Role of tumor necrosis factor alpha-induced protein 6(TNFAIP6)in tumors:a pan-cancer analysis
Yng Yng,Zhi-Gng Liu,Yn-Qi Yng,Zhi-Gng Zhng,Xio-Li Wng,Yu-Long Li,Rui-Fng Sun,*
Abstract Background:There is growing evidence that the gene named tumor necrosis factor α-induced protein 6(TNFAIP6)has an important role in various tumors.However,a systematic pan-cancer analysis of TNFAIP6 is lacking.Here we aimed to analyze the expression of TNFAIP6 across multiple cancers and verify its expression during the progression of colon cancer.Methods: We performed a comprehensive bioinformatics analysis to examine the expression of TNFAIP6 across 27 tumor types.GEPIA2 was used to evaluate the effect of TNFAIP6 on clinical cancer prognosis.c Bioportal was used to assess TNFAIP6 mutations.The correlation between TNFAIP6 and cancer immune infiltrates was explored using TIMER2.0.The CancerSEA database was used to perform functional analysis of TNFAIP6.Metascape was used to identify TNFAIP6-related gene enrichment pathways.Immunohistochemistry was performed to detect TNFAIP6 protein expression in the colon cancer.In addition,the Comparative Toxicogenomics Database was searched for known and possible antitumor drugs that may be associated with TNFAIP6.Results:We found that,in most of the cancers included in this analysis,TNFAIP6 was highly expressed,and there is a distinct relationship between TNFAIP6 expression and cancer prognosis.TNFAIP6 expression is associated with cancer-associated fibroblasts,neutrophils,and endothelial cells.TNFAIP6 and similar genes may also be involved in the PID_VEGF_VEGFR_pathway.Immunohistochemistry revealed an increasing trend of TNFAIP6 protein expression in normal,adenoma,and colon cancer tissues.Several known and possible antitumor drugs that may be associated with TNFAIP6 were identified in the Comparative Toxicogenomics Database.These results suggest that a number of drugs may target TNFAIP6 during cancer treatment,including cisplatin,irinotecan,resveratrol,U 0126,NSC689534,genistein,NSC668394,oxaliplatin,plerixafor,topotecan,vincristine,flutamide,doxorubicin,MRK 003,folic acid,demecolcine,tunicamycin,zoledronic acid,and schizandrin B.Conclusions:TNFAIP6 may function as an oncogene in certain cancers.Furthermore,this study provides evidence that TNFAIP6 is an important factor in colon cancer progression.
Keywords: Cancer;IHC;Mutation;Survival;TNFAIP6
1.Introduction
Tumor necrosis factor α (TNF-α)–induced protein 6 (TNFAIP6),also known as TSG-6,is a member of the TNFAIP family of proteins.TNFAIP6 is an inflammation-associated protein that plays an important roleinextracellular matrix formationand cellmigration.[1–4]Previousstudies have indicated that TNFAIP6 participates in the development of many tumors;however,its precise role in tumor formation and progression has yet to be elucidated.TNFAIP6 is associated with proliferation of gastric cancer(GC)cells and may serve as a novel indicator of GC prognosis.[5]The results of quantitative polymerase chain reaction have shown that TNFAIP6 is significantly overexpressed in renal cell carcinoma (RCC)compared with that in normal renal tissue.[6]TNFAIP6 may be a novel biomarker for clear cell RCC.[7]Upregulated TNFAIP6 expression is associated with aggressive pathological features in urothelial carcinomas and poor prognosis.[8]Transcript levels of TNFAIP6 are higher in head and neck cancer tissues than in normal tissues.[9]Currently,research on TNFAIP6 and its associated tumors is still in its infancy.Systematic TNFAIP6 analysis will shed new light on its role in various cancers.
In the present study,we comprehensively analyzed the expression of TNFAIP6 in various cancers using SangerBox.In addition,we analyzed survival outcomes,mutation status,tumor-infiltrating immune cells,and relevant cellular pathways of TNFAIP6 using GEPIA2,cBioportal,TIMER2.0,STRING,and Metascape.The results revealed a possible function of TNFAIP6 in the pathogenesis and clinical prognosis of various human cancers and may help to further distinguish other potential roles of this gene in the development of cancers.
2.Materials and methods
2.1.Gene expression analysis
TNFAIP6 expression across 27 tumors based on The Cancer Genome Atlas (TCGA) and GTEx data was analyzed using SangerBox[10](http://sangerbox.com).We entered TNFAIP6 in the“Gene”module of SangerBox and then chose“TCGA+GTEx.”In this way,we could assess differences in TNFAIP6 expression between different tumors and normal tissues.The UALCAN[11,12]tool(http://ualcan.path.uab.edu) was used to obtain the protein expression levels of TNFAIP6 across tumor types.GEPIA2[13](http://gepia2.cancer-pku.cn)was used to explore TNFAIP6 expression at different pathological stages across different cancers.
2.2.Survival prognosis analysis
GEPIA2 was used to detect the association of TNFAIP6 with overall survival(OS)and disease-free survival(DFS)in cancer cases,and the group cutoff was selected as median.In addition,we used the Kaplan-Meier survival analysis module to obtain the OS plots from cases of cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC),colon adenocarcinoma (COAD),kidney renal clear cell carcinoma(KIRC),brain lower-grade glioma(LGG),lung squamous cell carcinoma (LUSC),mesothelioma (MESO),and uveal melanoma(UVM)and DFS in CESC,KIRC,LGG,and UVM.
2.3.Mutation character analysis
The cBioPortal web(https://www.cbioportal.org)[14]was used to ascertain the genetic alteration characteristics of TNFAIP6;by searching for“TNFAIP6”in the“Quick Search”section,the alteration frequency of the tumors was obtained.The mutated site of TNFAIP6 was revealed by searching in the“Mutations”module.
2.4.Immune analysis
SangerBox was used to analyze the association of TNFAIP6 expression with microsatellite instability(MSI),tumor mutational burden(TMB),and immune checkpoint genes in human cancers.The“Immune”module of TIMER2.0[15](http://timer.cistrome.org)was used to investigate the correlation between TNFAIP6 and immune infiltrates in tumors.
2.5.Functional correlation analysis
CancerSEA(http://biocc.hrbmu.edu.cn/Cancer SEA/)can be used to analyze the functional specificity of cancer cells at the single-cell level.[16]An analysis was performed through CancerSEA to determine the functional state of TNFAIP6 across cancers.
2.6.TNFAIP6-related genes
The STRING website[17,18](https://string-db.org)was searched for genes related to TNFAIP6.The“Similar Gene Detection”module of GEPIA2,based on TCGA,was used to search for genes(associated with different cancers and tissue types)with an expression pattern or signature similar to that of TNFAIP6,and the top 5 genes obtained from this search were analyzed.TIMER2.0 was performed to determine the correlation between TNFAIP6 and the top 5 genes in various cancer types.Metascape[19](https://metascape.org)was used to obtain the top nonredundant enrichment clusters using TNFAIP6 and the top 100 genes similar to TNFAIP6.
2.7.Immunohistochemistry
The tissue samples were sourced from Shaanxi Provincial Cancer Hospital,and the consent was obtained from the patients.The tissue samples were fixed in formaldehyde,embedded in paraffin,and sectioned.Sections were deparaffinized in xylene and hydrated using graded alcohol.Citrate buffer was used to retrieve the antigen.Peroxide solution (3% H2O2) was used to block endogenous peroxidases and nonspecific binding for 20 minutes at room temperature.The cultures were then incubated with anti-TNFAIP6 primary antibodies(1:100;Santa Cruz Biotechnology,Santa Cruz,CA)at 4°C overnight,followed by incubation with a secondary antibody at 37°C for 30 minutes.The prepared samples were then stained with 3,3′-diaminobenzidine (DAB) for 3 minutes at room temperature and counter-stained with hematoxylin solution.
2.8.Toxicogenomic analysis
The Comparative Toxicogenomics Database (CTD)[20](http://ctdbase.org/downloads/)is a dataset used to investigate the relationships between chemicals,genes,and human diseases.These data were combined with functional and pathway data to help establish hypotheses regarding the mechanism of the environmental impact on diseases.The results aid in understanding the molecular mechanisms of potential antitumor drugs,as well as the complex interaction between genes and proteins.The antitumor drugs and carcinogens correlated with TNFAIP6 are listed in Tables 1 and 2,respectively.
3.Results
3.1.Analysis of TNFAIP6 expression
SangerBox was used to explore TNFAIP6 expression across tumor types.The expression of TNFAIP6 in 27 types of tumors and normal human tissues was analyzed using data sets from TCGA and GTEx databases.The expression level of TNFAIP6 (measured as TPM+1)was log-transformed to simplify presentation of the data.As shown in Figure 1A,the level of expression of TNFAIP6 was found to be higher,compared with control tissues,in the following 20 kinds of carcinomas:adrenocortical carcinoma,breast invasive carcinoma(BRCA),cholangiocarcinoma,COAD,esophageal carcinoma(ESCA),glioblastoma multiforme,head and neck squamous cell carcinoma (HNSC),KIRC,kidney renal papillary cell carcinoma(KIRP),acute myeloid leukemia,brain LGG,liver hepatocellular carcinoma (LIHC),lung adenocarcinoma (LUAD),LUSC,ovarian serous cystadenocarcinoma,pancreatic adenocarcinoma(PAAD),skin cutaneous melanoma (SKCM),stomach adenocarcinoma(STAD),thyroid carcinoma(THCA),and uterine carcinosarcoma(UCS).Analysis of the CPTAC dataset showed that TNFAIP6 protein expression was increased in breast cancer,clear cell RCC,LUAD,LUSC,HNSC,and PAAD (Figure 1B).GEPIA2 exhibited stage-specific changes in TNFAIP6 expression in BLCA,ESCA,KICH,KIRP,LIHC,LUAD,and THCA cells(Figure 1C).

Figure 1.Expression of TNFAIP6 in tumors and pathological stages.A,The expression of TNFAIP6 in different cancers was determined via TCGA analysis,and normal tissues from the GTEx database were included as controls.*P <0.05,**P <0.01,***P <0.001.B,The expression of the TNFAIP6 protein in breast cancer,clear cell RCC,lung adenocarcinoma,lung squamous cell carcinoma,head and neck squamous carcinoma,and pancreatic adenocarcinoma was analyzed using the CPTAC data set.C,TCGA data were used to analyze the expression levels of TNFAIP6 based on pathological stage of the different tumors.BLCA,bladder urothelial carcinoma;CPTAC,Clinical Proteomic Tumor Analysis Consortium;ESCA,esophageal carcinoma;KICH,kidney chromophobe;KIRP,kidney renal papillary cell carcinoma;LIHC,liver hepatocellular carcinoma;LUAD,lung adenocarcinoma;RCC,renal cell carcinoma;TCGA,The Cancer Genome Atlas;THCA,thyroid carcinoma.
3.2.TNFAIP6 expression and survival rate
GEPIA2 analysis showed that increased TNFAIP6 expression was linked to poor prognosis and reduced OS rate in certain cancers,including CESC,COAD,LGG,LUSC,MESO,and UVM.In KIRC,increased expression of TNFAIP6 was linked to good prognosis and better OS(Figure 2A).In addition,increased TNFAIP6 expression was associated with poor prognosis and decreased rates of DFS in CESC,LGG,and UVM;however,high expression of TNFAIP6 predicted good prognosis and increased DFS rates in KIRC(Figure 2B).

Figure 2.TNFAIP6 expression and prognosis in different types of cancer.GEPIA2 were used to obtain survival maps in terms of overall survival (OS) (A) and disease-free survival (DFS) (B).Kaplan-Meier curves with significantly different results are shown.
3.3.Genetic alteration of TNFAIP6
cBioPortal was used to analyze the alteration status of TNFAIP6.The frequency of TNFAIP6 alterations(>4%)was highest in prostate adenocarcinoma,with “deep deletion” as the primary type ofalteration(Figure 3A).We identified these mutations and their locations in the TNFAIP6 protein structure(Figure 3B).

Figure 3.Mutation status of TNFAIP6 in tumors.A,The frequency of alterations in TNFAIP6 was analyzed via the c BioPortal.B,The mutation site of TNFAIP6,as shown in c BioPortal.
Table 1Significantly enriched chemicals related with antitumor drugs identified by TNFAIP6 expression
3.4.Correlation of TNFAIP6 expression with TMB,MSI,and immune checkpoint genes
To evaluate whether TNFAIP6 plays an important role in immunity,we analyzed the association between TNFAIP6 expression and TMB and MSI in tumors.We found that TNFAIP6 expression was significantly correlated with TMB in the following types of tumors:ovarian serous cystadenocarcinoma,LUAD,ESCA,LIHC,SARC,BRCA,COAD,STAD,KIRC,HNSC,and LGG(Figure 4A).Furthermore,there were significant correlations between the expression of TNFAIP6 and MSI in LUAD,LUSC,COAD,SKCM,KIRC,THCA,HNSC,and READ(Figure 4B).The expression of TNFAIP6 was associated with several immune checkpoint(ICP)genes in many types of cancer(Figure 4C).

Figure 4.TNFAIP6 expression and TMB,MSI,immune check point genes.The relationship between TNFAIP6 expression and TMB (A),MSI (B),and immune checkpoint genes (C).*P <0.05;**P <0.01;***P <0.001.MSI,microsatellite instability;TMB,tumor mutational burden.
3.5.Correlation between immune infiltration and TNFAIP6 expression
We used TIMER2.0 to explore the potential relationship between the infiltration of different immune cells and TNFAIP6 expression in cancers.EPIC,MCPCOUNTER,XCELL,and TIDE analyses were performed to analyze the correlation between cancer-associated fibroblasts(CAFs)and TNFAIP6.A statistically significant positive correlation was observed between the immune filtration of CAFs and the expression of TNFAIP6 in the following tumor types: BLCA,BRCA,BRCA-basal,BRCA-lumA,BRCA-lumB,CESC,COAD,ESCA,HNSC,HNSCHPV.,LIHC,LUAD,LUSC,PAAD,PRAD,READ,SARC,STAD,TGCT,THCA,THYM,and UCS.However,there was a negative correlation with respect to KIRC(Figure 5A).TIMER,CIBERSORT,CIBERSORT-ABS,MCPCOUNTER,QUANTISEQ,and XCELL analyses were executed to explore the association between neutrophil infiltration and the expression of TNFAIP6.The TIMER results showed a significant positive correlation between the immune filtration of neutrophils and expression of TNFAIP6 in most cancers(Figure 5B).The results of the EPIC,MCPCOUNTER,and XCELL analysesrevealed associationsbetween endothelialcellsand theexpression of TNFAIP6.A significant positive correlation exists between endothelial cells and the expression of TNFAIP6 in the following types of cancer:BRCA,BRCA-lumA,CESC,COAD,glioblastoma multiforme,HNSC,HNSC-HPV-,HNSC-HPV+,KIRC,LGG,PAAD,PCPG,PRAD,READ,SKCM,SKCM-metastasis,and UCS(Figure 5C).

Figure 5.Expression of TNFAIP6 and infiltration of cancer-associated fibroblasts,neutrophils,and endothelial cells.Correlation between TNFAIP6 expression and cancer-associated fibroblasts(A),neutrophils(B),and endothelial cells(C).
3.6.TNFAIP6 and functional states in various cancers
We used the CancerSEA database to uncover the relevance of TNFAIP6 to functional states in several cancers(Figure 6A).TNFAIP6was negatively correlated with DNA repair(correlation=-0.43,P=0)in UM,which indicates that TNFAIP6 may participate in UM by affecting DNA repair(Figure 6B).

Figure 6.Relevance of TNFAIP6 across functional states in different cancers.A,The correlation between TNFAIP6 and 14 functional states in different cancers from CancerSEA.B,TNFAIP6 was negatively correlated with DNA repair in UM.UM,uveal melanoma.

Table 2Significantly enriched chemicals related with carcinogen identified by TNFAIP6 expression
3.7.TNFAIP6-related proteins
Using the STRING program,we searched for TNFAIP6-binding proteins (Figure 7A).GEPIA2 was used to obtain genes similar to TNFAIP6,and the results showed that the top 5 genes that correlated positively with TNFAIP6 expression were TM4SF18,FUT11,DAB2,SLC3A1,and VCAM1(Figure 7B).The corresponding heat map data revealed a positive correlation between TNFAIP6 and TM4SF18,FUT11,DAB2,and VCAM1 in most cancer types(Figure 7C).To explore the molecular mechanisms by which TNFAIP6 mediates tumorigenesis,Metascape was used to perform enrichment analysis of the top 100 genes similar to TNFAIP6,which revealed that they may be functional mediators of the PID VEGF-VEGFR pathway,organic anion transport,and cobalamin transport(Figure 7D).
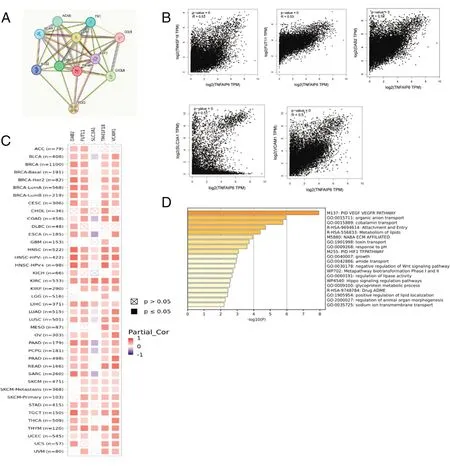
Figure 7.Enrichment analysis of TNFAIP6-related genes.A,The TNFAIP6-binding protein by STRING.B,GEPIA2 showed the relationship between the expression of TNFAIP6 and TM4SF18,FUT11,DAB2,SLC3A1,VCAM1.C,The corresponding heat map of the relationship between TNFAIP6 and TM4SF18,FUT11,DAB2,and VCAM1 in the majority of cancer types.D,Metascape showing the results of enrichment analysis.
3.8.TNFAIP6 expression is higher in colon cancer tissues
Bioinformatics analysis revealed that TNFAIP6 is highly expressed in colon cancer.Immunohistochemistry was performed to verify the expression of the gene in the colon cancer cells.Owing to the close relationship between adenoma and colon cancer,we collected pairs of tissues,including adjacent normal tissues,colon adenoma tissues,and colon cancer tissues,to detect TNFAIP6 expression in colon carcinogenesis.We found that TNFAIP6 expression in colon adenoma tissue increased,and TNFAIP6 expression in colon cancer tissues was upregulated compared with that in colon adenoma tissue,revealing an increasing trend in TNFAIP6 expression with the progression of colon cancer(Figure 8).

Figure 8.TNFAIP6 expression in colon cancer tissues.The expression of TNFAIP6 in colon adenoma and carcinoma,compared with gene expression in normal tissues,was measured through immunohistochemical analysis.
3.9.Carcinogens and antitumor drugs correlated with TNFAIP6
We present in Table 1 a list of antitumor drugs that are correlated with TNFAIP6 expression,including cisplatin,irinotecan,resveratrol,U 0126,and other drugs.Carcinogens that correlated with TNFAIP6 expression,such as asbestos,crocidolite,and benzo(a)pyrene,are presented in Table 2.
4.Discussion
Abnormal gene expression is known to cause tumor development.[21–23]Examining the similarities and differences between tumor types with respect to the tumor cell genome and cell changes is necessary to better understand gene functions in cancers.There is growing evidence that TNFAIP6 plays an important role in several cancers.[24–27]However,its role in most cancer types has not been extensively studied.Whether TNFAIP6 acts via certain common molecular mechanisms in the pathogenesis of different tumors requires further study.Pan-cancer analysis is useful for elucidating the role of TNFAIP6 in tumors at the macro level.In the present study,we used SangerBox to assess TNFAIP6 expression in cancer and normal tissues based on TCGA and GTEx databases.The results showed that TNFAIP6 expression was upregulated in most of the tumor types included in our analysis.These data provide insights into the prospective role of TNFAIP6 as a cancer biomarker.
Previous studies have predicted that TNFAIP6 is a biomarker for metastatic non–small cell lung cancer,[28]GC,[5]clear cell RCC,[7]and urothelial carcinomas.[8]Survival and prognostic analyses showed that TNFAIP6 expression was associated with poor prognosis and OS in CESC,COAD,LGG,LUSC,MESO,and UVM.Increased TNFAIP6 expression is associated with poor prognosis and DFS in CESC,LGG,and UVM.However,it is unclear why the high level of expression of TNFAIP6 in KIRC is correlated with good OS and DFS.Considering that the number of sample cases of KIRC was adequate(>200),we speculate that this correlation may be indicative of a dual role of TNFAIP6 in KIRC.However,further experimental research is required to determine the exact function of TNFAIP6 in KIRC.Overall,the results of our study demonstrate that TNFAIP6 is differentially linked to the prognosis of different tumor types.Future studies focusing on the relationship between TNFAIP6 expression and prognosis may contribute to the integration of TNFAIP6 as a prognostic biomarker of a number of cancers.
The tumor microenvironment (TME) comprises heterogeneous cell types,including immune cells,endothelial cells,stromal cells,fibroblasts,and cancer cells.[29,30]The TME is widely implicated in cancer development and progression;it influences the complex interactions of genetic and epigenetic changes in cells.[31,32]TNFAIP6 is a TME-related prognostic gene in colon cancer.[33]TNFAIP6,as an inflammation-related secretory protein,has various functions,including control of matrix molecules and immune regulators,as it binds to numerous ligands.[34]Cancer-associated fibroblasts are major components of the TME and participate in tumorigenesis,tumor progression,and prognosis.[35–39]In the present study,we found that TNFAIP6 expression was positively correlated with the immune filtration of CAFs in various types of tumors,namely,BLCA,BRCA,BRCA-basal,BRCA-lumA,BRCA-lumB,CESC,COAD,ESCA,HNSC,HNSC-HPV.,LIHC,LUAD,LUSC,PAAD,PRAD,READ,SARC,STAD,TGCT,THCA,THYM,and UCS.Previous studies have shown that ICP genes play important roles in immunotherapy.[40,41]We found that TNFAIP6 expression is related to several ICP genes in many types of cancers.
We analyzed TNFAIP6-similar genes correlated with TNFAIP6 expression in tumors using Metascape and found that the PID VEGF-VEGFR pathway,organic anion transport,and cobalamin transport were the top 3 enriched clusters.
We selected COAD to detect the expression of TNFAIP6,given that TNFAIP6 is highly expressed in COAD and reduced OS in COAD.The results showed that TNFAIP6 expression was significantly associated with TMB and MSI in COAD,suggesting that TNFAIP6 plays an important role in COAD.
Immunohistochemistry was performed to detect TNFAIP6 expression in normal colon adenomas and colon tissues.The results revealed an increasing trend in TNFAIP6 expression with colon cancer progression.This indicates that TNFAIP6 plays an important role in the pathogenesis of colon cancer.
Antitumor drugs and carcinogens correlated with TNFAIP6 were sorted using the CTD database.Among the drugs that may target TNFAIP6 are both confirmed and potential antitumor drugs,including cisplatin,irinotecan,resveratrol,U 0126,NSC689534,genistein,NSC668394,oxaliplatin,plerixafor,topotecan,vincristine,flutamide,doxorubicin,MRK 003,folic acid,demecolcine,tunicamycin,zoledronic acid,and schizandrin B.In addition,somecarcinogens—asbestos,crocidolite,benzo(a)pyrene,acrylamide,1,2-dimethylhydrazine,glycidol,2,6-dinitrotoluene,naphthalene,furan,dioxins,clofibrate,and aristolochic acid I—may be responsible for the upregulation of TNFAIP6 expression in tumors.Further mechanistic studies are required.
In summary,we performed a comprehensive bioinformatics analysis,utilizing multiple online databases to comprehensively analyze the significance of TNFAIP6 expression across tumor types.In the case of colon cancer,this association was further confirmed by immunohistochemistry of colon cancer tissues.This overview of the role of TNFAIP6 in cancer provides a preliminary scientific basis for future clinical biomarkers or molecularly targeted therapy research.However,the specific regulatory mechanisms of TNFAIP6 in pan-cancer require further investigation.
Acknowledgments
None.
Financial support and sponsorship
This study was funded by the Natural Science Basic Research Program of Shaanxi (no.2023-JC-QN-0876) and Key Research and Development Program of Shaanxi(no.2023-YBSF-447).
Conflicts of interest statement
The authors declare that they have no conflict of interest with regard to the content of this report.
Author contributions
Yang Y.,X.-L.W.,Y.-L.L.,and R.-F.S.conceived and designed the study.Yang Y.,Z.-G.L.,and Z.-G.Z.performed bioinformatics analysis.Yang Y.Q.did the IHC assay.Yang Y.,Y.-L.L.,and R.-F.S.wrote the manuscript.All the authors reviewed the manuscript.
Data availability statement
The data used in the bioinformatics analyses in this study are available at SangerBox(http://sangerbox.com),UALCAN(http://ualcan.path.uab.edu),cBioPortal web(https://www.cbioportal.org),TIMER2.0 database(http://timer.cistrome.org/),GEPIA2(http://gepia2.cancerpku.cn),CancerSEA(http://biocc.hrbmu.edu.cn/CancerSEA/),STRING website (https://string-db.org),Metascape (https://metascape.org),and CTD(http://ctdbase.org/downloads/).
Ethical approval
This study was approved by the Medical Ethics Committee of the College of Medicine at Xi'an Jiaotong University.Informed consent was obtained from all patients.All methods were performed in accordance with relevant guidelines and regulations.
 Oncology and Translational Medicine2024年1期
Oncology and Translational Medicine2024年1期
- Oncology and Translational Medicine的其它文章
- Wan-Yee Joseph Lau:the promoter of modernization and internationalization of Chinese surgery
- In vivo study of the effect of anlotinib on the stemness of the lenvatinib-resistant hepatocellular carcinoma cells and the underlying mechanisms
- Utilizing bioinformatics for integrated analysis of multiple genes in the diagnosis and pathogenesis of metastatic pheochromocytoma and paraganglioma
- Clinical and pathological characteristics and expression of related molecules in patients with airway disseminated lung adenocarcinoma
- Three novel rare TP53 fusion mutations in a patient with multiple primary cancers:a case report
