybean.
4.Discussion
As sessile organism, plants must cope with various environmental stresses such as drought.To withstand drought,plants have evolved sophisticated mechanism.Uncovering the drought response mechanism will facilitate the developing of droughtresistant crops to increase crop productivity.In the current study,we found that miR166 is an essential player in the regulation of drought response in soybean.Under drought stress conditions,expression of soybean miR166 was inhibited,leading to the upregulated expression of its target genes such as ATHB14-LIKE.ATHB14-LIKE can activate the expression of ABA signaling pathway genes.Knockdown the expression of miR166 makes plants more tolerant to drought stress.Thus, drought-triggered repression of miR166 might help plants promote their tolerance to drought stress, and this is achieved at least partially through the induced expression of ABA signaling pathway genes.
A feedback loop between ATHB14-LIKE and MIR166 operates under drought-stress conditions.Drought triggered the repression of MIR166, leading to the decreased expression of mature miR166,in turn causing the released repression of its target gene ATHB14-LIKE.The upregulated ATHB14-LIKE negatively regulates the expression of MIR166, further increasing the expression of ATHB14-LIKE to modulate downstream genes to respond to drought stress.We propose that this feedback regulatory loop helps plants increase their ability to adapt to drought stress.

Fig.5.The expression of ABA signaling pathway genes GmPYLs was modulated by the miR166 mediated regulatory module.(A)RT-qPCR measurement of relative expression of ABA receptor genes GmPYLs in leaves of WT and T3 GmSTTM166 transformants.(B)RT-qPCR measurement of relative expression of ABA receptor genes GmPYLs between empty vector (EV) control and MIR166c overexpression transgenic hairy soybean root.(C) RT-qPCR measurement of relative expression of ABA receptor genes GmPYLs between empty vector(EV)control and 35S:ATHB14-LIKE-GFP transgenic hairy soybean root.(D)RT-qPCR measurement of relative expression of ABA receptor genes GmPYLs between empty vector (EV) control and 35S:ATHB14-LIKEG202G-GFP transgenic hairy soybean root.The experiment was conducted with three independent biological replicates and means were compared by Student’s t-test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).Bars indicate means ± SD.
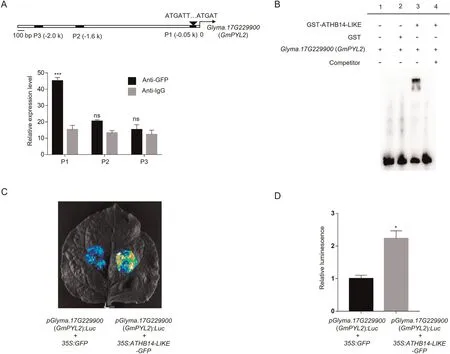
Fig.6.ATHB14-LIKE directly regulates the expression of Glyma.17G229900(GmPYL2).(A)Binding of ATHB14-LIKE to the promoter regions of Glyma.17G229900(GmPYL2)was validated by Chromatin immunoprecipitation(ChIP)qPCR analysis.Binding capacity of ATHB14-LIKE to the promoter regions of Glyma.17G229900(GmPYL2)was determined by ChIP-qPCR analysis with anti-green fluorescent protein (GFP) and IgG.GFP antibody was used to precipitate ATHB14-LIKE.The ‘‘IgG” samples were immunoprecipitated with IgG antibody and served as a negative control.Error bars represent±SD(n =3)and asterisks indicate differences from negative control(***, P<0.001;ns,not significant).(B) EMSA showing that ATHB14-LIKE directly binds to the promoter of Glyma.17G229900 (GmPYL2).Labeled probe of the Glyma.17G229900 (GmPYL2) promoter region was incubated with glutathione S-transferase(GST)-ATHB14-LIKE fusion protein.An unlabeled probe was added for specific competitors.GST protein only was used as a negative control.(C-D) Dual-LUC assay showing that ATHB14-LIKE positively regulated the expression of Glyma.17G229900 (GmPYL2).All values are mean ± SD(n= 3).*, P < 0.05 by Student’s t-test.
In wheat, overexpression of PYL increased drought tolerance,demonstrating a connection between PYL and drought tolerance[34].In soybean, PYL overexpression promoted the expression of stress-responsive genes and conferred higher tolerance to drought stress[35].In the present study,knockdown of miR166 in soybean increased plant resistance to drought stress and the transcript levels of the PYLs.These findings suggest that the increased drought tolerance associated with knockdown of miR166 expression may be attributed at least partially to the miR166 mediated regulation of ABA signaling pathway genes.In addition to the altered the expression of GmPYLs and ABA-responsive genes in miR166 knockdown lines (Figs.5, S4), other factors involved in the regulation of POD and CAT were also regulated (Fig.S5).The drought tolerance phenotype of GmSTTM166 might be attributed to the miR166 mediated regulation of all these factors, not merely the ABA signaling pathway genes.
Previous studies have shown that miR166 functions in drought tolerance in Arabidopsis,rice,and maize[20,23,36].Here,we report that miR166 is also a vital player in the regulation of drought tolerance in soybean.These suggest that miR166 might play a conserved role in plant drought tolerance.Given that the highly conserved miR166 functions in the regulation of plant growth and development, we speculated that the miR166 mediated regulatory pathway might be vital for balancing growth and stress tolerance.
CRediT authorship contribution statement
Chen Zhao:Project administration,Investigation,Visualization,Writing – original draft, Writing – review & editing.Jingjing Ma:Project administration,Investigation,Visualization,Writing–original draft, Writing – review & editing.Chen Yan:Investigation,Visualization.Yu Jiang:Investigation, Visualization.Yaohua Zhang:Investigation,Visualization.Yudan Lu:Investigation,Visualization.Ye Zhang:Investigation, Visualization.Suxin Yang:Supervision, Resources, Funding acquisition.Xianzhong Feng:Supervision,Resources,Writing–review&editing,Funding acquisition.Jun Yan:Supervision,Resources,Writing–review&editing,Funding acquisition.
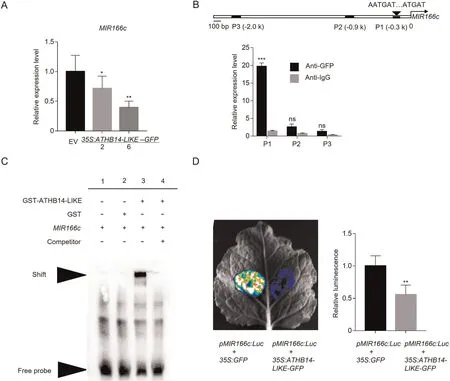
Fig.7.ATHB14-LIKE negatively regulates the expression of MIR166c.(A) RT-qPCR measurement of MIR166c abundance in 35S:ATHB14-LIKE-GFP transgenic hairy roots compared to empty vector(EV)control.Error bars represent±SD(n=3)and asterisks indicate differences from the negative control(*,P<0.05;**,P<0.01).(B,C)Chromatin immunoprecipitation(ChIP)qPCR and EMSA analysis showing that ATHB14-LIKE is associated with the promoter regions of MIR166c.Binding capacity of ATHB14-LIKE to the promoter regions of MIR166c was determined by ChIP-qPCR analysis with anti-green fluorescent protein (GFP) and IgG.GST-tagged ATHB14-LIKE fusion proteins were incubated with the promoter region of MIR166c for EMSA.Values are mean ± SD (n = 3).Means were compared by Student’s t-test (***, P < 0.001; ns, not significant).(D)ATHB14-LIKE activates the promoter activity of MIR166c as revealed from the pMIR166c-LUC activity in tobacco leaf assay.Left panel:fluorescence intensity in tobacco leaf.Right panel: quantitative analysis of LUC activity from tobacco leaf shown at left.Bars indicate means ± SD (n = 3).Student’s t-test: **, P < 0.01.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by grants from the Projects of Science and Technology of Shanghai (18PJ1402800, 20ZR1417900, and 22N11900400),the Strategic Priority Research Program of the Chinese Academy of Sciences(XDA24030303),and Hainan Yazhou Bay Seed Laboratory and China National Seed Group (B23YQ1502).
Appendix A.Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2023.12.005.
- The Crop Journal的其它文章
- Flowering-time regulation by the circadian clock: From Arabidopsis to crops
- Global characterization of OsPIP aquaporins reveals that the H2O2 transporter OsPIP2;6 increases resistance to rice blast
- The OsBSK1-2-MAPK module regulates blast resistance in rice
- Natural variation of an autophagy-family gene among rice subspecies affects grain size and weight
- Rice gene OsUGT75A regulates seedling emergence under deep-sowing conditions
- A telomere-to-telomere genome assembly of Zhonghuang 13,a widely-grown soybean variety from the original center of Glycine max

