Rice gene OsUGT75A regulates seedling emergence under deep-sowing conditions
Jia Zhao, Siyu Liu, Xiaoqian Zhao, Zhibo Huang, Shan Sun, Zixuan Zeng, Yongqi He*, Zhoufei Wang*
The Laboratory of Seed Science and Technology, Guangdong Key Laboratory of Plant Molecular Breeding, National Engineering Research Center of Plant Space Breeding, South China Agricultural University, Guangzhou 510642, Guangdong, China
Keywords:Abscisic acid Deep-sowing Jasmonic acid Rice Shoot length
ABSTRACT Poor seedling emergence is a challenge for direct seeding of rice under deep-sowing field conditions.Here we reveal that UDP-glucosyltransferase OsUGT75A promotes rice seedling emergence under deepsowing conditions by increasing shoot length.Expression of OsUGT75A was higher in the middle regions of the shoot and in shoots under deep-sowing conditions.Levels of free abscisic acid (ABA) and jasmonates (JA) were higher in shoots of OsUGT75A mutants than in those of wild-type plants, and OsUGT75A mutants were more sensitive to ABA and JA treatments.Reduced shoot length was attributed to higher ABA INSENSITIVE 3 (OsABI3) expression and lower JASMONATE-ZIM domain protein (OsJAZ)expression in shoots.Shoot extension by OsUGT75A is achieved mainly by promotion of cell elongation.An elite haplotype of OsUGT75A associated with increased shoot length was identified among indica rice accessions.OsUGT75A acts to increase seedling emergence under deep-sowing conditions.
1.Introduction
Direct seeding is applied in some rice-growing areas because of its low cost and convenience in comparison with conventional rice transplantation[1].But it is subject to poor seedling establishment,weed infestation,and high lodging[2].Deep seeding reduces damage from wildlife and increases lodging tolerance[3].Delayed and poor seedling emergence occurs in most modern rice cultivars,which are sensitive to sowing depth [4].Cultivars with vigorous seedling growth in the form of elongated mesocotyls, coleoptiles,and shoots are desired for increased seedling emergence, particularly under deep-sowing field conditions [5–7].
Several genetic factors affect rice shoot growth.Overexpression of the expansin gene OsEXP4 promotes mesocotyl and coleoptile elongation by wall stress relaxation and cell volumetric extension[8].KNOX genes, which encode homeodomain proteins and are expressed specifically in the shoot apical meristem, are essential for rice shoot development [9].The fatty acid elongase gene ONI2,encoding an enzyme that catalyzes the first step of the elongation of the carbon chain in very-long-chain fatty acids, is required for rice shoot development [10].GY1 inhibits the elongation of mesocotyls and coleoptiles in rice seedlings [11].GSK3(OsGK2) is a gene influencing mesocotyl length in rice, and determines mesocotyl by coordinating strigolactone(SL)and brassinosteroid (BR) signaling [12].However, the regulators of shoot elongation in rice under deep-sowing conditions are not fully known.
Plant hormones regulate rice mesocotyl and coleoptile elongation via cell division or cell elongation.Rice SLs negatively regulate mesocotyl elongation by inhibiting cell division[13],whereas cytokinin(CTK)increases mesocotyl elongation by promoting cell division [14].ABA inhibits rice shoot growth by arresting cell cycle progression by to induce the expression of OsKRP4, OsKRP5, and OsKRP6 [15].Mutations of jasmonate (JA) biosynthesis genes(OsAOS1,OsAOC,and OsJAR1)confer a longer coleoptile phenotype in rice[16–18].Ethylene(ETH)inhibits the expression of rice genes involved in JA biosynthesis,resulting in a reduction in JA level and then in mesocotyl and coleoptile length [11].
Glucosyltransferases catalyze the transformation of activated sugars to plant hormones during plant development and growth[19,20].Our previous study [21] showed that the UDPglucosyltransferase OsUGT75A regulates submergence tolerance in rice.The purpose of the present study was to investigate the influence of OsUGT75A on rice seedling emergence under deepsowing conditions.The induced OsUGT75A expression may promote seedling growth under deep-sowing condition.We hypothesized that OsUGT75A regulation of rice shoot length is mediated by the JA and ABA pathways.

Fig.1.OsUGT75A regulated rice seedling emergence under deep-sowing conditions.(A) Germination percentage after 7 and 10 d under normal condition.Representative images of(B)seedling emergence and(C,G)shoot length after 12 d of germination among OsUGT75A mutants and WT(Nipponbare)under nutritional soil depths of 5 cm and 10 cm in deep-sowing conditions.Scale bars,10 mm.Comparison of shoot,coleoptile,and mesocotyl length between OsUGT75A mutants and WT under(D–F)5 cm and(H–J)10 cm deep-sowing conditions.** indicates a difference from the WT at P < 0.01 by Student’s t-test.ns, not significant.
2.Materials and methods
2.1.Plant materials
Three homozygous OsUGT75A mutants(Osugt75a-1,Osugt75a-2,and Osugt75a-3)were previously generated in the japonica Nipponbare background[21].Mutants of Osabi3[21],Osjaz5,and Osjaz9 in were generated using the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9)system in the japonica Zhonghua 11 background and provided by Hangzhou Baige Biotechnology Co., Ltd., Hangzhou, Zhejiang,China.A set of 124 randomly selected rice accessions from the Rice Diversity Panel 1(https://www.ars.usda.gov/)were used for haplotype analyses (Table S1) [22].A near-isogenic line (NIL) derived from a cross between indica Kasalath and japonica Koshihikari was previously generated [21].Seeds were harvested at maturity in an experimental field at the Zhejiang Academy of Agricultural Sciences(Haining,Zhejiang,China).Well-filled seeds without seed dormancy were selected for experimental use.
2.2.Phenotype evaluation
After imbibing for 36 h, seeds of OsUGT75A mutants and their Nipponbare wild type (WT), and of the Osabi3, Osjaz5, and Osjaz9 mutants and their Zhonghua 11 WT were sown in nutritional soil(Jiffy Group, Zwijndrecht, Netherlands) at depths of 5 and 10 cm and grown for 12 d at 25 °C.Imbibed seeds of the OsUGT75A mutants and WT were grown in field soil at 3 and 5 cm depth for 20 d.Seed germination was considered complete when the shoot or radicle was longer than 2 mm, and germination percentage was then calculated.Seedling establishment was considered when the normal shoot emerged from the soils.Seedling percentage and shoot, coleoptile and mesocotyl lengths were recorded.Post-germinated seeds(with shoot length>1 mm)of WT rice were treated with ABA, JA and auxin (IAA) in various concentrations(0.05 to 10 μmol L-1) and germinated seeds of OsUGT75A mutants and WT were treated with 0.5 μmol L-1ABA and 0.5 μmol L-1IAA for 8 d at 25 °C, after which their shoot lengths were recorded.Water (H2O) was used as the control.The shoot lengths of the 124 randomly selected accessions(Table S1)after sowing in nutritional soil at 5 cm and grown for 9 d at 28°C were recorded.Three biological replicates were tested.
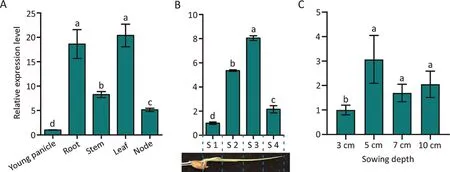
Fig.2.OsUGT75A expression in rice.Expression patterns of OsUGT75A in (A) five tissues and in (B) four regions of the shoot and in (C) the shoot under four deep-sowing depths after 9 d in Nipponbare as determined by quantitative RT-PCR.Expression of OsUGT75A was normalized to that of the OsActin control gene.Different letters indicate differences at P < 0.05 by one-way ANOVA.

Fig.3.Comparison of hormone levels in OsUGT75A mutants and WT(Nipponbare)rice shoots after 7 d of germination.(A)ABA;(B)JA;(C)MeJA;(D)JA-Ile;(E)IAA;(F)Me-IAA.* and ** indicate differences from WT at P < 0.05 and P < 0.01, respectively, by Student’s t-test.ns, not significant.
2.3.Histological observation
Shoots of WT(Nipponbare) rice and the OsUGT75A mutants 9 d after germination were collected and treated with 2.5% (v/v) glutaraldehyde.The outer surfaces of the shoots were observed under a NE610 microscope(Ningbo Yongxin Optics Co.,Ltd.,Ningbo,Zhejiang,China).The cell lengths and widths of the OsUGT75A mutants and WT were recorded.Three biological replications were tested.
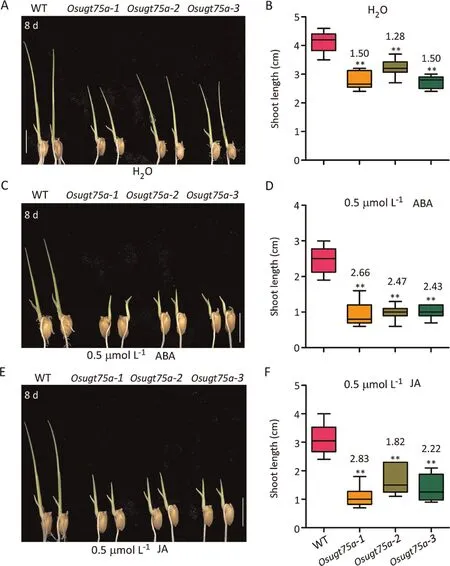
Fig.4.OsUGT75A regulated rice shoot length involving ABA and JA pathway.Representative images of shoot length under(A)H2O,(C)0.5 μmol L-1 ABA,and(E)0.5 μmol L-1 JA treatments after 8 d of OsUGT75A mutants and WT(Nipponbare).Scale bars,10 mm.Comparison of shoot length under(B)H2O,(D)0.5 μmol L-1 ABA,and(F)0.5 μmol L-1 JA treatments after 8 d of OsUGT75A mutants and WT.Numbers indicate the shoot length in WT relative to that of OsUGT75A mutants.**indicates a difference from the WT at P < 0.01 by Student’s t-test.
2.4.Gene expression analysis
Samples of young panicle,stem,node,leaf,and root of Nipponbare rice at the booting stage, as well as of shoots 9 d after germination at four deep-sowing depths (3, 5, 7, and 10 cm) at 25 °C in nutritional soil (Jiffy Group, Zwijndrecht, Netherlands) were harvested.Quantitative reverse transcription-polymerase chain reaction (RT-PCR) for each sample was performed under the following conditions: 95 °C for 2 min followed by 40 cycles of 95 °C for 5 s and 60°C for 10 s.The rice OsActin gene was used as an internal control.The primers used are listed in Table S2.The comparative 2–ΔΔCTmethod [23] was used to normalize the transcript levels.βglucuronidase (GUS) staining assay of tissues was performed as previously described[24].Three biological replicates were used.
2.5.Hormone quantification
Each harvested shoot(50 mg fresh weight)was rapidly frozen in liquid nitrogen, homogenized into powder, and extracted with methanol/water/formic acid(15:4:1,v:v:v).Hormone content was quantified with an LC-ESI-MS/MS system by Wuhan Metware Biotechnology Co.,Ltd.,Wuhan,Hubei,China.The analytical conditions followed Cao et al.[25].Three biological replicates were tested.
2.6.Identification of differentially expressed genes
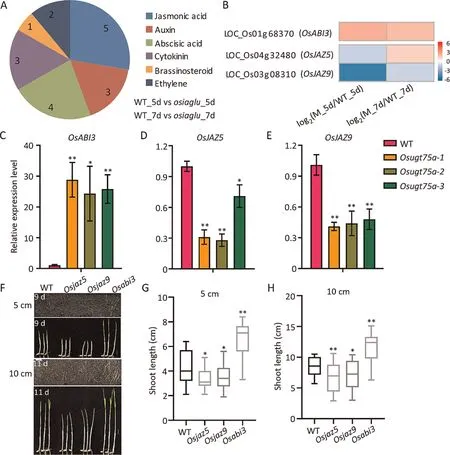
Fig.5.OsUGT75A regulated the expression of ABA and JA related genes in rice shoots.(A)Genes involved in hormone pathways differentially expressed in rice shoots of WT(Nipponbare)and OsUGT75A mutants,identified after both 5 and 7 d of germination.(B)Comparison of OsABI3,OsJAZ5,and OsJAZ9 expression according to RNA-seq.Red,upregulation;Blue,down-regulation.Comparison of(C)OsABI3,(D)OsJAZ5,and(E)OsJAZ9 expression in rice shoot after 5 d of germination determined by quantitative RT-PCR.Gene expression was normalized to that of the OsActin control gene.(F)Representative images of seedling emergence and shoot length under 5-cm deep sowing after 9 d and under 10 cm deep sowing after 11 d in nutritional soil.Scale bars,10 mm.Comparison of shoot length between WT(Zhonghua 11)and Osjaz and Osabi3 mutants under(G)5-cm deep sowing after 9 d and under (H) 10-cm deep sowing after 11 d.* and ** indicate differences from WT at P < 0.05 and P < 0.01, respectively, by Student’s t-test.
Shoots of WT and Osugt75a-2 seeds 5 and 7 d after germination were subjected to RNA sequencing (RNA-seq) by BGI-Wuhan Co.,Ltd.,Wuhan,Hubei,China.Gene expression levels were quantified in FPKM (fragments per kilobase of exon per million) [26].Differentially expressed genes (DEGs) with a Padj(P-adjusted) < 0.001 and a fold change ≥2 were selected for further metabolic pathways (https://mapman.gabipd.org).Three biological replicates were used.
2.7.Haplotype analyses
Haplotypes of OsUGT75A were identified using https://www.ricediversity.org/ [22].Haploview software version 4.2 (Daly Lab at the Broad Institute,Cambridge,USA)was used to assess the linkage disequilibrium (LD).The shoot lengths of 124 randomly selected accessions (Table S1), as described above, were used for the identification of elite haplotype associated with greater shoot length.Haplotypes representing at least 10 accessions were used for comparative analysis of shoot length.
2.8.Data analysis
The experimental data were analyzed using SAS software (SAS Institute, Cary, NC, USA).Significant differences among samples were compared using Student’s t-test or analysis of variance.
3.Results
3.1.OsUGT75A regulated seedling emergence under deep-sowing conditions

Fig.6.OsUGT75A altered shoot cell elongation in rice.(A)Representative images of shoot cells after 9 d of germination among WT(Nipponbare)and OsUGT75A mutants.Scale bars,20 μm.Comparison of shoot(B)cell length and(C)cell width between WT and OsUGT75A mutants after 9 d of germination.(d)Expansin genes differentially expressed in shoots of WT(Nipponbare)and OsUGT75A mutants identified after 5 and after 7 d of germination.Red,up-regulation;Blue,down-regulation.(E–H)Comparison of expansin expression in shoots between WT and OsUGT75A mutants after 5 d of germination determined by quantitative RT-PCR.Gene expression is normalized to that of the OsActin control gene.* and ** indicate differences from WT at the P < 0.05 and P < 0.01, respectively, by Student’s t-test.ns, not significant.
The germination rates (germination percentage after 7 and 10 d) did not differ between the wild-type (WT) Nipponbare and OsUGT75A mutants under normal condition (Fig.1A).However,the disruption of OsUGT75A reduced seedling emergence under deep-sowing (5 and 10 cm) conditions.Only a few seedlings (respectively 24% and 13% for 5- and 10-cm deep sowing) of the OsUGT75A mutant emerged from the soil after 12 d, whereas approximately half of WT seedlings emerged (Fig.1B).The shoots of the OsUGT75A mutant were significantly shorter than those of WT under deep-sowing conditions, but coleoptile and mesocotyl lengths did not differ(Fig.1C–J).Generally,the seedling emergence and seedling length of OsUGT75A mutants were significantly lower than those of WT under deep-sowing conditions after 20 days(Fig.S1).Similarly in field soil, seedling emergence and seedling length, but not coleoptile and mesocotyl length, of OsUGT75A mutants were significantly reduced under 3 and 5 cm-depth sowing conditions after 20 d (Fig.S2).
3.2.Expression pattern of OsUGT75A
Expression of OsUGT75A was higher in root and leaf than in young panicle, stem, or node at the booting stage (Fig.2A) and higher in the S2 and S3 than in the S1 and S4 regions of the shoot(Fig.2B).OsUGT75A expression in shoots was significantly higher under deep sowing (5 to 10 cm) than under shallow sowing(3 cm) (Fig.2C).
3.3.OsUGT75A regulated shoot length via JA and ABA pathways
Significantly higher levels of free ABA, JA, methyl jasmonate(MeJA), and jasmonic acid-isoleucine (JA-Ile) were observed in OsUGT75A mutants than in WT plants,whereas there was no difference in indole-3-acetic acid (IAA) or methyl-indole-3-acetic acid(Me-IAA) (Fig.3).The shoot length of WT plants was significantly reduced by JA and ABA(>0.5 μmol L-1)but not by IAA treatments,relative to the control (H2O) (Fig.S3).OsUGT75A mutant plants showed a more JA-and ABA-sensitive phenotype.The relative suppressions of shoot length (ratio of WT/mutant) were significantly higher under ABA and JA treatments than under the control condition (Fig.4).
3.4.OsUGT75A regulated shoot length via JA and ABA pathways
Respectively 1972 and 1404 DEGs (fold change > 2) were identified in shoots of Osugt75a-2 and WT after 5 and 7 d of germination, and 437 DEGs were identified at both stages (Fig.S4;Table S3).JA and ABA related DEGs, including OsABI3, OsJAZ5, and OsJAZ9, were identified at both stages (Fig.5A–B).Transcripts of OsABI3 were significantly increased in OsUGT75A mutants relative to those of WT plants, whereas OsJAZ5 and OsJAZ9 expression significantly decreased during seed germination (Fig.5C–E).The disruption of OsJAZ5 and OsJAZ9 (Fig.S5) reduced seedling emergence under deep sowing in soils, whereas the disruption of OsABI3 increased seedling emergence (Fig.5F).The shoots of the OsJAZ5 and OsJAZ9 mutants were significantly shorter than those of WT under deep-sowing conditions,whereas those of the OsABI3 mutant were longer (Fig.5G–H).
3.5.OsUGT75A regulated shoot length via cell elongation
Cells of the OsUGT75A mutants were significantly shorter than those of WT(Nipponbare)plants,whereas cell width did not differ(Fig.6A–C).Four expansin genes (LOC_Os04g15840,LOC_Os04g46630,LOC_Os09g29690,and LOC_Os09g29710)were differentially expressed in the OsUGT75A mutant and WT plants in shoots at both post-germination stages(Fig.6D).Expression levels of three genes (LOC_Os04g46630, LOC_Os09g29690, and LOC_Os09g29710) were significantly lower in shoots of OsUGT75A mutants than in those of WT plants after 5 d of germination(Fig.6E–H).Thus, OsUGT75A promoted shoot length mainly by increasing expansin expression leading to cell elongation in rice.
3.6.A natural allelic variation of OsUGT75A contributed to shoot length

Fig.7.Haplotype of OsUGT75A associated with shoot length under deep-sowing conditions in rice.(A) Comparison of shoot length between indica and japonica accessions under 5-cm deep-sowing conditions after 9 d in nutritional soil.(B)Haplotypes of OsUGT75A identified in ~2 kb up/downstream and coding regions of the gene.The red box represents an exon.(C) Functional SNP of OsUGT75A and linkage analysis of SNPs.As a measure of linkage disequilibrium, the color intensity of the box corresponds to the magnitude of r2,as shown in the legend.(D)Shoot length of accessions of two different haplotypes under 5-cm deep-sowing conditions after 9 d.(E)Representative images of shoot length in accessions of two different haplotypes under 10-cm deep-sowing conditions after 12 d in nutritional soil.Bars,10 mm.(F)Representative images of seedling emergence and shoot length among indica Kasalath (with the Hap 2 allele), japonica Koshihikari (with the Hap 1 allele) and NIL (with the Hap 2 allele) in Koshihikari background under 5-cm deep-sowing condition after 9 d in nutritional soil.Scale bars, 10 mm.(G) Comparison of shoot length among Kasalath, Koshihikari and NIL of germination under 5 cm deep-sowing condition after 9 d in nutritional soil.* and ** indicate differences at P < 0.05 and P < 0.01, respectively, by Student’s t-test.
Shoots of indica were longer than those of japonica accessions(Fig.7A) and two major OsUGT75A haplotypes (Hap 1 and Hap 2)were identified according to six SNPs(Fig.7B).In which,two SNPs(SNP1 and SNP2)were closely linked(Fig.7C).SNP2,located in the coding region of OsUGT75A,causes a single-nucleotide substitution(A to G),converting a tryptophan codon(CCA)to an arginine codon(CCG).The elite Hap 2 associated with greater shoot length was present mainly in indica accessions, whereas Hap 1 was present mainly in japonica accessions (Fig.7D).Several elite indica accessions carrying Hap 2 displayed longer shoots under deep sowing(10 cm) than did japonica accessions carrying Hap 1 (Fig.7E).Under deep sowing (5 cm), NIL plants carrying the transferred Hap2 (Kasalath) in the Koshihikari background displayed longer shoots than did Koshihikari(Fig.7F–G).The variations of OsUGT75A seemed to be associated with the variation of seedling emergence under deep-sowing conditions.
4.Discussion
Mesocotyl and coleoptile elongation facilitates rice seedling establishment from deeper soil layers [27,28,29].In the present study, shoot length, as regulated by OsUGT75A, was primarily responsible for seedling emergence, but not by influencing the elongation of the rice mesocotyl and coleoptile under deepsowing conditions.Why OsUGT75A promoted emergence by influencing seedling length (presumably via leaf sheath elongation)rather than mesocotyl and coleoptile length remains unclear.Because seedling growth is also regulated by environmental factors such as temperature,a future study could investigate the influence of high (35 °C) or low (15 °C) temperatures on OsUGT75A regulation of shoot, coleoptile, and mesocotyl length.
Our data showed that shoot length regulated by OsUGT75A was mainly involved in ABA and JA signaling pathways.In this study,increases in free ABA and JA were observed in the shoots of OsUGT75A rice mutants under deep-sowing conditions.Because it has been reported [11,15] that ABA and JA inhibit shoot growth,and mesocotyl and coleoptile elongation in rice, we hypothesize that increased accumulation of free ABA and JA accounts for the reduced shoot length of the OsUGT75A mutants.JAZ proteins have been reported[30]to interact with ABI5 modulating seed germination in bread wheat and Arabidopsis.Expression of OsABI3 is negatively associated with germination speed and shoot growth in rice[31].We speculate that the lower OsJAZ5 and OsJAZ9 expression and higher OsABI3 expression in the shoots of the OsUGT75A mutants account for their reduced shoot length.Expression of cell-wall-loosening expansin genes is induced by gibberellic acid(GA) [32] but suppressed by ABA [33] involving seed germination in Arabidopsis.Expansin gene expression is inhibited by JA, reducing mesocotyl and coleoptile elongation in etiolated rice seedlings[11].Because in the present study,the expression of three expansin genes was lower in shoots of OsUGT75A mutants than in those of the WT, we propose that OsUGT75A regulation of shoot length involves ABA- and JA-mediated expression of expansin genes.
In the present study, shoot length varies across rice genotypes and shoots of indica were longer than those of japonica accessions.To determine whether the natural variations of OsUGT75A contribute to shoot length, the allelic diversity of OsUGT75A was analyzed in this study.The elite haplotype Hap 2,which was positively associated with shoot length,was present mainly in indica and not in japonica accessions.Submergence also often occurs in rice direct seeding.In our previous study [21], the favorable haplotype of OsUGT75A for submergence tolerance during seed germination was from japonica.We cannot find a haplotype in the rice genetic resources that increases tolerance of both submergence and deep sowing.The genetic relationships between natural variation of shoot and coleoptile length with tolerance to submergence and deep sowing conditions await further investigation.Several elite indica such as Peh-Kuh-Tsao-Tu (shoot and coleoptile length,13.39 and 2.19 cm), DM 43 (16.03 and 2.09 cm), Santhi Sufaid(16.36 and 2.17 cm)displayed longer shoots and coleoptiles under both deep sowing and submergence conditions [21].These accessions might be useful resources for improving rice seedling emergence under these conditions.
In conclusion, OsUGT75A regulates rice seedling emergence under deep-sowing conditions.Shoot length regulation by OsUGT75A may involve the ABA and JA pathways.Cell elongation,also regulated by OsUGT75A, is the primary contributor to shoot length in rice.Other factors regulating OsUGT75A expression in rice shoots,as well as the mechanism by which OsUGT75A acts through the ABA and JA signaling pathways, await further investigation.
CRediT authorship contribution statement
Jia Zhao:Investigation, Writing – original draft.Siyu Liu:Data curation, Formal analysis.Xiaoqian Zhao:Data curation.Zhibo Huang:Resources, Software.Shan Sun:Methodology, Validation.Zixuan Zeng:Resources.Yongqi He:Data curation,Writing–original draft.Zhoufei Wang:Conceptualization, Funding acquisition,Supervision, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors thank USDA-ARS for seeds of the Rice Diversity Panel.This work was supported by the Key-Area Research and Development Program of Guangdong Province(2022B0202060006), the Natural Science Foundation of Guangdong Province (2023A1515012052, 2023A1515012092), the Science and Technology Project of Guangzhou (2023A04J0749,2023A04J1452), the Special Fund for Student Cultivation of Scientific and Technological Innovation of Guangdong Province(pdjh2021b0084),and the Double First-Class Discipline Promotion Project of South China Agricultural University (2021B10564001).
Appendix A.Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2023.10.010.
- The Crop Journal的其它文章
- Flowering-time regulation by the circadian clock: From Arabidopsis to crops
- Global characterization of OsPIP aquaporins reveals that the H2O2 transporter OsPIP2;6 increases resistance to rice blast
- Drought-triggered repression of miR166 promotes drought tolerance in soybean
- The OsBSK1-2-MAPK module regulates blast resistance in rice
- Natural variation of an autophagy-family gene among rice subspecies affects grain size and weight
- A telomere-to-telomere genome assembly of Zhonghuang 13,a widely-grown soybean variety from the original center of Glycine max

