In vivo study of the effect of anlotinib on the stemness of the lenvatinib-resistant hepatocellular carcinoma cells and the underlying mechanisms
Jing Zhn,Shu Hung,Bi Wei,Zo-Zo Hung,Sheng-Li Yng
Abstract Background:In vivo experiments were conducted to examine the effects of the targeted drug anlotinib on the stemness of hepatocellular carcinoma(HCC)cells and lenvatinib-resistant liver cancer cells and to explore the underlying molecular mechanisms.Methods: A subcutaneous xenograft model of Hep3B-derived HCC was established in nude mice,which were randomly divided into 2 groups(n=5 males per group):(1)intragastric administration of anlotinib(0.4 mg/kg)and(2)intragastric administration of normal saline.We constructed lenvatinib-resistant cell lines and randomly divided the mice into 3 groups(n=5 males per group):(1)intragastric administration of anlotinib,(2)intragastric administration of lenvatinib,and(3)intragastric administration of normal saline.After 2 weeks of treatment,tumor tissues were harvested,and mRNA and proteins were isolated from the tissues.Changes in the expression of cancer stemness markers(epithelial cell adhesion molecule[EpCAM],CD13,CD90,aldehyde dehydrogenase 1[ALDH1],CD44,and CD45),totipotency factors(sex-determining region Y-box 2[Sox2],Nanog,octamer-binding transcription factor 4[Oct4]),and genes related to the Notch signaling pathway were examined.Results:Compared with that in the control group,tumor size and weight were reduced in nude mice treated with anlotinib.These differences were statistically significant in both the types of nude mice.Anlotinib affected stemness markers and totipotency factors by downregulating the expression of CD133,CD90,and G-protein-coupled receptor 5(LGR5)and upregulating the expression of intercellular adhesion molecule 1 (ICAM-1) and Sox2.In addition,lenvatinib-resistant cell lines increased Notch signaling pathway,whereas anlotinib inhibited Notch signaling pathway.Conclusions:The antitumor effect of anlotinib on HCC and lenvatinib-resistant HCC cells may occur through inhibition of the Notch signaling pathway.Anlotinib may be the drug of choice for sequential therapy in lenvatinib-resistant liver cancer.
Keywords: Anlotinib;HCC;Lenvatinib-resistant;Notch signaling pathway
1.Introduction
Hepatocellular carcinoma (HCC),one of the most prevalent malignant carcinomas,is the sixth most common cancer and has the fourth highest rate of tumor-related deaths,with more than 830,000 deaths every year globally.[1]Patients with HCC have a short life span from diagnosis to death,with a median survival time of 9 months and a 3-year survival rate of 12.7%.[2]Several options are available for the treatment of HCC,including surgery,chemotherapy,radiotherapy,liver transplantation,radiofrequency ablation,transcatheter arterial chemoembolization,and selective internal irradiation.However,the therapeutic windows for some treatments such as liver transplantation and radical surgery are narrow.Other treatment options have poor therapeutic efficacies,and patients are highly prone to recurrence and resistance.[3]Therefore,the treatment of liver cancer remains challenging.
Cancer stem cells(CSCs)are involved in persistent proliferation,drug resistance,and tumor recurrence.Liver CSCs are the origins of liver cancer malignance.[4]Cancer stem cells have been identified in human liver cancer tissues and are considered effective therapeutic targets to treat liver cancer.[5]Liver CSCs exhibit a stemness appearance,with increased expression of stemness markers(CD133,CD13,CD90,CD24,CD44,and EpCAM)and potent factors(sexdetermining region Y-box 2[Sox2],Nanog,c-Myc,octamer-binding transcription factor 4[Oct4],sal-like protein 4[SALL4],and forkhead box protein D3[Foxd3]),as well as activation of stemness pathways,including the Notch,Hedgehog,and Hippo signaling pathways.[5]Studies have shown that inhibition of these stemness pathways effectively inhibits tumor proliferation,metastasis,recurrence,and drug resistance.[6]Therefore,these genes and signaling pathways are promising targets for liver cancer treatment.
With growing knowledge of the relationship between genes and diseases,targeted drugs against liver cancer have recently been developed.Lenvatinib,a multityrosine kinase inhibitor,is a first-line targeted drug approved for the treatment of liver cancer.The target genes of lenvatinib include vascular endothelial growth factor receptors 1–3 (VEGFR1–3),fibroblast growth factor receptors 1–4 (FGFR1–4),platelet-derived growth factor receptor a(PDGFRa),RET,and KIT.The patients'survival and progression-free survival(PFS)can be significantly improved by lenvatinib.[7]Kudo et al recruited 1492 patients for a multicenter randomized controlled trial and revealed that lenvatinib has a high overall survival(OS)rate in the first-line treatment of advanced HCC with manageable adverse effects.[8]In addition,this phase III clinical trial demonstrated that the median PFS and time to progression of lenvatinib were longer than those of sorafenib,the first-line targeted drug for HCC,in patients with advanced HCC.[8]Lenvatinib has been approved as a first-line treatment drug in the United States,European Union,Japan,and China since 2018.
However,drug resistance to lenvatinib has been reported,and treatment of patients with lenvatinib-resistant is a challenge.[9]Anlotinib is a novel small-molecule,multitarget tyrosine kinase inhibitor that effectively inhibits multiple targets (eg,VEGFR,PDGFR,FGFR,and c-Kit),tumor angiogenesis,and tumor growth.In China,anlotinib was approved as a third-line treatment for refractory advanced non–small cell lung cancer in May 2018 and as a second-line treatment for advanced soft-tissue sarcoma in June 2019.Studies have demonstrated that anlotinib has encouraging efficacy,a manageable profile,and promising antitumor activity against selected advanced tumor types such as medullary thyroid(PFS rate [at 48 weeks],85.5%;partial response,56.90%),renal cell (median PFS,17.5 vs 16.6 months;median OS,30.9 vs 30.5 months),and esophageal squamous cell carcinoma (median PFS,3.02 vs 1.41 months;median OS:6.11 vs 7.20 months;disease control rate,64.22%vs 18.18%).[10]In this study,we investigated the efficacy of anlotinib against lenvatinib-resistant liver cancer.We also studied whether the stemness of liver cancer cells is a potential target of anlotinib.These experiments were performed in nude mice subcutaneously inoculated with human liver cancer cells(Hep3B-derived HCC cells)with or without lenvatinib resistance.
2.Materials and methods
2.1.Materials
Twenty-five BALB/c nude mice(12 females and 13 males,4–5 weeks old,14–21 g) were housed under specific pathogen-free conditions.All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee of Huazhong University of Science and Technology,Wuhan,China.
The human hepatoma cell line Hep3B (hereafter referred to as Hep3B cells) was purchased from the China Center for Type Culture Collection and stored in liquid nitrogen.The major instruments used in this study included a carbon dioxide(CO2)incubator(Harris Corporation,Melbourne,Florida,USA),fluorescence quantitative polymerase chain reaction (PCR) instrument (Ambion Inc,Austin,Texas,United State),electrophoresis apparatus(Bio-Rad Laboratories,Shanghai,China),and high-speed refrigerated centrifuge(Heraeus Instruments,Wuhan,China).Anlotinib hydrochloride(12 mg/tablet)was provided by Nanjing Sanhome Pharmaceutical Co.Ltd (Nanjing,China).Lenvatinib was purchased from Selleck Chemical (Houston,Tex).RPMI 1640 medium,fetal calf serum,and trypsin were obtained from HyClone Laboratories,Inc(Logan,Utah,USA).Dimethyl sulfoxide (DMSO)was purchased from MP Biomedicals.The TRIzol and SYBR Green PCR Master Mix were purchased from Life Technologies.All PCR primers were designed and synthesized by Life Technology(Carlsbad,California,USA).
2.2.Establishment of a subcutaneous xenograft nude mouse model with human HCC cells
Lenvatinib-resistant Hep3B cells were generated as described previously.[11]After resuscitation,Hep3B cells or lenvatinib-resistant Hep3B cells were cultured in RPMI 1640 medium containing 10%fetal calf serum under an environment of 37°C and 5%CO2.The medium was changed once every 2 to 3 days.The cells were passaged at a 1:3 ratio when they reached 80%to 90%confluence.After 2 passages,cells in the logarithmic growth phase were collected,trypsinized,and counted.Phosphate-buffered saline was added to prepare a cell suspension at a density of 1×107cells/mL.The cell suspension (0.2 mL) was injected into the right side of nude mice using an insulin syringe.
2.3.Animal treatment
After nude mice were inoculated with either Hep3B cells or lenvatinibresistant Hep3B cells,their general condition,liquid absorption,and tumor formation at the inoculated site were observed daily for 19 days.Ten male nude mice inoculated with Hep3B cells were randomly assigned to a control(DMSO 100 μL/20 g)or anlotinib(0.4 mg/kg)group(n=5 per group).Fifteen nude mice inoculated with lenvatinib-resistant Hep3B cells were assigned to a control(DMSO 100 μL/20 g),lenvatinib(30 mg/kg),or anlotinib(0.4 mg/kg)group(n=5 per group).The doses were administered via oral gavage once daily for 18 consecutive days.The first day of dose administration was designated as day 0.
2.4.Clinical observations and tumor size measurements
After inoculation,the animals were monitored daily for clinical signs,food and water intake,and activity levels.The maximum(A)and minimum(B)diameters of the tumors were measured with calipers on days 0,3,6,9,12,15,and 18,as well as 24 hours after the last dose(day 19).Tumor volume was calculated using the following formula:
Tumor volume=(A×B^2)/2.(1)
The growth curves of the transplanted tumors were plotted using GraphPad Prism 7 (GraphPad Software,Inc,San Diego,Calif).Body weight was measured on days 0,3,6,9,12,15,and 18.
2.5.Quantitative PCR analysis of m RNA expression of stemness-related genes in the subcutaneous HCC xenografts
Nude mice were killed on day 19,and HCC xenografts were collected.Total RNA was extracted from HCC tumor tissues using the TRIzol reagent according to the manufacturer's instructions.RNA concentration and purity were determined using an ultraviolet spectrophotometer (Ambion Inc).Equal amounts of mRNA from each animal were reverse-transcribed into complementary DNA.Quantitative PCR was performed to evaluate mRNA expression using the primers described in our previous study.[12]
The quantitative PCR conditions were as follows:predenaturation at 95°C for 30 seconds,40 cycles of denaturation at 95°C for 30 seconds,annealing at 60°C for 30 seconds,elongation at 72°C for 60 seconds,and a final extension at 72°C for 10 minutes.Glyceraldehyde 3-phosphate dehydrogenase was used as an internal reference.Each reaction was performed in triplicates.The relative expression of the target genes was calculated using the 2-ΔΔCtmethod.
2.6.Statistical methods
Data were expressed as mean±SD.Statistical analyses were performed using SPSS 18.0 statistical analysis software(SPSS Inc,Chicago,Ill).Analysis of variance was used to compare 2 groups.Statistical significance was set at P <0.05.
3.Results
3.1.Growth of subcutaneous HCC xenografts
Approximately 3 days after inoculation,the injected materials were well absorbed at the subcutaneous inoculation site(right axillary).Tumor nodules of 3 to 5 mm were observed at the inoculation site.The tumor formation rate was 100%.The small nodules under the skin gradually became irregular,and the surface became increasingly concave or convex.As the tumor volume increased,nude mice became lethargic and showed decreased activity.Their appetite decreased significantly.One week after inoculation,the tumor volume increased to approximately 100 mm3.
In nude mice inoculated with normal Hep3B cells,the tumor volumes were smaller in the anlotinib group than the controls during the treatment period(Figure 1A),indicating that the growth of subcutaneous xenografts in the anlotinib group was slower than that in the controls.In addition,the anlotinib group had steady body weights during the treatment period,but their body weights were significantly lower than those of the control group(Figure 1B).The final tumor size(Figure 1C)and weight(Figure 1D)in the anlotinib group were significantly smaller than those in the control group.

Figure 1.In the nude mice inoculated with normal Hep3B cells,the tumor volumes in the anlotinib group were significantly smaller than controls during the treatment period(A),indicating that the growth of the subcutaneous xenografts in the anlotinib group was slower than in the controls.Also,the anlotinib group had steady body weights during the treatment period,but their body weights were significantly lower compared with controls (B).The final tumor size (C) and weight (D) in the anlotinib group were significantly smaller compared with controls.*P <0.05.
In nude mice inoculated with lenvatinib-resistant Hep3B cells,the results showed that the growth of subcutaneous xenografts in mice treated with lenvatinib was slightly slower than that in controls(Figures 2A,B),but the difference was not statistically significant.Compared with those in the control and lenvatinib groups,the tumor volumes in the anlotinib group were significantly smaller during the treatment period(Figure 2B),indicating that the growth of the subcutaneous xenografts in the anlotinib group was slower.In the final excised tumors(Figure 2C),tumor weights in the anlotinib group were significantly lower than those in the control and lenvatinib groups(Figure 2D).

Figure 2.In nude mice inoculated with lenvatinib-resistant Hep3B cells,the anlotinib group showed slower growth of subcutaneous xenografts (A) and significantly smaller tumor volumes (B).In the final excised tumors (C),tumor weights in the anlotinib group were significantly lower than those in the control and lenvatinib groups(D).*P <0.05.
3.2.mRNA expression of stemness-related genes in the subcutaneous HCC xenografts in nude mice
In the nude mice inoculated with Hep3B cells,the effects of anlotinib treatment on the mRNA expression of liver CSC surface marker–related genes are shown in Figure 3.Compared with that in the controls,anlotinib increased the mRNA expression of ICAM-1 and decreased the mRNA expression of CD133,CD90,and LGR5.No changes were observed in the expression of CD13,CK19,CD34,Notch ligand 1(DLK1),CD45,CD24,CD47,ATP-binding cassette subfamily G member 2(ABCG2),CD44,EpCAM,ALDH1A1,or cytokeratin 7(CK7)after anlotinib treatment.

Figure 3.In nude mice inoculated with Hep3B cells,the administration of anlotinib(0.4 mg/kg)significantly increased the mRNA expression of ICAM-1 and decreased the mRNA expression of CD133,CD90,and LGR5 compared with the controls.No changes were observed in the expression of ABCG2,ALDH1A1,CD13,CD24,CD34,CD44,CD45,CD47,CK7,CK19,Notch ligand 1(DLK1),or Ep CAM after anlotinib treatment.*P <0.05.ABCG2,ATP-binding cassette subfamily G member 2;CK7,cytokeratin 7;CK19,cytokeratin 19;Ep CAM,epithelial cell adhesion molecule;ICAM-1,intercellular adhesion molecule 1.
In the nude mice inoculated with Hep3B cells,anlotinib on the mRNA expression of totipotency-related genes is shown in Figure 4.Anlotinib treatment resulted in the upregulation of Sox2 compared with controls.No changes were observed in the expression of SALL4,Nanog,c-Myc,Foxd,or Oct4 following anlotinib treatment.

Figure 4.In the nude mice inoculated with Hep3B cells,administration of anlotinib(0.4 mg/kg)significantly increased the mRNA expression of Sox2 compared with controls.No changes were observed in the expression of c-Myc,Foxd,NANOG,Oct4,or SALL4 following anlotinib treatment.*P <0.05.Foxd,forkhead box protein D3;Oct4,octamer-binding transcription factor4;SALL4,sal-likeprotein 4;Sox2,sex-determiningregion Y-box2.
Anlotinib treatment resulted in the upregulation of Sox2,with no changes in the expression of SALL4,Nanog,c-Myc,forkhead box protein D3(Foxd),or Oct4(P <0.05).
The effects of anlotinib on the mRNA expression of genes related to the Notch signaling pathway are shown in Figure 5.Anlotinib treatment led to decreased expression of Notch3,Notch4,Hes1,Hey1,and Hey5;increased expression of HeyL and Jagged (Jag)1;and no expression changes in Delta-like(DLL)1,DLL3,DLL4,Ngn,NOTCH1,NOTCH2,Jag2,or Hes5.
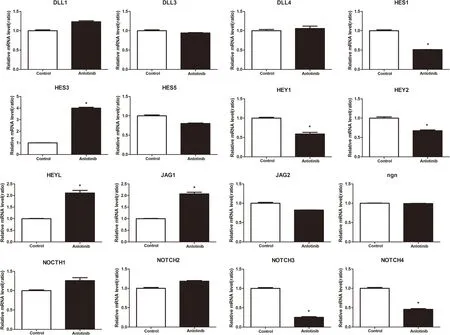
Figure 5.Anlotinib treatment led to decreased expression of Notch3,Notch4,Hes1,Hey1,and Hey5,increased expression of HeyL and Jagged(Jag)1,and no expression changed in Delta-like(DLL)1,DLL3,DLL4,Ngn,NOTCH1,NOTCH2,Jag2,or Hes5.*P <0.05.
Real-time PCR expression of HCC stem cell surface marker–related genes in lenvatinib-resistant Hep3B cells is shown in Figure 6.Compared with the control group,lenvatinib resistance resulted in upregulated expression of CD133,CD13,CK19,CD34,ICAM-1,LGR5,and EpCAM;downregulation of CD90 and ABCG2;and no changes in the expressionof DLK1,CD45,CD24,CD47,CD44,ALDH1A1,and CK7.
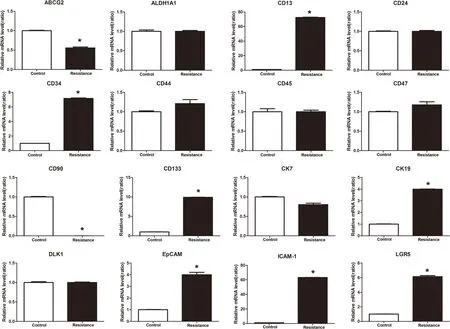
Figure 6.Compared with the control group,lenvatinib resistance resulted in upregulated expression of CD133,CD13,CK19,CD34,ICAM-1,LGR5,and Ep CAM;downregulation of CD90 and ABCG2;and no changes in the expression of DLK1,CD45,CD24,CD47,CD44,ALDH1A1,and CK7.*P <0.05.ALDH1A1,aldehyde dehydrogenase 1A1;EpCAM,epithelial cell adhesion molecule;ICAM-1,intercellular adhesion molecule 1;LGR5,G-protein-coupled receptor 5.
The mRNA expression levels of totipotency-related genes in the lenvatinib-resistant group are shown in Figure 7.Compared with the control group,the lenvatinib-resistant group showed upregulated Sox2 and downregulated SALL4 expression,with no statistically significant difference in the expression of Nanog,c-Myc,Foxd,and Oct4 between the 2 groups.

Figure 7.Compared with the control group,the lenvatinib-resistant group showed upregulated Sox2 and downregulated SALL4 expression,and there was no statistically significant difference in the expression of Nanog,c-Myc,Foxd,and Oct4 between the 2 groups.*P <0.05.Foxd,forkhead box protein D3;Oct4,octamer-binding transcription factor 4;Sox2,sex-determining region Y-box 2.
For the NOTCH signaling pathway–related genes,which is shown in Figure 8,compared with the control group,the lenvatinib-resistant group can downregulate the Hey2 gene and upregulate NOTCH1,NOTCH2,NOTCH3,NOTCH4,DLL1,DLL3,DLL4,Hes1,Hes3,Hes5,HeyL,Ngn,and Jag2,and the expression of jag1,Hey1 was not significantly different from that in the control group.
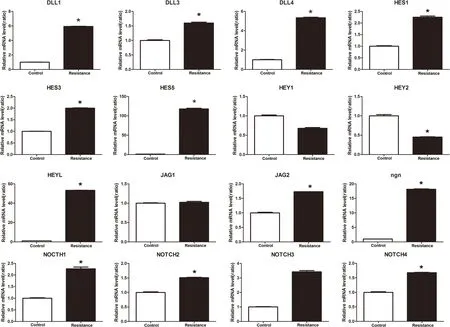
Figure 8.Compared with the control group,the lenvatinib-resistant group can downregulate the Hey2 gene and upregulate NOTCH1,NOTCH2,NOTCH3,NOTCH4,DLL1,DLL3,DLL4,Hes1,Hes3,Hes5,HeyL,Ngn,and Jag2,and the expression of jag1,Hey1 was not significantly different from the control group.*P <0.05.DLL,Delta-like.
4.Discussion
Anlotinib is a novel tyrosine kinase inhibitor with significant antitumor effects.ALTER-0303 is a randomized,double-blind,placebo-controlled,multicenter,phase III trial in which lenvatinib improved the progression of second-line drugs in patients with cell lung cancer compared with placebo.Progression-free survival was 5.37 versus 1.40 months (P <0.0001),and OS was 9.63 versus 6.30 months(P <0.0001).Therefore,anlotinib has been approved by the China Food and Drug Administration as a monotherapy for the third-line treatment of patients with advanced non–small cell lung cancer.[13]A randomized phase IIB trial showed that anlotinib significantly prolonged the PFS and OS in patients with advanced soft-tissue sarcoma.[14]Anlotinib is effective in patients with advanced medullary thyroid and metastatic renal cell carcinoma.[15]He et al[16]showed that anlotinib induces apoptosis in liver cancer cells and is a promising drug for liver cancer treatment.This is consistent with the results of our study,which showed that anlotinib inhibits liver cancer proliferation.
With the development of tyrosine kinase inhibitors,the emergence of resistance to tyrosine kinase inhibitors has become a clinically challenging problem.Cancer stem cells are the root cause of proliferation,metastasis,recurrence,and drug resistance in liver cancer.Cancer stem cells are endogenously resistant to chemotherapy,radiation,and drugs that have not been used in cancer therapy and may develop resistance.[17]There is no doubt that resistance is currently the main cause of failure in the treatment of liver cancer.Cancer stem cells participate in liver cancer resistance in the following ways:(1)epithelial-to-mesenchymal transition(EMT):cells undergoing EMT may enter a quiescent state,at which time the cells no longer divide,and antitumor drugs act on the proliferating cells;(2) ATP-binding cassette transporter: drugs are transported from the cell membrane to the extracellular membrane;(3)hypoxia:hypoxia can maintain the dry characteristics of CSCs and promote the expression of EMT.Wnt Hedgehog and Notch signaling pathways,as well as stem cell markers,such as CD133,Nanog,and Sox2,are elevated under hypoxic conditions.[18]Lenvatinib is the first-line treatment for liver cancer,and we speculate that its resistance mechanism is similar to that of other tyrosine kinase inhibitors.
Notch signaling is a breast oncogene,a prognostic biomarker,which promotes the activation of genes required for EMT and contributes to the resistance of breast cancer treatments.[19]NOTCH1-activating mutations occur in 60%of the cases,and high expression of Notch signaling is associated with worse outcomes.NOTCH-related resistance includes chemotherapy and targeted therapies.[20]These results are consistent with those of other studies.In this study,the Notch signaling pathway was activated in the subcutaneous xenograft tissues of the lenvatinib-resistant group.Many studies directed that anlotinib can treat HCC.[21,22]However,there have been few studies on the mechanism of action of anlotinib in hepatoma.It has been observed that anlotinib induces HCC apoptosis and inhibits proliferation via the ERK and Akt pathways.[16]
In our study,subcutaneous HCC xenografts were inhibited by anlotinib treatment.However,in our study,lenvatinib-resistant subcutaneous xenografts of hepatic carcinoma treated with anlotinib exhibited good antitumor effects.Jin et al[23]indicated that the inhibition of the epidermal growth factor receptor is synthetically lethal with lenvatinib in liver cancer by treating 12 patients with advanced HCC who were unresponsive to lenvatinib treatment with a combination of lenvatinib and gefitinib,resulting in meaningful clinical responses.Lenvatinib is a small-molecule multitarget tyrosine kinase inhibitor,which can effectively inhibit VEGFR,FGFR,PDGFR,and c-Kit.It can also inhibit tumor angiogenesis and growth.Hepatocellular carcinoma cells with high FGFR1 expression were resistant to lenvatinib.Overexpression of FGFR1 induces the activation of AKT/mTOR/ERK signaling in HCC cells,leading to lenvatinib resistance,whereas downregulation of FGFR1 and its downstream AKT/mTOR/ERK expression inhibitors restores the sensitivity of HCC cells to lenvatinib.[24]Several studies have shown that HCC CSCs led to lenvatinib resistance.[25–27]In our study,we found that the genes of the Notch signaling pathway,NOTCH3,NOTCH4,Hes1,Hey1,and Hey2,were downregulated;only 1 gene,Heyl,was upregulated,and other genes showed no significant differences between the 2 groups.This indicated that anlotinib exerts its antitumor effects mainly by inhibiting the Notch signaling pathway.To date,no study has indicated that the antitumor effects of anlotinib are achieved through the Notch signaling pathway.
In summary,this study found that anlotinib resistance and lenvatinib resistance act together on a part of liver CSC“stemness”-related genes and Notch signaling pathway,which is targeted for liver cancer.The clinical application of sequential therapy has laid the theoretical foundation and indicated the direction for the follow-up treatment of tyrosine kinase inhibitor lenvatinib,which is already well known.
Acknowledgments
Not applicable.
Financial support and sponsorship
This study was supported by the Natural Science Foundation of the Hubei Province(no.2023 AFB894)and Open for the Key Laboratory of Biological Targeted Therapy in 2021(no.2021swbx019).
Conflicts of interest statement
S.L.Yang is an editorial board member of Oncology and Translational Medicine.This article is subject to the journal's standard procedures,with peer review handled independently of the relevant editorial board member and his/her research groups.
Author contributions
S.L.Yang and S.Huang designed this research;J.Zhan completed most of the experiments and wrote the manuscript;Z.Z.Huang's job involved gathering and analyzing data;and B.Wei was responsible for language modification.
Data availability statement
All data generated or analyzed in this study are included in the published article.
Ethical approval
Studies involving animals were reviewed and approved by the ethics committee of Union Hospital.
 Oncology and Translational Medicine2024年1期
Oncology and Translational Medicine2024年1期
- Oncology and Translational Medicine的其它文章
- Wan-Yee Joseph Lau:the promoter of modernization and internationalization of Chinese surgery
- Utilizing bioinformatics for integrated analysis of multiple genes in the diagnosis and pathogenesis of metastatic pheochromocytoma and paraganglioma
- Role of tumor necrosis factor alpha-induced protein 6(TNFAIP6)in tumors:a pan-cancer analysis
- Clinical and pathological characteristics and expression of related molecules in patients with airway disseminated lung adenocarcinoma
- Three novel rare TP53 fusion mutations in a patient with multiple primary cancers:a case report
