Global characterization of OsPIP aquaporins reveals that the H2O2 transporter OsPIP2;6 increases resistance to rice blast
Gousi Li, Jinglun Hn, Chen Yi, Ho Luo,b, Yuzhu Wng,b, Fengpin Wng, Xioyu Wng,Letin Chen,b,*, Yling Zhng,b,c,*
a State Key Laboratory for Conservation and Utilization of Subtropical Agro-Bioresources, College of Life Sciences, South China Agricultural University, Guangzhou 510642,Guangdong, China
b Guangdong Laboratory for Lingnan Modern Agriculture, Guangzhou 510642, Guangdong, China
c National Key Laboratory of Plant Molecular Genetics, Shanghai Center for Plant Stress Biology, CAS Center for Excellence in Molecular Plant Sciences, Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai 200032, China
Keywords:Aquaporin Plant immunity Rice blast H2O2 transport
ABSTRACT Plasma membrane intrinsic proteins (PIPs) are conserved plant aquaporins that transport small molecules across the plasma membrane to trigger instant stress responses and maintain cellular homeostasis under biotic and abiotic stress.To elucidate their roles in plant immunity to pathogen attack,we characterized the expression patterns, subcellular localizations, and H2O2-transport ability of 11 OsPIPs in rice(Oryza sativa), and identified OsPIP2;6 as necessary for rice disease resistance.OsPIP2;6 resides on the plasma membrane and facilitates cytoplasmic import of the immune signaling molecule H2O2.Knockout of OsPIP2;6 increases rice susceptibility to Magnaporthe oryzae, indicating a positive function in plant immunity.OsPIP2;6 interacts with OsPIP2;2,which has been reported to increase rice resistance to pathogens via H2O2 transport.Our findings suggest that OsPIP2;6 cooperates with OsPIP2;2 as a defense signal transporter complex during plant–pathogen interaction.
1.Introduction
Aquaporins (AQPs) are tetrameric channel proteins that facilitate the transport of water,gases,nutrients,and other small molecules across biological membranes in almost all organisms [1].Plants, as sessile organisms, often possess more AQPs than other organisms, allowing cellular homeostasis to be maintained in the face of environmental stress[2].AQPs are found in nearly all plant organs and participate in an array of activities during vegetative and reproductive development [1].
Plant AQPs are typically divided into five subfamilies according to sequence similarity and subcellular localization: plasma membrane intrinsic proteins (PIPs); tonoplast intrinsic proteins (TIPs);nodulin-26 like intrinsic proteins (NIPs); small basic intrinsic proteins(SIPs);and uncharacterized X intrinsic proteins(XIPs),which are missing in monocots[3].PIPs are further divided into the PIP1 and PIP2 subgroups, which differ in N- and C-terminal sequence lengths and water transport capacities [4].Much research [3,4]on the function of PIPs in plant adaptation to multiple abiotic stresses, including drought, high salinity, and extreme temperatures,has been conducted.However,little is known about the role of PIPs in plant immune response to pathogen attack.
Recent research[5,6]suggests that PIPs are necessary for pathogen defense in plants.In Arabidopsis,both AtPIP1;4 and TaPIP1 positively regulate defense response, with AtPIP1;4-triggered resistance relying on H2O2transport [7,8].AtPIP2;1-facilitated H2O2import is required for pathogen-triggered stomatal closure[9].Cellular uptake of H2O2mediated by OsPIP2;2, TaPIP2;10,and NbPIP2;2 confers resistance to fungal and/or bacterial diseases in rice[10],wheat[11],and tobacco[12].Thus PIP-mediated H2O2transport appears to function in plant immune response.
Of 11 OsPIPs identified in rice to date [13], only OsPIP2;2 has been confirmed [10] to function in pathogen defense.The roles of the other OsPIPs remain largely unknown.The purpose of this study was to functionally characterize each of the 11 OsPIPs and clarify their roles in plant immunity by expression and subcellular localization analysis,H2O2-transport ability test,and fungal inoculation of OsPIP-knockout lines.
2.Materials and methods
2.1.Plant materials and pathogen strains
OsPIP knockout lines were generated from japonica rice cultivar Nipponbare and Tohhoku IL-6.The Magnaporthe oryzae(M.oryzae)strain 08-T13 was used for pathogenicity testing.
2.2.Heat map and real time quantitative PCR (RT-qPCR) analysis of gene expression
A heat map of OsPIP expression was created based on gene microarray data obtained from the RiceXPro database (https://ricexpro.dna.affrc.go.jp/).
For RT-qPCR analysis, total RNA was extracted from roots,stems,leaves,and seeds of rice cultivar Nipponbare.A commercial kit (Promega (Beijing) Biotech Co., Ltd., M1701) was used to synthesize cDNA.RT-qPCR assays were performed with iQ SYBR Green Supermix (Bio-Rad Laboratories (Shanghai) Co., Ltd., China) using primers listed in Table S1.Relative expression of OsPIP genes was normalized to OsActin.
2.3.Subcellular localization of proteins and bimolecular fluorescence complementation (BiFC) assays
The full-length cDNAs of OsPIPs:GFP (for subcellular localization) and OsPIPs:Vn/Vc (for BiFC) driven by the 35S promoter were transiently expressed in rice protoplasts by PEG/Ca2+-mediated transfection.PM(OsRac3-mCherry)and ER(mCherry-HEDL)markers carrying mCherry were co-expressed with OsPIPs:GFP for subcellular localization observation of OsPIPs [14].Transfected rice protoplasts were incubated at 28 °C for 12–16 h in the dark and then observed under a confocal microscope.
2.4.H2O2 transport assays
Full-length OsPIP cDNAs were individually cloned into the pYES2 vector for expression in yeast strain BY4742(MATα,his3Δ1,leu2Δ0, lys2Δ0, ura3Δ0).For the yeast growth assay, yeast transformants were diluted to specific OD600values (0.7, 0.07, and 0.007) and spotted uniformly on SD-Ura + Gal solid medium containing either 0 or 1.5 mmol L-1H2O2.The viability of yeast transformants was recorded after 3 d of incubation at 30 °C.To create the growth curves, yeast transformants were adjusted to OD600=0.07 and cultured in SD-Ura+Gal liquid medium containing either 0 or 1.5 mmol L-1H2O2at 30°C with 200 r min-1shaking.OD600was recorded every 4 or 8 h.
For detection of H2O2uptake,yeast transformants grown in SDUra+Gal liquid medium for 15–18 h were collected by centrifugation and resuspended in 0.2 mmol L-1PBS buffer to OD600= 0.7.The yeast transformants were incubated with 10 μmol L-1H2O2-probing dye (Amplex Red and CM-H2DCFDA, separately) for 30 min, after which the yeast cells were treated with either 0 or 1.5 mmol L-1H2O2.Fluorescence was measured every 5 min using a microplate reader.
2.5.M.Oryzae inoculation
The fungal strain activated by potato dextrose agar (PDA) was cultured on oatmeal agar for spore production (26 °C, 12 h dark/12 h blue-light,2–3 d).Spores were prepared as a suspension(2×105spores mL-1with 0.02%Tween 20)for rice inoculation.Leaf spray inoculation was performed as described previously[15],and inoculated leaves were photoed 7 d post-inoculation.For leafpunch inoculation, rice plants were grown for 5–7 weeks and the young leaves of each plant were punched and inoculated with a piece of M.oryzae spore colony.The lesion area was measured 12–15 d post-inoculation.
2.6.Yeast two-hybrid (Y2H) assay
Full-length OsPIP cDNAs were subcloned into pGADT7/pGBKT7 vectors for co-expression in yeast strain Y2H Gold.Yeast transformants were diluted to OD600= 1 and spotted equally on SD/-Leu-Trp and SD/-Ade-His-Leu-Trp solid media.Yeast transformants spotted on solid media were photoed after 3–5 d.
2.7.Co-immunoprecipitation (Co-IP) assay
Full-length OsPIP2;2 and OsPIP2;6 were fused with HA and GFP tags, respectively.HA-OsPIP2;2 and OsPIP2;6-GFP were coexpressed in rice protoplasts.Co-IP assays were performed with a commercial kit(Beyotime Biotechnology,Shanghai,P2185)using lysates of protoplasts prepared with lysis buffer in the kit.Protein samples precipitated with IgG beads were used as a negative control.
2.8.Sequence alignment and modeling of protein complex
Multiple alignment of OsPIPs was performed with ClustalW(https://www.clustal.org/clustal2/) and GeneDoc (https://nrbsc.org/gfx/genedoc).ColabFold (https://github.com/sokrypton/Colab-Fold) was used to model the structure of the OsPIP2;6-OsPIP2;1 complex.The highest-ranked model was used to visualize the interface between OsPIP2;6 and OsPIP2;1 with PDBePISA(Proteins,Interfaces,Structures and Assemblies,https://www.ebi.ac.uk/msdsrv/prot_int/).The predicted model was visualized using Swiss–PdbViewer (https://spdbv.unil.ch/).
3.Results
3.1.Expression profiles of OsPIPs
To compare the tissue-specific expression of the 11 previouslyidentified OsPIP genes,we created an expression heatmap of these genes and found that OsPIP1;1, OsPIP1;2, OsPIP2;1, and OsPIP2;6 were highly and constitutively expressed in rice(Fig.1A).OsPIP1;3,OsPIP2;2, and OsPIP2;7 were expressed preferentially in roots,stems, and/or leaves (Fig.1A), suggesting that these genes are active primarily during vegetative growth.OsPIP2;3, OsPIP2;5, and OsPIP2;8 were expressed at relatively low levels(Fig.1A),suggesting that either they serve redundant functions or their transcription is induced only under specific conditions.The expression of these genes in roots, stems, leaves, and seeds detected by RTqPCR was consistent with the microarray data (Fig.1B).
3.2.Subcellular localization of OsPIPs
To explore the possible function of each OsPIP,we observed the subcellular localization of OsPIPs in rice protoplasts.OsPIP1s were localized specifically to the endoplasmic reticulum (ER), whereas OsPIP2s were localized to both the ER and the plasma membrane(PM) (Fig.2A, B).Multiple alignment of OsPIPs suggested that the differences in subcellular localization between OsPIP1s and OsPIP2s are due to variation in their N-terminal structures(Fig.S1).Because localization to the PM is usually required for appropriate function of aquaporin,we further co-expressed OsPIP1 and OsPIP2 in rice protoplasts.The localization pattern of OsPIP1 changed from the ER to the PM when it was co-expressed with OsPIP2 (Fig.2C), suggesting that PIP1 forms a complex with PIP2 for proper localization to the PM.
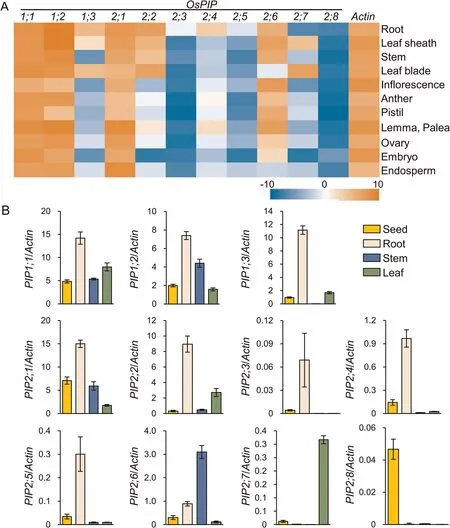
Fig.1.Tissue-specific OsPIP expression.(A)Heat map of OsPIP expression in various tissues based on raw signal intensity from RiceXpro.Red and blue colors represent high and low expression, respectively.(B) OsPIP expression in rice roots, stems, leaves, and seeds detected by RT-qPCR.OsActin (Os03g0718100) was used as a reference gene.
3.3.OsPIP2;6 transports H2O2 in yeast
To test whether the OsPIPs could transport H2O2, each OsPIP was individually expressed in yeast cultured with or without H2O2.The growth of yeast transformants expressing OsPIP2;2,OsPIP2;3,OsPIP2;4,OsPIP2;5,or OsPIP2;6 was inhibited to various degrees by exogenous application of H2O2.Notably,yeast expressing OsPIP2;6 showed severe growth inhibition, indicating the strongest activity of OsPIP2;6 to transport H2O2(Fig.3A).Examination of the growth curves of these yeast transformants with or without H2O2showed similar growth inhibition, with transformants expressing OsPIP2;7 and OsPIP2;8 also identified as transporting H2O2, albeit with lower activity (Fig.3B).
To confirm the ability of OsPIP2;6 to transport H2O2, we detected real-time changes in intracellular H2O2levels in yeast transformants after H2O2treatment.Increasing intracellular H2O2levels were detected in each yeast transformant treated with H2O2,with greater H2O2accumulation in OsPIP2;6-expressing yeast than in yeast carrying the empty vector (Fig.4).These results showed that OsPIP2;6 can efficiently mediate the transport of extracellular H2O2into the cytoplasm across the PM,implying that OsPIP2;6 may contribute to pathogen response via H2O2signal transduction.
3.4.Knockout of OsPIP2;6 increases rice susceptibility to Magnaporthe oryzae
To verify our guess, we knocked out OsPIP2;6 in japonica rice cultivar Nipponbare and Tohhoku IL-6, using a CRISPR/Cas9 genome editing system [16] and the associated bioinformatic toolkit CRISPR-GE [17].Several edited lines with different mutations in OsPIP2;6 coding region were generated and two OsPIP2;6 knockout lines were chosen for further study (Figs.5, S2).To test whether OsPIP2;6 functions in pathogen response, we inoculated OsPIP2;6-KO and wild-type(WT) plants with the M.oryzae.OsPIP2;6 knockout increased rice susceptibility to M.oryzae at both the seedling and jointing stages (Figs.5, S2), indicating that OsPIP2;6 functions in defense against rice blast.
3.5.OsPIP2;6 may form a complex with OsPIP2;2 for H2O2 transport and pathogen resistance

Fig.2.Subcellular localization of OsPIPs.(A)GFP signals from OsPIP-GFPs and mCherry signals from the endoplasmic reticulum(ER)marker(mCherry-HEDL)overlapping in rice protoplasts.(B)GFP signals from OsPIP2-GFPs,but not OsPIP1-GFPs,overlapped with mCherry signals from the plasma membrane(PM)marker(OsRac3-mCherry)in rice protoplast.(C) GFP signals from OsPIP1;1-GFP overlapped with mCherry signals on the PM, when co-expressed with OsPIP2;1-mCherry.Scale bars, 10 μm.
Recently,OsPIP2;2 was found to increase rice resistance to fungal and bacterial pathogens, including M.oryzae, by transporting pathogen-induced H2O2[10].In this study, OsPIP2;6 also contributed to rice innate immunity by way of H2O2signal transduction.Because according to previous studies [2], aquaporins function primarily as tetramers, we hypothesized that OsPIP2;6 and OsPIP2;2 cooperate to import apoplastic H2O2as a heterotetramer.To test this hypothesis, we tested for interaction among OsPIP2;2,OsPIP2;6, and other OsPIPs.Interestingly, we found that OsPIP2;2 interacted only with OsPIP2;6 and OsPIP1s, while OsPIP2;6 was capable of forming a complex with each of the 11 OsPIPs,including itself (Fig.6A).It was predicted that two amino acid residues(Ile141,Gln145)in OsPIP2;6 contributed to the interaction specificity among OsPIPs(Figs.S1,S3).The interaction between OsPIP2;6 and OsPIP2;2 was further confirmed by BiFC and Co-IP assays in rice protoplasts (Fig.6B, C).The interactions between OsPIP2;6 itself and other OsPIPs were confirmed by BiFC (Fig.S4).
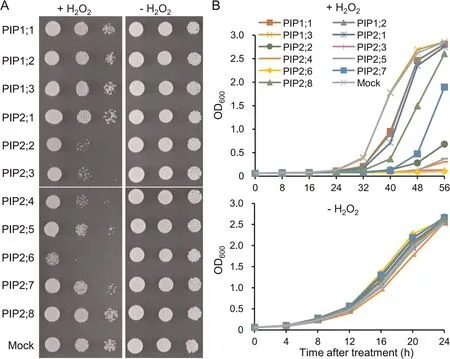
Fig.3.H2O2-transport ability of OsPIPs in yeast.(A)Growth phenotype of yeasts expressing OsPIPs cultured on solid medium with or without H2O2.(B)Growth curve of yeasts expressing OsPIPs cultured in liquid medium with or without H2O2.Values are mean±standard deviation(SD)of three replicates.Yeast transformed with the empty pYES2 plasmid was used as the control.
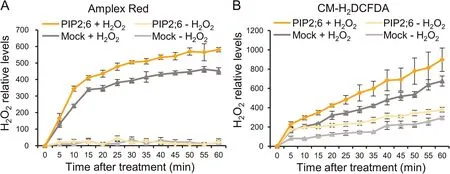
Fig.4.OsPIP2;6 imports extracellular H2O2 into yeast cells.Relative H2O2 levels in yeast after H2O2 treatment(+H2O2)or ddH2O treatment(-H2O2),as quantified by H2O2-probing dyes Amplex Red (A) and CM-H2DCFDA (B), respectively.Values are mean ± SD of three replicates.
Although four OsPIP2s (OsPIP2;3, OsPIP2;4, OsPIP2;5, and OsPIP2;7) transported H2O2and also interacted with OsPIP2;6,knocking out these OsPIP2s did not affect rice resistance against M.oryzae (Fig.S5).These results not only support the cooperative relationship between OsPIP2;6 and OsPIP2;2, but suggest that OsPIP2;6 performs multiple functions by forming homotetramers or heterotetramers with various OsPIPs during rice development.
We propose that OsPIP2;6 acts in a coordinated fashion with OsPIP2;2 as a H2O2transporter to support rice immune response to pathogen attack (Fig.7).
4.Discussion

Fig.5.OsPIP2;6 increases rice resistance against M.oryzae.(A) Schematic diagram and sequencing of the CRISPR/Cas9-editing target sites in the OsPIP2;6-KO line.WT,reference sequence of wild-type plants.Black dots represent the abridged sequence between two targets.Red dashed lines indicate the sequence deleted by gene editing.(B)Leaf phenotypes of OsPIP2;6-KO and wild type (WT) seedings were recorded 7 d post-spray inoculation with M.oryzae.(C) Phenotypes of OsPIP2;6-KO and WT plants were recorded 12 d post punch inoculation with M.oryzae.(D)Average lesion areas of OsPIP2;6-KO and WT plants were measured.Values are mean±standard error(SE)of at least three replicates.Bars with different letters indicate statistically significant differences according to Student’s t-test (P < 0.05).
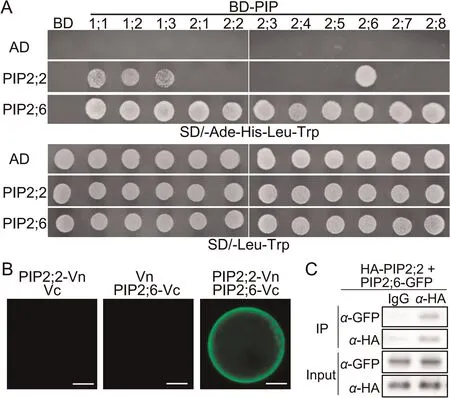
Fig.6.OsPIP2;6 interacts with OsPIP2;2.(A) AD-OsPIP2;2 or AD-OsPIP2;6 was coexpressed with BD-OsPIPs in yeast grown on SD/-Ade-His-Leu-Trp and SD/-Leu-Trp media,respectively.The interactions were observed after 3 days of culture.(B)The interaction between OsPIP2;2-Vn and OsPIP2;6-Vc was verified by BiFC assay in rice protoplast.Scale bars, 10 μm.(C) In vivo interaction between HA-OsPIP2;2 and OsPIP2;6-GFP was confirmed by Co-IP assay in rice protoplasts.Lysates of protoplasts expressing HA-OsPIP2;2 and OsPIP2;6-GFP were incubated with IgG and anti-HA(α-HA)beads,respectively,and the OsPIP complex was detected using anti-GFP (α-GFP) antibody.
H2O2,a relatively stable reactive oxygen species,is produced in response to early pathogen perception and serves as an immune signal in plants [18].Although H2O2can cross the PM by passive diffusion,more efficient H2O2transport is required for rapid induction of the defense response.AQPs have been reported [19,20] to mediate transmembrane H2O2transport in different organisms,making these proteins ideal transporters for H2O2signaling.Indeed, the ability of several AQPs to transport H2O2, including AtPIP2;1, OsPIP2;2, TaPIP2;10, and NbPIP2;2, has been demonstrated and linked to plant defense against diverse pathogens [9–12].The findings that OsPIP2;6 transports H2O2and increases rice immunity to rice blast supports the notion that PIPs act as a bridge between extracellular H2O2production and intracellular H2O2signaling in plants.We suggest that further studies be performed to verify whether other H2O2-transporting PIPs, such as ZmPIP2;5[21], also contribute to pathogen defense.
Our conclusion of PIPs subcellular localization was based on known organelle markers.We used mCherry fused ER retention signal peptide HEDL (mCherry-HEDL) and plasma membrane anchored protein OsRac3 (OsRac3-mCherry) [14] as markers to determine the ER and/or PM localization of PIPs (Fig.2).However,this method failed to distinguish the precise organelle structures.Electron microscopy or high-resolution microscopy may provide more convincing evidence for the subcellular localization of specific proteins in vivo.
With the exception of OsPIP2;1,all OsPIP2s have been shown to mediate H2O2import in rice(Fig.3),likely because of their homology [13,22].However, the OsPIP2s differ in their transport efficiency (Fig.3).According to our results and the previous study[10], both OsPIP2;2 and OsPIP2;6 were capable of H2O2transport and were highly expressed in leaves, especially after pathogen attack or H2O2treatment, suggesting a function as inducers of defense against multiple pathogens (Figs.1, 3).Indeed, both OsPIP2;2 and OsPIP2;6 have been shown to enhance the defense response in rice (Figs.5, S2) [10].Therefore, we propose that they work together as a complex in H2O2signaling for pathogen defense.
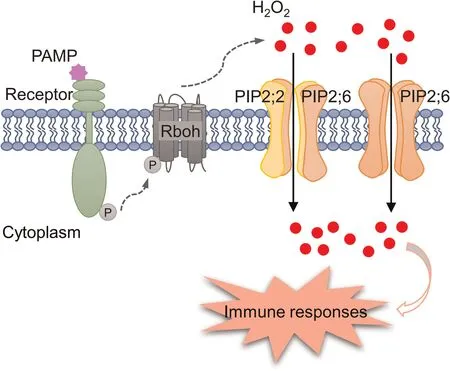
Fig.7.Model of rice immune response triggered by the OsPIP2 complexes for H2O2 transport.During pathogen attack, pathogen-associated molecular patterns(PAMPs) are detected by receptors in the PM, which mediates the phosphorylation of Rboh and subsequent H2O2 production.The apoplastic H2O2 is then transported into the cytoplasm by the OsPIP2;6-OsPIP2;2 heterotetramer or/and the OsPIP2;6 homotetramer,triggering a series of defense responses to enhance plant resistance.
Although our model requires further verification,mounting evidence suggests that PIPs often interact with one another to perform different functions.PIP1s generally interact with PIP2s to increase transportation efficiency, and co-expression of PIP1s and PIP2s often redirects PIP1s from the ER to the PM [23–26].PIP2s also selectively cooperate with PIP2s to regulate cellular development, flowering, and alkalinity tolerance, among other processes,in various plants[27–29].It should be noted that OsPIP2;2 ensures the nuclear localization of OsmaMYB to promote pathogen defense,whereas OsPIP2;6 lacks this ability [10].We speculate that although OsPIP2;2 and OsPIP2;6 transport H2O2cooperatively,they also function individually during the rice immune response.
The findings that OsPIP2;6 was highly expressed in multiple tissues and that OsPIP2;6 interacted with each of the 11 OsPIPs(Figs.1, 6, S4) suggests that OsPIP2;6 performs other functions besides pathogen response.A number of PIPs have been reported to participate in multiple processes and activities.For example,OsPIP2;2 facilitates water transport to increase drought tolerance as well as exporting H2O2to increase alkalinity tolerance [29,30].TaPIP2;10 contributes to photosynthesis by transporting CO2, as well as functioning similarly to OsPIP2;2 in pathogen response[11,31].Overexpression of TaPIP2;10 increased both grain yield and disease resistance in wheat [31].Therefore, comprehensive research into the functions of OsPIP2;6 will advance its utilization in crop breeding.
5.Conclusions
OsPIP2;6 facilitates cytoplasmic import of H2O2.Loss of function of OsPIP2;6 results in susceptibility to M.oryzae, presumably owing to its ability to import apoplastic H2O2and trigger a defense response coordinately with OsPIP2;2 as a heterologous complex.
CRediT authorship contribution statement
Gousi Li:Data curation, Investigation, Funding acquisition,Writing–original draft.Jingluan Han:Data curation, Investigation,Methodology,Validation.Chen Yi:Investigation.Hao Luo:Investigation, Validation.Yuzhu Wang:Investigation.Fengpin Wang:Conceptualization, Supervision.Xiaoyu Wang:Resources.Letian Chen:Conceptualization, Resources, Supervision, Project administration.Yaling Zhang:Conceptualization, Funding acquisition,Writing–review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We are grateful to Prof.Ping Yin and Dr.Qiang Wang from Huazhong Agricultural University for technical support on the structural analysis of OsPIP proteins.This work was supported by the Guangdong Basic and Applied Basic Research Foundation(2020A1515111101, 2022A1515110431).
Appendix A.Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2023.11.004.
- The Crop Journal的其它文章
- Flowering-time regulation by the circadian clock: From Arabidopsis to crops
- Drought-triggered repression of miR166 promotes drought tolerance in soybean
- The OsBSK1-2-MAPK module regulates blast resistance in rice
- Natural variation of an autophagy-family gene among rice subspecies affects grain size and weight
- Rice gene OsUGT75A regulates seedling emergence under deep-sowing conditions
- A telomere-to-telomere genome assembly of Zhonghuang 13,a widely-grown soybean variety from the original center of Glycine max

