The BEL1-like transcription factor GhBLH5-A05 participates in cotton response to drought stress
Jing-Bo Zhng, Yo Wng, Shi-Peng Zhng, Fn Cheng, Yong Zheng, Yng Li,*, Xue-Bo Li,*
a Hubei Key Laboratory of Genetic Regulation and Integrative Biology, School of Life Sciences, Central China Normal University, Wuhan 430079, Hubei, China
b College of Life Sciences, Xinjiang Normal University, Urumqi 830017, Xinjiang, China
Keywords:Cotton (Gossypium hirsutum)BEL1-like transcription factor Drought stress Transcriptional regulation Drought tolerance
ABSTRACT Drought stress impairs crop growth and development.BEL1-like family transcription factors may be involved in plant response to drought stress, but little is known of the molecular mechanism by which these proteins regulate plant response and defense to drought stress.Here we show that the BEL1-like transcription factor GhBLH5-A05 functions in cotton (Gossypium hirsutum) response and defense to drought stress.Expression of GhBLH5-A05 in cotton was induced by drought stress.Overexpression of GhBLH5-A05 in both Arabidopsis and cotton increased drought tolerance, whereas silencing GhBLH5-A05 in cotton resulted in elevated sensitivity to drought stress.GhBLH5-A05 binds to cis elements in the promoters of GhRD20-A09 and GhDREB2C-D05 to activate the expression of these genes.GhBLH5-A05 interacted with the KNOX transcription factor GhKNAT6-A03.Co-expression of GhBLH5-A05 and GhKNAT6-A03 increased the transcription of GhRD20-A09 and GhDREB2C-D05.We conclude that GhBLH5-A05 acts as a regulatory factor with GhKNAT6-A03 functioning in cotton response to drought stress by activating the expression of the drought-responsive genes GhRD20-A09 and GhDREB2C-D05.
1.Introduction
Since 1950s, global temperature has increased by 0.13 °C per 10 years, and rainfall has also decreased in many regions of the world,increasing the incidence of crop drought stress[1].Drought stress impairs plant growth and development,reducing crop yield.Under drought stress, plants sense environmental signals and transmit them into the cell, causing changes in the expression of a series of response genes and leading to physiological changes that may mitigate stress [2].Transcription factors, by modulating gene expression, act in plant response and defense to drought stress.Dehydration-responsive element binding (DREB) transcription factors regulate the expression of stress-responsive genes in plants and increase plant tolerance to abiotic stress by interacting with DRE/CRT cis-acting elements located in the promoter regions of abiotic stress-induced genes[3].CBF4-overexpressing rice plants showed increased resistance to freezing and drought stresses with increased expression of target genes responding to cold acclimation and drought[4].Overexpression of the rice R1R2R3 MYB gene OsMYB3R-2 in Arabidopsis increased drought tolerance in the transgenic plants[5].Overexpression of GbMYB5 conferred drought tolerance in transgenic tobacco, and silencing it reduced drought tolerance in cotton (Gossypium hirsutum) plants [6].Overexpression of SNAC1 (STRESS-RESPONSIVE NAC1) in rice increased plant drought tolerance [7].OsbZIP71-overexpressing plants showed increased drought tolerance[8].ZmPTF1,a bHLH transcription factor,increased drought tolerance in maize by promoting root development and abscisic acid synthesis [9].
Cotton, a world fiber crop, is grown in many regions subject to drought, such as Xinjiang in China, central Asia, and South Africa.Investigating the genetic basis of cotton response to drought stress and using genetic engineering to develop new cotton cultivars with high tolerance is desirable.Some genes involved in cotton drought response have been identified.GhWRKY59 is phosphorylated by a GhMAP3K15-GhMKK4-GhMPK6 canonical MAPK cascade and then interacts with the promoter of GhDREB2 to activate its expression for plant response to drought stress[10].Overexpression of GhABF2 in both Arabidopsis and cotton conferred drought and salt stress tolerance[11].Transgenic cotton overexpressing a Na+/H+antiporter, K2-NhaD, displayed increased drought and salt tolerance [12].Some stress-responsive genes: AtABF3, AtEDT1/HDG1125,
AtNHX129,AVP130,TsVP31,SNAC132,and AhCMO34 from Arabidopsis, Thellungiella halophila, rice, tobacco and Atriplex hortensis were introduced into cotton for increasing drought tolerance [13,14].Cotton genes have been transferred to other crops for the same purpose.Ectopic expression of GhDREB increased plant tolerance to drought,high salt and freezing stresses in transgenic wheat[15].
The TALE (Three Amino acid Loop Extension) protein family in plants comprises two subfamilies, BEL1-like (BELL) and KNOTTED1-like (KNOX) proteins [16].The Arabidopsis BELL subfamily has 13 members, and the potato genome also contains 13 BEL1-like genes [17].BLH2 and BLH4 positively regulate homogalacturonan de-methylesterification in Arabidopsis seed coat mucilage by activating the expression of PECTIN METHYLESTERASE 58 [18].BLH2/SAW1 and BLH4/SAW2 also redundantly regulate the morphogenesis of leaf margins by repressing the expression of class I KNOX gene BREVIPEDICELLUS (BP) [19].In rice, BEL1-like transcription factors, RI and RI-LIKE1 (RIL1), function in constructing inflorescence architecture and meristem maintenance[20].In potato,StBEL5 (a BELL transcription factor) and its protein partner POTH1 control tuber formation by modulating hormone levels[21].In tomato,SlBEL11 negatively regulates fruit chloroplast development by down-regulating expression of genes involved in chlorophyll biosynthesis: protochlorophyllide reductase (POR),magnesium chelatase H subunit(CHLH)and chlorophyllide an oxygenase (CAO) [22].SlBL4 regulates chlorophyll accumulation and chloroplast formation by inhibiting the expression of protoporphyrinogen oxidase (SlPPO), magnesium chelatase H subunit(SlCHLD), pectinesterase (SlPE), protochlorophyllide reductase(SlPOR),chlorophyll a/b binding protein(SlCAB-3B)and homeobox protein knotted 2(TKN2)in tomato[23].Overexpression of TaqSH1(a wheat BEL1-like gene) in Arabidopsis affects floral organ abscission by down-regulating the expression of the abscission related genes IDA, HAESA, KNAT1/6 and SHP1/2 [24].Ectopic expression of cotton BLH6-A13 influenced stem morphological structure and inhibits lignocellulose synthesis in Arabidopsis [25].BEL1 and KNOX proteins often form corresponding heterodimers to control transcription of the target genes [26].The BLH6-KNAT7 heterodimer regulates secondary cell wall development of Arabidopsis inflorescence stem by repressing expression of transcription factor REV/IFL1 [27].Arabidopsis BELL protein PENNYWISE (PNY) interacts with the KNOX proteins SHOOTMERISTEMLESS (STM) and BREVIPEDICELLUS(BP).The PNY-STM heterodimer is required for normal development of the shoot apical meristem, whereas the PNY-BP heterodimer functions in establishing early internode patterning events [28].Most studies of the function of BEL1-like genes in plants have focused on plant growth and development, and little is known about the function of BELL genes in plant response to abiotic stress.
2.Materials and methods
2.1.Plant materials and transformation
2.1.1.Arabidopsis transformation
Arabidopsis thaliana (Columbia ecotype) seeds were surfacesterilized with 10% NaClO for five minutes and then washed several times with sterile water.The sterilized seeds were plated on Murashige and Skoog (MS) medium, placed in darkness at 4°C for 72 h,and germinated at 22°C in an incubator(16 h light/8 h dark) for 7 d.The seedlings were transplanted into soil and grown at 22 °C in a growth chamber (16 h light/8 h dark).
The coding sequence of GhBLH5-A05 was inserted into the PK2GW7.0 vector to produce the 35S:GhBLH5-A05 construct and then introduced into Arabidopsis by the floral dip method.GhBLH5-A05 homozygous seeds (T3generation) were used for drought tests.Four-week-old seedlings were subjected to 14 d of water-deficit treatment and their phenotypes were observed under drought stress.Three independent biological replicates were tested for each experiment.
2.1.2.Cotton transformation
Seeds of upland cotton(Gossypium hirsutum cv.Coker 312)were germinated and grew in a growth chamber at 28°C(16 h light/8 h dark),and then the seedlings at three-leaf stage were subjected to water-deficit treatment for 18 d.Leaf samples were collected from both well-watered and drought-stressed seedlings.For genetic transformation, cotton seeds were surface-sterilized with 70%(v/v) ethanol for 1 min and 30% (v/v) H2O2for 2 h, and washed several times with sterile water.The sterilized cotton seeds were germinated on 1/2 MS medium for 6 d(28°C,16 h light/8 h dark).Then, their hypocotyls were cut from the sterile seedlings excised for cotton transformation.
The 35S:GhBLH5-A05 construct was transferred into cotton by Agrobacterium-mediated transformation as described previously[29].Transgenic cotton plants were identified by PCR and kanamycin resistance.For drought stress, the transgenic seedlings and wild-type controls were subjected to 18 d of water-deficit treatment and phenotypes were recorded.Three independent biological replicates were tested for each of the experiments.
2.1.3.Virus-induced gene silencing in cotton
Virus-induced gene silencing(VIGS)in cotton was performed as described previously [30].A 400-bp fragment of the GhBLH5-A05 gene was cloned into the TRV2 vector to produce a TRV2:-GhBLH5-A05 construct, which was introduced into Agrobacterium tumefaciens(strain GV3101).A solution mixture of the transformed Agrobacterium with the TRV1 and TRV2:00 or TRV2:GhBLH5-A05 was injected into seedling cotyledon cells.The expression of GhBLH5-A05 in the seedlings was detected by RT-qPCR, and then these seedlings were subjected to 14 d of water-deficit treatment.Three independent biological replicates were tested for each of the experiments.
2.2.Reverse transcription-quantitative polymerase chain reaction(RTqPCR) analysis
Total RNA was isolated and purified from leaves of both transgenic plants and wild type using the RNeasy kit (Qiagen, Dusseldorf, Germany).Expression analysis of genes was performed by RT-qPCR using SYBR Green dye (a fluorescent intercalating dye)as described previously [31].Two housekeeping genes each were used as internal controls in RT-qPCR analysis.The housekeeping genes GhUBI1 and GhHIS3 were used for cotton, while AtACTIN2 and AtUBQ5 were used for Arabidopsis.Three independent biological replicates were used for each of the experiments, and relative gene expression levels were estimated.The gene-specific primers used in the experiments are listed in Table S1.
2.3.Assay of physiological parameters associated with drought stress
Because malondialdehyde(MDA)condenses with thiobarbituric acid to form a red product with maximum absorption at 532 nm,MDA concentration can be calculated from absorbance at this wave length.MDA content was detected in leaves (0.1 g fresh tissue)with an MDA Quantification Assay Kit(Nanjing Jiancheng Bioengineering Institute, Nanjing, Jiangsu, China).Proline reacts with acidic ninhydrin to produce a stable red product with a absorption maximum at 515 nm,and its absorption value shows a linear relationship with proline content.For proline content detection, 0.1 g fresh leaf samples of cotton and Arabidopsis were prepared with a Proline Quantification Assay Kit(Nanjing Jiancheng Bioengineering Institute).Chlorophyll a and b have maximum absorptions at 645 nm and 663 nm,respectively,and the light absorption is used to calculate total chlorophyll content.Chlorophyll content was measured in stressed and control plants with a Chlorophyll Assay Kit (Solarbio Life Sciences).Stomatal aperture was observed and photographed in leaves of cotton and Arabidopsis under a microscope, and stomatal length-to-width ratio was determined for more than 50 stomata in each sample.
2.4.Yeast one-hybrid assay
The coding sequence of GhBLH5-A05 was cloned into the onehybrid vector pGADT7 with the GAL4 activation domain for generating the pGADT7-GhBLH5-A05 prey plasmid.The 2-kb promoter sequences of GhRD20-A09 and GhDREB2C-D05 were cloned separately into pAbAi vector to generate bait plasmids.The bait plasmid was introduced into Y1H Gold yeast strain for generating a baitspecific reporter yeast strain that was plated on SD/-leu medium with a proper concentration of aureobasidin A(AbA)to ensure that their growth was inhibited.The PGADT7 empty vector and PGADT7-GhBLH5-A05 were introduced into a bait-specific reporter strain.The yeast cells were cultured on selective SD/-leu medium containing AbA for detecting yeast growth.
2.5.Electrophoretic mobility shift assay (EMSA)
Biotin-labeled probes and unlabeled probes were synthesized for EMSA (Table S1).EMSA experiments employed a LightShift Chemiluminescent EMSA Kit (Thermo Fisher Scientific).Two mg of purified His-GhBLH5-A05 fusion protein and the probe were used to prepare a binding reaction held at 25°C for 30 min to allow the protein to bind to the probe.The binding protein–DNA mixture was separated by electrophoresis on 6% polyacrylamide gel and then transferred to a nylon membrane(Merck Millipore).An Easy-See Western Blot Kit(Thermo Fisher Scientific)was used to detect binding protein–DNA signals.
2.6.Transcriptional activation assay (dual-LUC assay)
The dual-luciferase reporter assay system employes a combination of luciferase (LUC) and Renilla luciferase (REN).REN shows constitutive expression driven by the 35S promoter, and LUC expression is driven by the promoter to be detected.If a transcription factor can bind to the promoter to be detected and activate its transcription activity,it will increase the expression of LUC and the activity of LUC/REN.The 2 kb promoter fragments of GhRD20-A09 and GhDREB2C-D05 were inserted separately into the pGreenII 0800-LUC vector to generate reporter constructs.The coding sequence of GhBLH5-A05 was cloned into the PK2GW7.0 vector to generate an effector construct.The reporter and effector constructs were introduced separately into Agrobacterium tumefaciens(strain GV3101)cells that were cultured to an optical density of 1.0 at 600 nm.The Agrobacterium cells were then mixed in equal volumes, injected into tobacco (Nicotiana benthamiana) leaves, and cultivated for 72 h.LUC and REN activities were measured with a Dual-Luciferase Reporter Kit (Promega).
2.7.Yeast two-hybrid assay
The open reading frame (ORF) of GhBLH5-A05 was cloned into the pGADT7 vector to generate a pGADT7-GhBLH5-A05 construct,which was introduced into yeast strain AH109.The ORF sequence of GhKNAT6-A03 was cloned into the pGBKT7 vector to generate a pGBKT7-GhKNAT6-A03 construct, which was transferred into yeast strain Y187.The AH109 yeast haploid strain was mated with the Y187 yeast haploid strain, and the mated diploid strains were incubated on a yeast double dropout medium without Leu or Trp for 72 h (30 °C).Positive interactions were identified on a yeast quadruple dropout medium without Leu, Trp, His, or Ade.
2.8.Luciferase complementation imaging (LCI) assay
LCI was performed as described previously [32].The ORF sequences of GhKNAT6-A03 and GhBLH5-A05 were inserted into JW771 and JW772 vectors, respectively.In the vectors, GhBLH5-A05 was inserted in front of the amino terminus of luciferase(nLUC) for generating GhBLH5-A05-nLUC, whereas GhKNAT6-A03 was inserted after the carboxy terminus of LUC(cLUC)for generating cLUC-GhKNAT6-A03.cLUC and nLUC were used as negative controls.The vectors were introduced separately into A.tumefaciens strain GV3101.Transformed Agrobacterium cells were suspended in infiltration buffer (10 mmol L-1MgCl2, 10 mmol L-1MES (2-(N-morpholino) ethanesulfonic acid) pH 5.7, 150 mmol L-1acetosyringone), and then injected into leaves of tobacco for 72 h co-cultivation.Luciferase activity in tobacco leaves was detected with a chemiluminescence image analysis system(Tanon 4600SF, Shanghai, China).
2.9.In vitro pull-down assay
The coding sequences of GhBLH5-A05 and GhKNAT6-A03 were inserted into PET32a and PGEX4T-1 vectors,respectively,to generate protein expression constructs.Then the constructs were transformed into Escherichia coli strain rosetta(BL21)cells,respectively.The transformed cells with recombinant His-GhBLH5-A05 that was fused to a N-terminal tag containing six His were cultured at 37°C with 0.3 mmol L-1isopropyl-β-d-thiogalactoside (IPTG) for 6 h to induce the expression of the recombinant proteins.The Histagged proteins were extracted and purified from E.coli cells according to the manufacturer’s instructions (Novagen, San Diego,CA, USA).The transformed cells with recombinant GST-GhKNAT6-A03 that was fused to a N-terminal tag containing glutathione S-transferase(GST)were cultured at 37°C with 0.5 mmol L-1IPTG for 4 h to induce the expression of the recombinant proteins.The GST-tagged proteins were incubated with an appropriate amount of glutathione-agarose resin for 30 min,and the unbound proteins were washed away with 10 volumes of phosphate buffer.The purified His-GhBLH5-A05 and GST-GhKNAT6-A03 proteins were then used for pulldown assay following Liu et al [33].The pulleddown proteins were determined by inmunoblotting with anti-GST and anti-His antibodies (CWBIO, China).
For immunoblotting, the protein samples were separated by SDS-PAGE and transferred to nitrocellulose membrane (Amersham) using a Trans-Blot Semi-DryElectrophoretic Transfer Cell(Bio-Rad).The membrane was incubated with primary antibody(anti-GST or anti-His)(CWBIO)overnight at 4°C,and washed three times for 15 min each time with TBST solution.Then the membrane was incubated with peroxidase-coupled secondary antibody(CWBIO,China)for 1.5 h,followed by washing three times(15 min per time)with Tris Buffered Saline with Tween 20(TBST)solution.The eECL Western Blot Kit(EpZyme)was used for detection of target proteins, and a chemiluminescence imaging system (LI-COR)was used for visualization of the target proteins.
All primers and probes used are listed in Table S1.
3.Results
3.1.Identification of drought stress-responsive BLH genes in cotton
To investigate the role of the BLH transcription factor in cotton under drought response,we first performed a genome-wide search for BLH genes in the public cotton(G.hirsutum)genome databases(https://cottonfgd.org/browse/), and identified 50 BLH genes.To identify drought stress-responsive BLH genes in cotton, we analyzed expression patterns of BLH genes in cotton tissues under drought stress and PEG treatment, using previous transcriptome data from our lab and the public databases.The transcriptome data showed that the expression of some BLH genes was altered in cotton under drought stress and PEG treatment.Among these drought stress-responsive BLH genes, GhBLH5-A05 was induced by drought stress and PEG treatment (Fig.1).Because RT-qPCR analysis confirmed that GhBLH5-A05 was drought-induced (Fig.S1),GhBLH5-A05 was selected for further study.First, we analyzed the expression pattern of GhBLH5-A05 in cotton using transcriptome data from a public database (https://cotton.zju.edu.cn/).GhBLH5-A05 was expressed at high levels in stems and leaves but at very low level in ovules and fibers (Fig.S2).GhBLH5-A05-GFP fluorescence signals were detected mainly in the cell nucleus(Fig.S3).
3.2.Suppression of GhBLH5-A05 resulted in reduced drought tolerance
To investigate the function of GhBLH5-A05 in cotton drought response, virus-induced gene silencing (VIGS) was employed to suppress the expression of GhBLH5-A05.An albino phenotype in the plants infected with TRV2:GhCLA indicated that the target gene had been effectively silenced(Fig.S4A).Expression of GhBLH5-A05 in the TRV2:GhBLH5-A05 plants was decreased relative to control TRV2:00 plants(Fig.S4B).There was no difference in growth status between TRV2:00 and TRV2:GhBLH5-A05 plants under normal growth condition (Fig.2A).The TRV2:00 and TRV2:GhBLH5-A05 plants were subjected to water-withholding treatment for 14 d.Upon drought treatment, growth status of the TRV2:00 plants was superior to that of the TRV2:GhBLH5-A05 plants.After rewatering for 3 d, all TRV2:00 plants returned to normal growth,but TRV2:GhBLH5-A05 plants still showed poor growth status(Fig.2A).Proline, chlorophyll and relative leaf water content were lower,but MDA content was higher,in leaves of the TRV2:GhBLH5-A05 plants than in those of TRV2:00 plants (Fig.2B–E).Under normal growth condition, stomatal apertures did not differ between TRV2:00 and TRV2:GhBLH5-A05 plants.After 14 d of drought treatment,the stomatal apertures were wider in TRV2:GhBLH5-A05 than in TRV2:00 plants (Fig.2F–G).Thus, silencing GhBLH5-A05 expression reduced drought tolerance.
3.3.Heterologous expression of GhBLH5-A05 in Arabidopsis increased plant drought tolerance
To further investigate the role of GhBLH5-A05 in plant response to drought stress, we generated transgenic Arabidopsis plants expressing GhBLH5-A05.Three independent T3generation transgenic lines with increased expression of GhBLH5-A05 were chosen(Fig.3A).In four-week-old transgenic plants and wild type under normal growth condition, there was no difference between GhBLH5-A05-overexpressing lines (OE7, OE13, and OE15) and the wild type.After drought treatment for 15 d, however, phenotypic differences appeared.The growth status of GhBLH5-A05-overexpressing plants was better than that of wild type under drought stress.After 3 d of rehydration, the transgenic lines had returned to normal growth, but the growth status of wild type plants was still poor (Fig.3B).After water-withholding for 15 d,GhBLH5-A05-overexpressing plants accumulated more proline but less MDA than the wild type (Fig.3C–D).Transgenic plants also had higher chlorophyll content and relative leaf water content than the wild type under drought stress (Fig.3E–F).Smaller stomatal openings were observed in GhBLH5-A05-overexpressing plants than in wild-type plants (Fig.3G–H).Thus, overexpression of GhBLH5-A05 increased drought tolerance in transgenic Arabidopsis.
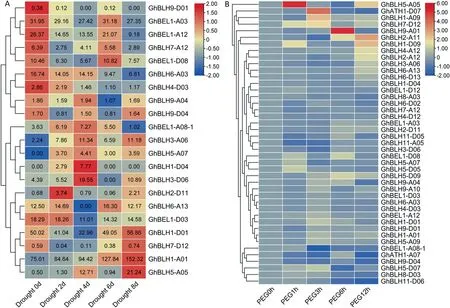
Fig.1.Expression patterns of GhBEL-like genes in cotton under drought stress and PEG treatment.(A)Drought stress responsive transcriptomic data were obtained from our lab, and visualization was performed with the pheatmap package.(B) PEG treated transcriptomic data were obtained from NCBI database, and its normalization and visualization were performed using the pheatmap package.
3.4.Overexpression of GhBLH5-A05 in cotton increased plant drought tolerance
To obtain a deeper understanding of the role of GhBLH5-A05 in cotton drought response, we transformed the GhBLH5-A05 gene into cotton by Agrobacterium-mediated cotton transformation,generating GhBLH5-A05-overexpressing transgenic cotton plants.Expression levels of GhBLH5-A05 were significantly higher in eight independent GhBLH5-A05-overexpressing lines(L1–L8)than in the wild type (Fig.4A), and Southern blot analysis showed that the transgenes were inserted at a single site (or in a single copy) in the genomes of these lines (Fig.S5).We chose two independent GhBLH5-A05-overexpressing lines (L4 and L8) for further study.First, we recorded the phenotypes of the transgenic cotton plants(T2–T3generations).Before drought treatment, GhBLH5-A05-overexpressing lines and the wild type displayed no phenotypic differences (Fig.4B).After drought treatment, the GhBLH5-A05-overexpressing lines showed stronger drought tolerance than the wild type.Leaf wilting in the wild type was more severe than in GhBLH5-A05-overexpressing lines (Fig.4B).There were no differences in MDA,proline,chlorophyll and relative leaf water contents in GhBLH5-A05-overexpressing lines under normal growth conditions, relative to the wild type.After drought treatment, however,proline was more abundant in GhBLH5-A05-overexpressing lines than in the wild type.Chlorophyll and relative leaf water contents in the GhBLH5-A05-overexpressing lines were also higher,but MDA content was lower in GhBLH5-A05-overexpressing than in wildtype plants (Fig.4C–F).As shown in Fig.4G–H, stomatal aperture did not differ between the transgenic lines and wild type under normal conditions, whereas apertures in wild-type leaves opened wider than those in the transgenic lines under drought stress.The drought-tolerance trait of the transgenic plants was stably inherited in the next generations (Fig.S6).As shown in Fig S7, no difference in single boll weight,boll number per plant,mature fiber length, thousand-seed weight, or mature seed size between the transgenic lines and wild type was observed under normal growth conditions.Thus, GhBLH5-A05-overexpressing plants displayed increased drought tolerance to drought stress.
3.5.GhBLH5-A05 activates expression of GhRD20-A09 and GhDREB2CD05
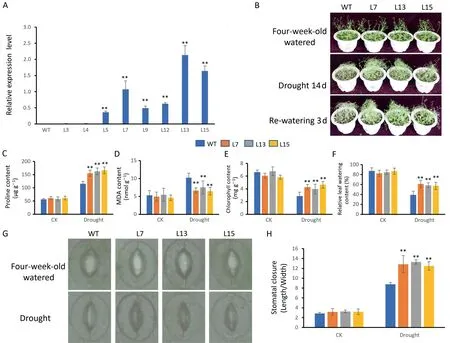
Fig.3.Phenotypic analysis of GhBLH5-A05-overexpressing transgenic Arabidopsis under drought stress.(A) RT-qPCR analysis of GhBLH5-A05 expression in GhBLH5-A05-overexpressing lines and wild type.(B)Phenotypes of 4-week-old wild type and GhBLH5-A05-overexpressing lines(OE7,OE13 and OE15)grown in soil under drought stress imposed by withholding water for 15 d.(C–F)Several physiological indexes measured in wild type and GhBLH5-A05-overexpressing lines(OE7,OE13 and OE15)under normal growing condition (control) and drought treatment.(C) Proline content.(D) MDA content.(E) Chlorophyll content.(F) Relative leaf water content.(G) Images of stomata in wild type and GhBLH5-A05-overexpressing lines(OE7,OE13 and OE15)under normal growing condition(control)and drought treatment.(H)Comparative stomatal aperture measurements(ratio of length to width).Experiments were repeated three times,and error bars denote standard deviation calculated from three independent experiments.Asterisks represent differences by Student’s t-test: *, P < 0.05; **, P < 0.01.WT, wild type; L3–L15, GhBLH5-A05-overexpressing transgenic lines.
Transcription levels of GhRD20-A09 and GhDREB2C-D05 were significantly upregulated in GhBLH5-A05-overexpressing transgenic cotton lines relative to the wild type (Fig.5A).GhRD20-A09 and GhDREB2C-D05 were induced by dehydration treatment, and shared similar expression patterns with GhBLH5-A05 in cotton under drought stress (Fig.S1).The promoter regions of GhRD20-A09 and GhDREB2C-D05 harbored TGAC cis elements that can be recognized and bound by BLH proteins (Fig.S8).We speculated that GhRD20-A09 and GhDREB2C-D05 were target genes of GhBLH5-A05.We conducted yeast one-hybrid (Y1H) assay to determine whether GhBLH5-A05 could bind directly to promoters of GhRD20-A09 and GhDREB2C-D05.Yeast strains containing pGADT7-GhBLH5-A05 and pAbAi-promoter constructs grew normally on selective medium supplemented with AbA, whereas strains containing pGADT7 and the pAbAi promoter did not grow(Fig.5B).To further confirm the results of Y1H assay, EMSA was performed to determine the physical interaction between GhBLH5-A05 protein and TGAC cis-elements in promoters of GhRD20-A09 and GhDREB2C-D05.As shown in Fig.5C, GhBLH5-A05 bound to the TGAC cis elements in promoters of GhRD20-A09 and GhDREB2C-D05.The binding signals were dramatically reduced when unlabeled competitor probes were added (Fig.5C).Finally,the 2 kb promoter fragments of GhRD20-A09 and GhDREB2C-D05 were fused with a luciferase (LUC) reporter gene and coexpressed with PK2GW7 empty vector or PK2GW7-GhBLH5-A05 in leaves of tobacco(N.benthamiana).Luciferase activity in tobacco leaves co-expressing PK2GW7-GhBLH5-A05 was higher than in leaves co-expressing empty vector (Fig.6E).
To further confirm the roles of GhRD20-A09 and GhDREB2C-D05 in cotton drought response, we conducted VIGS to suppress the expression of GhRD20-A09 and GhDREB2C-D05.RT-qPCR analysis showed that the expression of GhRD20-A09 and GhDREB2C-D05 was effectively inhibited in the VIGS plants (Fig.S9A).Then, the TRV2:00 (control), TRV2:GhRD20-A09 and TRV2:GhDREB2C-D05 plants were subjected to water-withholding treatment.After 14 d, growth status of the TRV2:00 plants was superior to that of the TRV2:GhRD20-A09 and TRV2:GhDREB2C-D05 VIGS plants.After three d of rehydration treatment, growth status of the TRV2:00 plants remained better than that of the TRV2:GhRD20-A09 and TRV2:GhDREB2C-D05 VIGS plants (Fig.S9B).Thus, the above data demonstrated that GhBLH5-A05 functions in cotton response to drought stress by regulating the expression of GhRD20-A09 and GhDREB2C-D05.
3.6.GhBLH5-A05 interacts with GhKNAT6-A03 to increase activation of GhRD20-A09 and GhDREB2C-D05
To identify proteins interacting with GhBLH5-A05 in cotton,we first identified gene orthologs of AtKNAT3 and AtKNAT6 in the cotton genome.AtKNAT3 has six orthologs (Ghir_A05G005930,Ghir_A06G010730, Ghir_A13G019410, Ghir_D05G005990,Ghir_D06G011070 and Ghir_D07G003500) and AtKNAT6 four(Ghir_A03G015310, Ghir_A05G032420, Ghir_D02G016670 and Ghir_D05G032380).We conducted LCI to verify that they interact with GhBLH5-A05, and found only GhKNAT6-A03(Ghir_A03G015310) interacted with GhBLH5-A05 (Figs.6A, S10).The GhKNAT6-A03 protein was located in the cell nucleus and membrane (Fig S11).Likewise, Y2H assay also revealed GhKNAT6-A03 interacted with GhBLH5-A05 in yeast cells(Fig.6B).In a pull-down assay (Fig.6C), a specific binding band appeared in the presence of GhKNAT6-A03 and GhBLH5-A05, but no binding signal was found in the lane containing GhBLH5-A05 and GST proteins, indicating that GhBLH5-A05 interacts with GhKNAT6-A03 in vitro.Expression analysis of GhKNAT6-A03 in cotton suggested that GhKNAT6-A03 is also involved in plant response to drought stress (Figs.6D, S1A).

Fig.4.Phenotypic analysis of GhBLH5-A05-overexpressing transgenic cotton under drought stress.(A)RT-qPCR analysis of GhBLH5-A05 expression in wild type and GhBLH5-A05-overexpressing cotton plants.(B) Phenotypes of 4-week-old wild type and GhBLH5-A05-overexpressing cotton plants (L4 and L8) grown in soil under drought stress imposed by withholding water for 18 d.(C–F) Several physiological indexes measured in wild type and GhBLH5-A05-overexpressing cotton lines (L4 and L8) under normal growing condition (control) and drought treatment.(C) Proline content.(D) MDA content.(E) Chlorophyll content.(F) Relative leaf water content.(G) Images of stomata in wild type and GhBLH5-A05-overexpressing cotton lines (L4 and L8) under normal growing condition (control) and drought treatment.(H) Comparison of stomata opening(ratio of length to width) between wild type and GhBLH5-A05-overexpressing transgenic cotton lines (L4 and L8) under normal growing condition (control) and drought treatment.The experiments were repeated three times, and error bars denote standard deviation calculated from three independent experiments.Asterisks represent differences by Student’s t-test: *, P < 0.05; **, P < 0.01.WT, wild type; L4–L8, GhBLH5-A05-overexpressing transgenic cotton lines.
Luciferase activity under the control of GhRD20-A09 and GhDREB2C-D05 promoters was increased when GhBLH5-A05 was co-expressed in tobacco leaves.When both GhBLH5-A05 and GhKNAT6-A03 were co-expressed with the promoter of GhRD20-A09 or GhDREB2C-D05, luciferase activity was further increased(Fig.6E).EMSA revealed that GhKNAT6-A03 bound to the promoters of GhRD20-A09 and GhDREB2C-D05(Fig S12).These results suggested that GhBLH5-A05 and GhKNAT6-A03 synergistically increase the transcription activity of GhRD20-A09 and GhDREB2CD05.
4.Discussion
Previous studies revealed that homeobox-containing BEL1-type transcription factors play important roles in plant development,but comparatively little is known about their biological functions in plant response to drought stress.In this study, we showed that a BEL1-type transcription factor (GhBLH5-A05) functions in regulating cotton response and defense to drought stress.Overexpression of GhBLH5-A05 strengthened the drought tolerance of transgenic Arabidopsis and cotton plants, whereas silencing GhBLH5-A05 weakened cotton drought tolerance.Proline, as an osmotic regulator,functions in maintaining plant cell homeostasis under abiotic stress [34].When plants face drought, they tend to accumulate more proline to improve their own osmotic adjustment ability [35,36].The finding that under drought stress,GhBLH5-A05-overexpressing lines accumulated more proline than the wild type, indicates that overexpression of GhBLH5-A05 increased osmotic adjustment ability.Drought can induce the rapid closing of stomata for preventing water loss by transpiration[37,38].Under drought stress, the GhBLH5-A05-overexpressing plants maintained narrower stomatal aperture than wild type,thus lowering the rate of water loss.Because chlorophyll is essential for photosynthesis, which is necessary for plant development, chlorophyll content under drought conditions can be used as an indicator of drought resistance [39].The finding that GhBLH5-A05-overexpressing plants displayed higher chlorophyll content than the wild type after drought treatment suggests that transgenic plants maintain a better growth state than the wild type.We infer that GhBLH5-A05 acts as a positive regulator in cotton response to drought stress.

Fig.5.GhBLH5-A05 regulated expression of GhRD20-A09 and GhDREB2C-A05 genes in cotton.(A) Expression of GhRD20-A09 and GhDREB2C-A05 in WT and GhBLH5-A05-overexpressing transgenic cotton lines.(B)Yeast one hybrid assay of the interaction between GhBLH5-A05 and promoters of GhRD20-A09 and GhDREB2C-A05.Yeast cells were cultured on SD/-Leu medium containing 0(control),150 ng mL-1 AbA,or 250 ng mL-1 AbA for 3 d(C)Electrophoretic mobility shift assay of GhBLH5-A05 protein binding to promoters of GhRD20-A09 and GhDREB2C-A05.Biotin-labeled probes from promoters of GhRD20-A09 and GhDREB2C-A05 were incubated with His-GhBLH5-A05 protein.Cold probe was added to compete with the labeled probe.Experiments were repeated three times, and error bars denote standard deviation calculated from three independent experiments.Asterisks represent differences by Student’s t-test: *, P < 0.05; **, P < 0.01.WT, wild type; L4 and L8, GhBLH5-A05-overexpressing transgenic cotton lines.
Some transcription factors function in plant response to drought by modulating expressions of drought-induced genes.GhWRKY59 regulates cotton drought signaling by regulating the expression of GhDREB2 [40].GhWRKY21 regulates the expression of a type 2C protein phosphatase(PP2C)gene GhHAB to participate in drought response of cotton[41].TaBZR2 increases plant drought tolerance by binding to the promoter of TaGST1 to activate its transcription [42].GhirNAC2 (a NAC transcription factor) functions in drought tolerance by regulating expression of GhNCED3a/3c [43].JUNGBRUNNEN1 increased tomato drought tolerance by stimulating expression of SlDREB1, SlDREB2 and SlDELLA [44].ZmNAC49 increased maize drought tolerance by suppressing ZmMUTE expression [45].RD20 and DREB genes function in plant drought response [46,47].ARAG1, an ABA-responsive DREB gene, functions in drought tolerance of rice[48].Expressions of the two DREB/CBF genes,TaDREB3 and TaCBF5L,increased drought and frost tolerance in transgenic barley and frost tolerance in transgenic wheat seedlings [49].In the present study, transcripts of GhRD20-A09 and GhDREB2C-D05 were increased in GhBLH5-A05-overexpressing cotton relative to the wild type.In contrast, suppressing the expression of GhRD20-A09 and GhDREB2C-D05 in cotton reduced drought tolerance(Fig.S9).GhBLH5-A05 bound to the promoters of GhRD20-A09 and GhDREB2C-D05, activating their transcription,indicating that GhBLH5-A05 increases cotton drought tolerance by promoting the expression of these drought-induced genes.
Plant transcription factors often form corresponding homodimers and heterodimers to regulate transcription of target genes[50–52].Cotton GhHOX3 interacted with GhHD1 to increase the transcriptional activation of a downstream target gene [53].Coexpression of Populus NF-YB21 and FUS3 increased the promoter activity of PdNCED3 [54].In soybean, GmWRKY27 interacted with GmMYB174 to cooperatively inhibit GmNAC29 expression [55].AtBLH1 interacted with AtKNAT3, and the BLH1-KNAT3 complex increased expression of the ABA-responsive reporter gene [56].In our study,GhBLH5-A05 interacted with GhKNAT6-A03 to increase the transcription activity of GhRD20-A09 and GhDREB2C-D05.
Fig.7 illustrates a proposed mechanism by which GhBLH5-A05 regulates cotton drought tolerance.After drought-stress signals promote GhBLH5-A05 expression,the activated GhBLH5-A05 interacts with GhKNAT6-A03 to form a protein complex.The GhBLH5-A05-GhKNAT6-A03 protein complex binds to the TGAC elements in promoters of GhRD20-A09 and GhDREB2C-D05, activating their expression.The increased expression of GhRD20-A09 and GhDREB2C-D05 strengthens tolerance to drought stress.Thus,BEL1-like proteins function in cotton response to drought stress.Their encoding genes are candidates for genetic manipulation for improving cotton drought resistance.
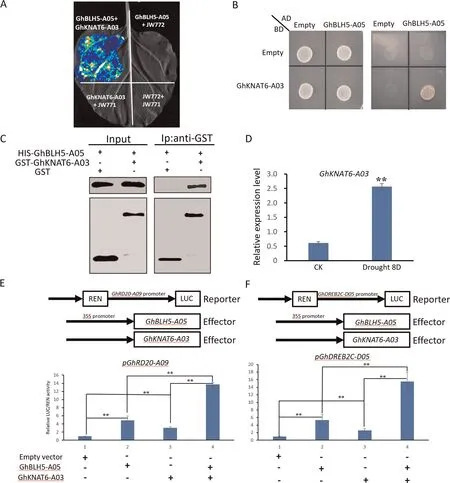
Fig.6.GhBLH5-A05 interacted with GhKNAT6-A03 to increase transcriptional regulation of its target genes.(A)Luciferase complementation imaging(LCI)assay of GhBLH5-A05 interacting with GhKNAT6-A03.cLUC, carboxy terminus of luciferase; nLUC, amino terminus of luciferase.(B) Yeast two-hybrid assay of GhBLH5-A05 interaction with GhKNAT6-A03.BD,GAL4 DNA binding domain;AD,GAL4 activation domain.Left panel:yeast cells grown on 2-dropout medium.Right panel:yeast cells grown on selective 4-dropout medium.(C)Pulldown assay of GhBLH5-A05 interacting with GhKNAT6-A03 in vitro.Purified His-GhBLH5-A05 protein was incubated with GST or GST-GhKNAT6-A03 proteins, and proteins recovered from pulldown assays were detected by immunoblotting using anti-His and anti-GST antibodies.(D) Expression of GhKNAT-A03 in cotton leaves under drought stress.(E)Transcriptional activation assay of GhRD20-A09 and GhDREB2C promoters by GhBLH5-A05 with GhKNAT6-A03.The coding sequences of GhBLH5-A05 and GhKNAT-A03 were inserted into corresponding vector to generate effector.GhRD20-A09 and GhDREB2C promoters were fused separately to the LUC reporter and promoter activities were measured by transient dual-LUC assay in leaves of tobacco(Nicotiana benthamiana).Experiments were repeated three times,and error bars denote standard deviation calculated from three independent experiments.Asterisks represent differences by Student’s t-test: *, P < 0.05; **, P < 0.01.
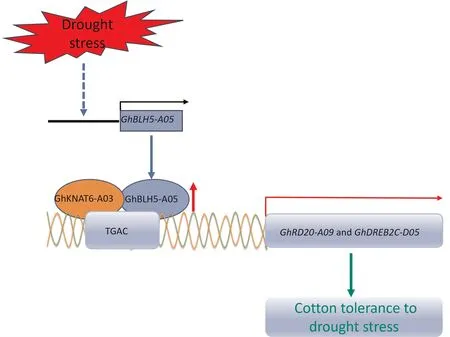
Fig.7.A schematic model depicting the mechanism by which the GhBLH5-A05 complex with GhKNAT6-A03 activates genes for regulating cotton tolerance to drought stress.
CRediT authorship contribution statement
Jing-Bo Zhang:Investigation, Data curation, Writing.Yao Wang: Investigation.Shi-Peng Zhang: Investigation.Fan Cheng:Investigation.Yong Zheng:Data curation.Yang Li:Investigation,Data curation.Xue-Bao Li:Conceptualization,Data curation,Writing, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the Project from the Ministry of Agriculture of China for Transgenic Research (2014ZX0800927B)and the National Natural Science Foundation of China(31871667).
Appendix A.Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2023.10.011.
- The Crop Journal的其它文章
- Flowering-time regulation by the circadian clock: From Arabidopsis to crops
- Global characterization of OsPIP aquaporins reveals that the H2O2 transporter OsPIP2;6 increases resistance to rice blast
- Drought-triggered repression of miR166 promotes drought tolerance in soybean
- The OsBSK1-2-MAPK module regulates blast resistance in rice
- Natural variation of an autophagy-family gene among rice subspecies affects grain size and weight
- Rice gene OsUGT75A regulates seedling emergence under deep-sowing conditions

