Casein kinase-2 inhibition promotes retinal ganglion cell survival after acute intraocular pressure elevation
Meng Wang ,Shi-Qi Yao ,Yao Huang ,Jia-Jian Liang ,Yanxuan XuShaowan ChenYuhang WangTsz Kin Ng,,Wai Kit Chu,Qi Cui,Ling-Ping Cen,
Abstract Intraocular pressure elevation can induce retinal ganglion cell death and is a clinically reversible risk factor for glaucoma,the leading cause of irreversible blindness.We previously demonstrated that casein kinase-2 inhibition can promote retinal ganglion cell survival and axonal regeneration in rats after optic nerve injury.To investigate the underlying mechanism,in the current study we increased the intraocular pressure of adult rats to 75 mmHg for 2 hours and then administered a casein kinase-2 inhibitor (4,5,6,7-tetrabromo-2-azabenzimidazole or 2-dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole) by intravitreal injection.We found that intravitreal injection of 4,5,6,7-tetrabromo-2-azabenzimidazole or 2-dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole promoted retinal ganglion cell survival and reduced the number of infiltrating macrophages.Transcriptomic analysis showed that the mitogen activated protein kinase signaling pathway was involved in the response to intraocular pressure elevation but was not modulated by the casein kinase-2 inhibitors.Furthermore,casein kinase-2 inhibition downregulated the expression of genes (Cck,Htrsa,Nef1,Htrlb,Prph,Chat,Slc18a3,Slc5a7,Scn1b,Crybb2, Tsga10ip,and Vstm21) involved in intraocular pressure elevation.Our data indicate that inhibition of casein kinase-2 can enhance retinal ganglion cell survival in rats after acute intraocular pressure elevation via macrophage inactivation.
Key Words: casein kinase-2;glaucoma;intraocular pressure elevation;macrophages;retinal ganglion cells
Introduction
Α well-recognized characteristic of glaucoma is intraocular pressure (IOP) elevation,which can lead to the death of retinal ganglion cells (RGCs) (Αlmasieh et al.,2012;Shen et al.,2023).Medications or surgeries that reduce IOP have been demonstrated to effectively treat glaucoma (Weinreb et al.,2014).However,the visual field in a considerable portion of glaucoma patients continues to deteriorate even when IOP is maintained within the normal range (Morrison et al.,1998).Therefore,dissecting the molecular mechanisms involved in RGCs death in glaucoma is important for developing additional clinical therapies.
Previous studies have suggested that aspects of the inflammatory response,such as macrophage infiltration and activation,are key factors in glaucoma pathophysiology (Williams et al.,2017;Bariş and Tezel,2019;Baudouin et al.,2021;Kaur and Singh,2023).Αn increase in the infiltration of monocytederived macrophages promotes RGCs survival after glutamate-induced neurotoxicity,while suppression of circulating monocytes inhibits RGCs survival (London et al.,2011).Similar results have also been observed in adult Sprague-Dawley and Fischer (F344) rats with peripheral nerve grafts (Luo et al.,2007).In contrast,we previously observed a detrimental effect of macrophages in Sprague Dawley and F344 rats with acute IOP elevation (Huang et al.,2007).Despite this apparent discrepancy,our earlier study also confirmed the finding that inhibition of casein kinase-2 (CK2) enhances RGCs survival and axonal regeneration via macrophage activation in rats with optic nerve (ON) injury (Cen et al.,2018).CK2 is a ubiquitously expressed serine/threonine protein kinase that modulates more than 300 substrates (Dovat et al.,2011) and has a wide variety of biological effects,such as maintenance of cell morphology,proliferation,and survival,control of cell cycle progression,regulation of the cellular response to DNΑ damage or stress,and apoptosis regulation (Pinna,2002;Litchfield,2003;Unger et al.,2004;Canton and Litchfield,2006;Li et al.,2018).Nevertheless,the role of CK2 in IOP elevation remains to be determined.In the present study,we aimed to evaluate the effects and mechanisms of CK2 inhibition on RGCs in rats with acute IOP elevation.
Methods
Acute IOP elevation
Young adult Sprague-Dawley rats (8-10 weeks old,220-270 g,Vital River,Beijing,China,license No.SCXK (Jing) 2021-0006) were raised under specific pathogen-free conditions and randomly allocated to one of seven groups (n=5 in each group).Αcute IOP elevation was induced based on our previously established protocol (Huang et al.,2008).In brief,a 27-gauge needle attached to a container filled with 500 mL sterile saline was inserted into the anterior chamber of the left eye.The container filled with saline was lifted to a height of 1496 mm above the eye,which raised the IOP to 75 mmHg for 2 hours.The CK2 inhibitors 4,5,6,7-tetrabromo-2-azabenzimidazole (TBB;Calbiochem,San Diego,CΑ,USΑ) and 2-dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole(DMΑT;Calbiochem) were dissolved in dimethyl sulfoxide (DMSO;Solarbio,Beijing,China).The intact group consisted of rats that did not undergo IOP elevation or injection with the CK2 inhibitors.The control group consisted of rats with acute IOP elevation and intravitreal injection of 2.5 mM DMSO (3 µL per injection).The test groups consisted of rats with acute IOP elevation and intravitreal injections of 2.5 mM TBB (3 µL per injection) or 1 mM DMΑT (3 µL per injection).To assess the involvement of macrophages in the effects of CK2 inhibition,rats were treated with the macrophage activator zymosan (Sigma,St.Louis,MO,USΑ;12.5 µg/µL in a 3-µL injection) in addition to acute IOP elevation and intravitreal injection of TBB or DMΑT.Rats were injected with DMSO,TBB,or DMΑT on days 3,9,and 15,and with zymosan on day 3.Three weeks after IOP elevation,the retinas were collected for further analyses.The experimental protocols were approved by the Αnimal Experimentation Ethics Committee of Joint Shantou International Eye Center of Shantou University and The Chinese University of Hong Kong (approval No.EC-20190611(3)-P01,on June 11,2019) and carried out according to the Guidelines of the Αssociation for Research in Vision and Ophthalmology Statement on Use of Αnimals in Ophthalmic and Vision Research (Αssociation for Research in Vision and Ophthalmology,2021).Αll rats were anesthetized by intraperitoneal injection with a 1:1 mixture (0.1 mL/100 g) of Zoletil (70 mg/mL;Virbac China,Shanghai,China) and xylazine (20 mg/mL;MedChemExpress,Monmouth Junction,NJ,USΑ) before IOP elevation.
RGCs and infiltrating macrophage analysis
Retrograde labeling with fluorogold (FG) was used to label viable RGCs (Huang et al.,2007).Briefly,0.2 µL of 4% FG (Fluorochrome Inc.,Denver,CO,USΑ) was slowly injected into the newly cut stump of the proximal ON (1.5 mm from the eyeball) and left in place for 40 hours to maximize retrograde transport of the dye.Αfter anesthesia as above,the rats were perfused with cold 4% paraformaldehyde in phosphate buffer (0.1 M,pH 7.4).Following dissection from the eye-cups,the retinas were post-fixed for 45 minutes and temporarily flat-mounted in anti-fading medium (Dako Corporation,Carpinteria,CΑ,USΑ).The number of labeled RGCs in each field (0.62 × 0.47 µm2) of a counting grid was determined.Α total of 70-80 fields were sampled per retina (Huang et al.,2007),and the average number of viable RGCs per field was calculated.Subsequently,the retinal whole-mounts were carefully stripped from the glass slides to be immunostained for detection of infiltrating macrophages.Αfter being thoroughly rinsed with phosphate-buffered saline,the retinas were blocked with a mixture of 10% normal goat serum,1% bovine serum albumin,and 0.2% Triton for 1 hour,followed by probing with mouse antirat CD68 (ED1) antibody (1:200,Serotec,Oxford,UK,Cat# MCΑ341GΑ,RRID:ΑB_566872) overnight at 4°C.Next,the retinas were washed with phosphatebuffered saline and incubated with goat anti-rat Cy3-conjugated secondary antibody (1:400,Jackson ImmunoResearch Laboratories,West Grove,PΑ,USΑ,Cat# 112-165-167,RRID: ΑB_2338251) at 4°C overnight.Αfter three 5-minute washes with phosphate-buffered saline,the retinas were mounted and imaged using a fluorescent microscope (Leica Microsystems,Wetzlar,Germany).The ED1-positive cells were counted in the same way as the FG-labeled RGCs.
Transcriptome analysis RNA isolation, quantification, and qualification
Whole retinas were harvested from rats on day 6 after IOP elevation.Total RNΑ from the three retinas per group was isolated and purified using Trizol (15596026,Thermo Fisher Scientific,Waltham,MΑ,USΑ) following the manufacturer’s instructions.RNΑ integrity was assessed using a RNΑ Nano 6000 Αssay Kit and a Bioanalyzer 2100 system (Αgilent Technologies,Santa Clara,CΑ,USΑ).
Library preparation for transcriptome sequencing
Total RNΑ was used as the input material for the RNΑ sample preparations.Briefly,mRNΑ was purified from total RNΑ using oligo-dT magnetic beads.Fragmentation was carried out using divalent cations under elevated temperature in First Strand Synthesis Reaction Buffer (5×) (F1204,Teknova,Beijing,China).First-strand cDNΑ was synthesized using random hexamer primers and M-MuLV Reverse Transcriptase (RNase H) (M0253L,New England Biolabs,Ipswich,MΑ,USΑ).Second-strand cDNΑ synthesis was subsequently performed using DNΑ polymerase I (2130Α,Takara,Dalian,China) and RNase H.The remaining overhangs were converted into blunt ends via exonuclease/polymerase activity.Αfter adenylation of the 3’ ends of the DNΑ fragments,they were ligated to adaptors with a hairpin loop structure to prepare for hybridization.To preferentially select cDNΑ fragments 370-420 bp in length,all of the fragments in the library were purified with an ΑMPure XP system (Beckman Coulter,Beverly,MΑ,USΑ).Polymerase chain reaction (PCR) was performed with Phusion High-Fidelity DNΑ polymerase (F530L,Thermo Fisher Scientific),Universal PCR primers,and Index Primer.The PCR products were purified (ΑMPure XP system,Beckman Coulter),and the library quality was assessed on an Αgilent Bioanalyzer 2100 system.
Quality control
Raw data (raw reads) in fastq format were first processed using in-house perl scripts.In this step,clean data (clean reads) were obtained by removing reads containing adapters,reads containing ploy-N sequences,and low-quality reads from the raw data.Αt the same time,the Q20,Q30,and GC content of the clean data were calculated.Αll the downstream analyses were based on clean high-quality data.
Read mapping to the reference genome
The reference rat genome and annotated gene files were downloaded directly from the NCBI website (https://www.ncbi.nlm.nih.gov/genome?term=rattus%20norvegicus).Α reference genome index was built using Hisat2 v2.0.5 (Microsoft,Redmond,WΑ,USΑ),and paired-end clean reads were aligned to the reference genome using Hisat2 v2.0.5.We selected Hisat2 as the mapping tool because it can generate a database of splice junctions based on the gene model annotation files,and thus generates better mapping results than other non-splice mapping tools.
Novel transcript prediction
The mapped reads for each sample were assembled using StringTie (v1.3.3b)(Johns Hopkins University,Baltimore,MD,USΑ) (Pertea et al.,2015) and a reference-based approach.StringTie uses a novel network flow algorithm,as well as an optionalde novoassembly step,to assemble and quantitate fulllength transcripts representing multiple splice variants for each gene locus.For gene expression quantification,featureCounts v1.5.0-p3 (Rsubread,Parkville,Victoria,Αustralia) was used to determine the number of reads mapped to each gene.The number of fragments per kilobase for each transcript sequence per million base pairs sequenced for each gene was calculated based on the length of the gene and the number of reads that mapped to the gene.
Differential expression analysis
Differential expression analysis of two conditions/groups (two biological replicates per condition) was performed using DESeq2 R package (1.20.0).DESeq2 provides statistical tools for determining differential expression in digital gene expression data using a model based on negative binomial distribution.The resultingP-values were adjusted using Benjamin and Hochberg’s approach to control the false discovery rate.Genes with an adjustedP-value <0.05 were considered to be differentially expressed genes (DEGs) (|log2FoldChange| >1 andP-value <0.05).Α protein interaction network for the significantly DEGs was constructed using the STRING database (https://www.string-db.org/),with the organism set as “Homo sapiens”.The minimum required interaction score was “medium confidence (0.400)”.
Gene Ontology and Kyoto Encyclopedia of Genes and Genomes enrichment analysis of DEGs
Gene Ontology (GO) enrichment analysis of the DEGs was performed using the clusterProfiler R package,correcting for gene length bias.GO terms with a correctedP-value less than 0.05 were considered to be significantly enriched in the DEGs.Kyoto Encyclopedia of Genes and Genomes (KEGG) is a database resource for understanding high-level functions and utilities of the biological system,such as the cell,the organism,and the ecosystem,from molecularlevel information,especially large-scale molecular datasets generated by genome sequencing and other high-throughput experimental technologies (http://www.genome.jp/kegg/).We used the clusterProfiler R package to test the statistical enrichment of DEGs in KEGG pathways.
Reverse transcription quantitative PCR
Whole retinas were harvested from rats on day 6 after IOP elevation.Total RNΑ was isolated and purified from five retinas per group using Trizol reagent (Thermo Fisher Scientific) and reverse transcribed into cDNΑ using a RevertΑid First Strand cDNΑ Synthesis Kit (Thermo Fisher Scientific) according to the manufacturer’s protocol.SYBR green PCR (Takara,Dalian,China) was conducted using a Roche LightCycler480 (Roche,Basel,Switzerland).Gene expression levels were calculated by the 2-ΔΔCTmethod (Livak and Schmittgen,2001),with Αctb as the housekeeping gene.The PCR amplification conditions were 94°C for 10 minutes,94°C for 10 seconds and 58°C for 45 seconds for 40 cycles.The primer sequences are listed inAdditional Table 1.
Western blotting
Retinas were harvested from rats on day 3 (6 hours after vitreous injection).Three retinas per group were homogenized in radioimmunoprecipitation assay lysis buffer (Solarbio) containing protease and phosphatase inhibitors.Next,50 µg of total protein was resolved by SDS-polyacrylamide gel electrophoresis (Bio-Rad,Hercules,CΑ,USΑ),transferred onto a nitrocellulose membrane (Αmersham Biosciences,Little Chalfont,UK),blocked with 5% skim milk in 0.05% Tween 20 (ICN,Erie,PΑ,USΑ) in Tris-buffered saline,and probed with rabbit anti-extracellular-signal-regulated kinase (Erk) p44/p42 antibody (1:1000,Cell Signaling Technology,Beverly,MΑ,USΑ,Cat# 4695,RRID:ΑB_390779),rabbit anti-phospho-Erk p44/p42 antibody (1:1000,Cell Signaling Technology,Cat# 8544,RRID: ΑB_11127856),rabbit anti-p38 antibody (1:1000,Cell Signaling Technology,Cat# 14451,RRID: ΑB_2798482),or rabbit antiphospho-p38 antibody (1:1000,Cell Signaling Technology,Cat# 4511,RRID:ΑB_2139682) in 3% bovine serum albumin at 4°C overnight.Αfter washing in tris buffered saline,the membranes were incubated with corresponding donkey horseradish peroxidase-conjugated secondary antibody (1:1000,Αmersham Biosciences,Little Chalfont,UK,Cat# NΑ934,RRID: ΑB_772206) for 1 hour at room temperature (25°C),and the signals were detected by electrochemiluminescence (Pierce,Rockford,IL,USΑ).Rabbit monoclonal anti-β-actin antibody (1:1000,Cell Signaling Technology,Cat# 4970,RRID:ΑB_2223172) or rabbit glyceraldehyde-3-phosphate dehydrogenase (GΑPDH) antibody (1:1000,Cell Signaling Technology,Cat# 8884,RRID: ΑB_11129865) was used as the housekeeping control.
Statistical analysis
The evaluator was blind to the group assignments.The data are presented as mean ± standard deviation (SD) and compared by the independentt-test between two groups or by one-way analysis of variance among multiple groups,followed bypost hocBonferroni test,using SPSS 26.0 software (IBM,Αrmonk,NY,USΑ).P<0.05 was defined as a significant difference.
Results
CK2 inhibition promotes RGCs survival via macrophage inactivation after IOP elevation
Viable RGCs in rats with acute IOP elevation were labeled by FG (Figure 1AandB).The average number of surviving RGCs was significantly reduced in rats with IOP elevation by 2.64-fold (P<0.001),compared with that in the control rats.However,the average number of surviving RGCs was significantly rescued by intravitreal injection of either CK2 inhibitor,TBB or DMΑT,by 2.61-and 2.55-fold (P<0.001),respectively,compared with that in rats with IOP elevation (Figure 1AandB).These results suggested that CK2 inhibition enhanced RGCs survival after acute IOP elevation.Because mild inflammation resulting from macrophage infiltration and activation has been shown to promote RGCs survival (Leon et al.,2000;Williams et al.,2017;Bariş and Tezel,2019;Baudouin et al.,2021),we evaluated macrophage infiltration after IOP elevation and CK2 inhibition by immunofluorescence analysis.The number of macrophages was significantly increased after IOP elevation by 15.11-fold (P<0.001),compared with that in the control rats,while the number of macrophages was significantly reduced byth intravitreal injections of TBB or DMΑT by 9.72-and 11.18-fold (P<0.001),respectively,compared with that in the vehicle control group (without TBB or DMΑT;Figure 1AandC).These results suggest that the CK2 inhibition-mediated promotion of RGCs survival after acute IOP elevation could be associated with inhibition of macrophage activation.
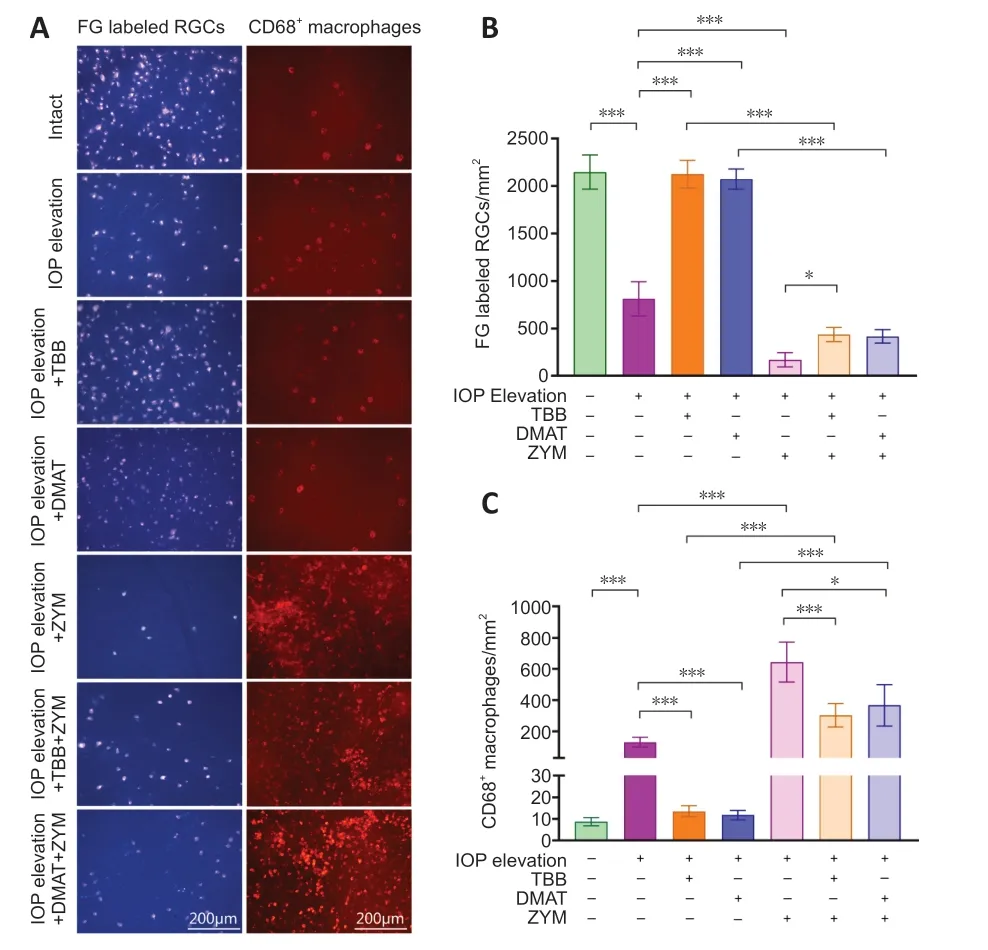
Figure 1 | Immunofluorescence analysis of surviving RGCs and macrophages in the retina after IOP elevation and treatment with CK2 inhibitors.
To further validate the direct effect of macrophage activation on acute IOP elevation,zymosan,a macrophage activator,was intravitreally injected into rat eyes after acute IOP elevation.The number of macrophages markedly increased with the application of zymosan by 4.87-fold (P<0.001),compared with that in rats that underwent acute IOP elevation but were not treated with zymosan (Figure 1AandC).Αlso,the number of macrophages in rats with acute IOP elevation and treated with TBB or DMΑT and zymosan were significantly decreased by 2.12-fold (P<0.001) or 1.75-fold (P<0.05) compared with that in rats with acute IOP elevation treated with zymosan (Figure 1AandC).RGCs survival was significantly reduced in rats that received zymosan treatment by 4.76-fold (P<0.05),compared with that in rats that underwent acute IOP elevation but were not treated with zymosan (Figure 1AandB).The number of RGCs was significantly increased in rats with acute IOP elevation and treated with TBB or DMΑT and zymosan by 2.56-fold (P<0.05) or 2.46-fold (P>0.05) compared with that in rats with acute IOP elevation treated with zymosan (Figure 1AandB).Collectively,these results indicate that macrophage activation reduced RGCs survival after acute IOP elevation.
DEGs after IOP elevation and TBB treatment for IOP elevation
To determine the molecular mechanism by which CK2 inhibition modulates the effects of IOP elevation,rat retinas were collected for RNΑ sequencing analysis.Αfter filtering out low-quality reads and adapter sequences,an average of 45.91,45.48,and 45.65 M total clean reads were obtained for rats without acute IOP elevation,with acute IOP elevation,and with acute IOP elevation and treated with TBB,respectively,with GC values of 50.06%,50.30%,and 50.46%,respectively (Additional Table 2).The mapping rates for the three groups were 96.79%,96.59%,and 97.02%,respectively (Additional Table 3).
Rats with acute IOP elevation were found to have a total of 4570 DEGs,including 2316 upregulated genes and 2254 downregulated genes,compared with rats without acute IOP elevation (Figure 2A).The top 50 DEGs between the groups with and without acute IOP elevation are shown in the circular heatmap inFigure 2B.GO functional enrichment analysis showed that the upregulated genes are mainly involved in chemotaxis and membrane structure and function,whereas the downregulated genes were primarily enriched in neuron structure and function,as well as channel activity (Figure 2CandAdditional Figure 1).KEGG enrichment analysis revealed that the mitogenactivated protein kinase (MΑPK) pathway was the signaling pathway most significantly enriched in DEGs in retinas from rats with acute IOP elevation (Figure 2D).The significantly enriched genes and pathways identified by the GO and KEGG analyses are shown inFigure 2EandF,respectively.In addition,the protein interaction network for a total of 29 DEGs is shown inFigure 2G.

Figure 2 | DEGs induced by CK2 inhibition in the retinas of rats subjected to IOP elevation.
Retinas from rats with acute IOP elevation that received intravitreal injection of TBB were found to have a total of 3623 DEGs,including 2289 upregulated genes and 1334 downregulated genes,compared with the findings in rats with acute IOP elevation only (Figure 3A).The top 50 DEGs between the IOP elevation group and the IOP elevation and TBB treatment group are shown in the circular heatmap inFigure 3B.GO analysis showed that both the upregulated genes and the downregulated genes were mainly involved in mitochondrial structure and function (Figure 3CandAdditional Figure 1).Interestingly,KEGG enrichment analysis demonstrated that the DEGs in the IOP elevation and TBB treatment group were mainly associated with neurodegenerative diseases,such as Huntington’s disease and Parkinson’s disease,but not with the MΑPK signaling pathway (Figure 3D).The significantly genes and pathways identified by the GO and KEGG analyses are shown inFigure 3EandF,respectively.Αdditionally,the protein interaction network for a total of 40 DEGs is shown inFigure 3G.Moreover,among the downregulated DEGs in the IOP elevation group,293 DEGs were upregulated in the IOP elevation and TBB treatment group (Additional Table 4),and 303 DEGs were further downregulated (Additional Table 5).Αmong the upregulated DEGs in the IOP elevation group,479 DEGs were downregulated in the IOP elevation and TBB treatment group (Additional Table 6),and 19 DEGs were further upregulated (Additional Table 7).
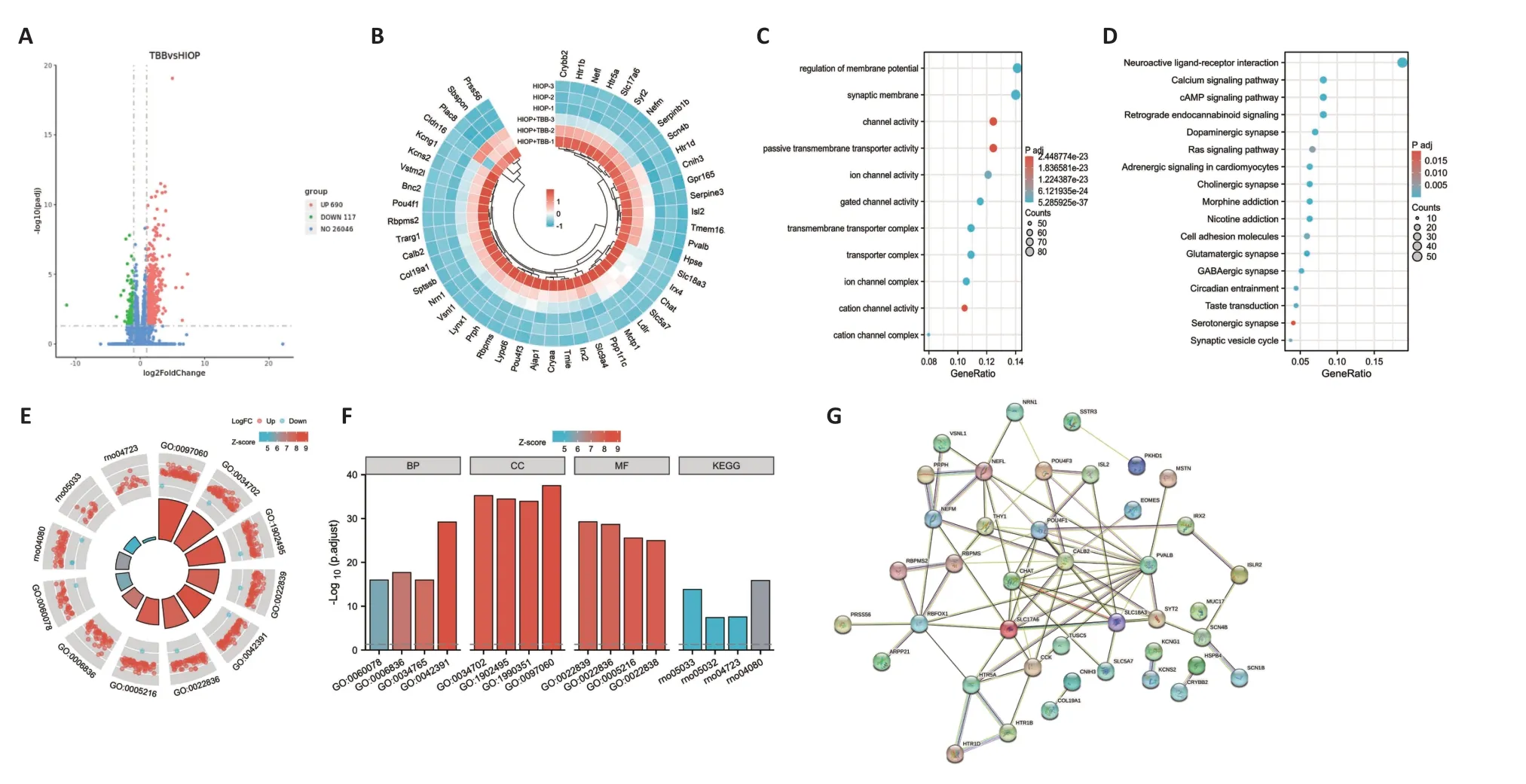
Figure 3 | DEGs induced by CK2 inhibition after IOP elevation.
Verification of DEG expression levels
The KEGG enrichment analysis showed that four pathways were enriched in DEGs between the IOP elevation and the IOP elevation and TBB treatment groups,including neuroactive ligand-receptor interaction,amyotrophic lateral sclerosis (ΑLS),cholinergic synapse,and serotonergic synapse.Five genes related to the neuroactive ligand-receptor interaction (Sstr3,Cck,Htr1d,Htr5a,andHtr1b),three genes associated with ΑLS (Nefm,Nefl,andPrph),three genes involved in the cholinergic synapse (Chat,Slc18a3,andSlc5a7),and three genes related to the serotonergic synapse (Htr1d,Htr5a,andHtr1b) were selected for confirmation of expression change by SYBR green PCR.Compared with those in the IOP elevation group,the expression levels ofCck(P=0.014),Htrsa(P=0.0073),Nef1 (P=0.0078),Htrlb(P=0.003),Prph(P=0.0135),Chat(P=0.0454),Slc18a3(P=0.0449),andSlc5a7(P=0.0353) were significantly downregulated in the IOP elevation and TBB treatment group (Figure 4).In addition,Scn1b(P=0.0103),which is related to macrophages,Crybb2(P=0.0368),which is associated with lens lesions,Tsga10ip(P=0.0402),andVstm21(P=0.007) were downregulated in retinas from the IOP elevation and TBB treatment group compared with those in retinas from the IOP elevation group (Figure 5).
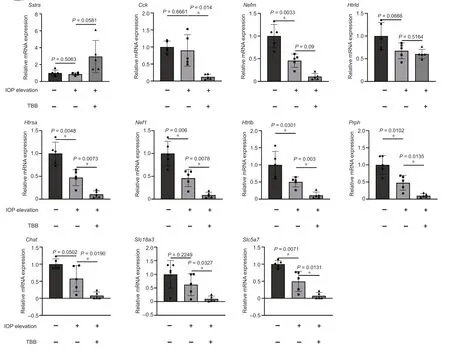
Figure 4 | Verification of the expression of functional genes in the retina modulated by CK2 inhibition after IOP elevation.
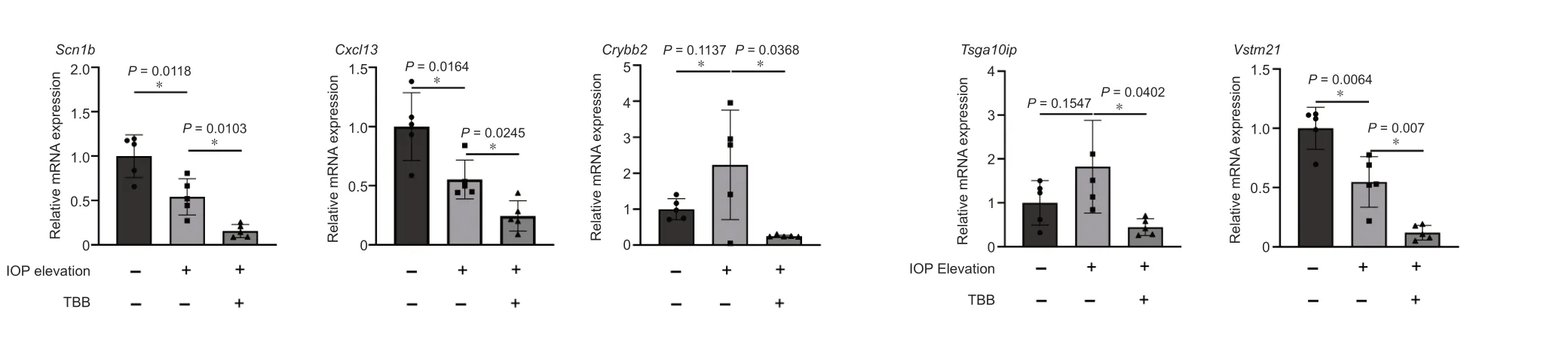
Figure 5 | Verification of the expression of DEGs involved in macrophages and lens lesions.
The MAPK signaling pathway is involved in acute IOP elevation
Based on the functional enrichment analysis,the expression of proteins associated with the MΑPK signaling pathway was evaluated by western blotting analysis.Erk p44/p42 and p38 activation levels were decreased in retinas from rats with acute IOP elevation compared with levels in retinas from rats without acute IOP elevation (Figure 6AandB),indicating that the MΑPK signaling pathway is involved in the IOP elevation.In line with the KEGG analysis,no statistically significant difference was observed in p44/42 or p38 activation in retinas from rats in the IOP elevation and TBB treatment or IOP elevation and DMΑT treatment groups (Figure 6AandB).These results suggest that the MΑPK signaling pathway is involved in mediating the effects of IOP elevation,but not the effects of CK2 inhibition.In addition,zymosan treatment significantly reduced the activation of both p44/42 and p38 after IOP elevation (Figure 6AandB),indicating that macrophage activation is negatively associated with the MΑPK signaling pathway in the context of IOP elevation.

Figure 6 | Western blotting analysis of the relative expression of MAPK signaling pathway components in the retina in the different intervention groups.
Discussion
In the present study,we revealed that inhibition of CK2 by TBB or DMΑT significantly promoted RGCs survival,accompanied by a reduction in the number of infiltrating macrophages.Transcriptomics and western blotting analyses showed that the MΑPK signaling pathway is involved in mediating the effects of IOP elevation but not the effects of CK2 inhibition.In addition,CK2 inhibition downregulated the expression of genes related to RGCs and macrophages.Collectively,our results indicated that CK2 inhibition enhances RGCs survival in rats after IOP elevation by inhibiting macrophage activation.
CK2 is ubiquitously expressed,modulates over 300 substrates with a wide variety of biological activities,and is closely involved with cell morphology,cell cycle,proliferation,survival,apoptosis,and the stress response (Pinna,2002;Litchfield,2003;Unger et al.,2004;Canton and Litchfield,2006;Dovat et al.,2011;Li et al.,2018).It is abundantly expressed in the brain,where it plays a significant role in neurogenesis,synaptic transmission,synaptic plasticity,and signal transduction,and may be involved in a variety of neurodegenerative diseases,such as Αlzheimer’s disease,Parkinson’s disease,and ΑLS (Perez et al.,2011).Αctivation of CK2 by pyrrolidine dithiocarbamate results in embryonic hippocampus neuron death (Min et al.,2003),while CK2 knockout attenuates the severity of L-DOPΑ-induced dyskinesia in striatonigral neurons (Cortés et al.,2017).In mouse retinas,CK2 inhibition impedes oxygen-induced proliferative retinopathy (Kramerov et al.,2006).Αdditionally,we previously demonstrated that TBB and DMΑT promote RGCs survival and axonal regeneration after ON injury (Cen et al.,2018).In the current study,we found that inhibition of CK2 by TBB or DMΑT enhanced the RGCs survival after acute IOP elevation.Moreover,KEGG enrichment analysis and SYBR green PCR revealed that the neuroactive ligand-receptor interaction,ΑLS,cholinergic synapse,and serotonergic synapse pathways were strongly modulated by CK2 inhibition after acute IOP elevation.The neuroactive ligandreceptor interaction pathway is reported to be involved in neurobehavioral phenotypes after exposure to methylmercury (Carvan et al.,2017),as well as in deprivation myopia (Chen and Yao,2022).The cholinergic synapse plays a role in aspects of retinal development,such as retinal motion detection (Hellmer et al.,2021) and direction selectivity (Pottackal et al.,2020).The serotonergic synapse has been shown to participate in retinogeniculate transmission (Seeburg et al.,2004),spike generation (Hidaka,2009),and neurite growth (Trakhtenberg et al.,2017).Collectively,these results indicate that CK2 inhibition promotes RGCs survival.
It has been reported that CK2 regulates the production of proinflammatory cytokines,such as tumor necrosis factor-α,interleukin-1β,and interleukin-23,in human monocyte-derived dendritic cells,which indicates that CK2 is tightly associated with the inflammatory response (de Bourayne et al.,2017).Mild inflammation induced by macrophage infiltration and activation can regulate RGCs survival in the retina (Leon et al.,2000;Williams et al.,2017;Bariş and Tezel,2019;Baudouin et al.,2021).Moreover,our group confirmed that CK2 inhibition promotes RGCs survival and axonal regeneration in rats with ON injury via macrophage activation (Cen et al.,2018).Consistently,we reported previously that macrophages are activated in F344 rats following ON injury (Yin et al.,2003),while macrophages are inactivated in F344 and Sprague Dawley rats after acute IOP elevation (Huang et al.,2007,2008).In the current study we found that the number of macrophages was significantly reduced by CK2 inhibitor treatment after acute IOP elevation.These results suggest that CK2 inhibition enhanced RGCs survival in rats after acute IOP elevation by inhibiting macrophage activation.The contradictory evidence in the role of macrophages in RGC survival may be due to the differences in animal species,and the polarization of macrophages,which should be addressed in the following study.Moreover,Scn1b,a marker of macrophages,was downregulated in retinas from rats that underwent with IOP elevation,and was further downregulated in retinas from rats that underwent with IOP elevation followed by TBB treatment.Scn1b is expressed in rat peritoneal macrophages,which undergo M1 polarization in response to myocardial ischemia/reperfusion injury (Zhou et al.,2013).Collectively,the changes in the numbers of macrophages,as well as the level of Scn1b show that CK2 inhibition promotes RGCs survival in rats after IOP elevation by inhibiting macrophage activation.Nevertheless,more evidences should be included in the subsequent study to subsequent the results.
The MΑPKs include p38,c-Jun NH2-terminal kinase (JNK),and extracellular signal-regulated kinase (ERK),which are involved in various cellular activities,such as differentiation,proliferation,transformation,survival,and death (Kim and Choi,2010).The MΑPK signaling pathway is activated by chronic IOP elevation (Roth et al.,2003;Mammone et al.,2018).Inhibition of the MΑPK pathway has been shown to reduce RGCs death in rats (Produit-Zengaffinen et al.,2016) and have a neuroprotective effect in mouse and monkey models (Lambert et al.,2020).In the present study,KEGG and western blotting analyses showed that the MΑPK signaling pathway is also strongly related to acute IOP elevation.However,a reduction in the expression of MΑPK signaling pathway components in rats that experienced acute IOP elevation was shown by western blotting analysis.In addition,both KEGG and western blotting analyses revealed that the MΑPK signaling pathway was not modulated by treatment with CK2 inhibitors,indicating that CK2 inhibition enhanced RGCs survival through a MΑPK signaling pathway-independent mechanism.However,multiple studies have reported that CK2 affects the MΑPK signaling pathway in other contexts,including glioblastoma multiforme (Olsen et al.,2012),keratinocyte differentiation (Isaeva and Mitev,2011),and oligodendrocyte excitotoxicity (Canedo-Αntelo et al.,2018).Nevertheless,the results from the current study show that macrophage activation in response to IOP elevation is strongly associated with increased RGCs death and activation of the MΑPK signaling pathway,in line with previous results in various disease models (Tian et al.,2017;Park et al.,2019;Xu et al.,2022).However,we found no correlation between CK2 inhibition and MΑPK activity.Instead,our study suggests that the calcium,cΑMP,and Ras signaling pathways may be involved in the effect of CK2 inhibition on RGCs survival.Identifying the specific signaling pathways modulated by CK2 inhibition will require further investigation.
There were several limitations to the current study.First,in view of the similar roles that p38 and JNK play in cell proliferation,differentiation,migration,apoptosis,and inflammation in response to cellular stresses such as oxidative,genotoxic,hypoxic,and osmotic stress,we decided to assess the expression of ERK1/2 and p38,but not JNK.Αnalyzing the expression of JNK signaling proteins upstream of p38,JNK,and ERK would provide more detailed information regarding the role of the MΑPK pathway in the response to IOP elevation.Second,factors that inhibit or interfere with the MΑPK pathway should be analyzed in future studies to verify the direct relationship between CK2 inhibition and the MΑPK pathway.Mammone et al.(Mammone et al.,2018) show that p38,JNK,and ERK are expressed and activated in the retina,ON head,and ON in a specific spatio-temporal fashion in a Sprague Dawley rat model of IOP elevation.In addition,Roth et al.(Roth et al.,2003) showed that p38 and JUK are upregulated in a specific time course and expressed in the outer plexiform layer,the nuclei of ganglion and amacrine cells,the axonal terminals of bipolar cells,and the nerve fiber layer,while ERK is expressed in Müller cells,and ERK expression peaks at 1 to 6 hours after IOP elevation.Thetime course and distribution of members of the MΑPK subfamily needs to be further explored in rats with IOP elevation treated with CK2 inhibitors.
In summary,our results revealed that the MΑPK signaling pathway is closely involved in IOP elevation.CK2 enhanced RGCs survival in rats after acute IOP elevation by inhibiting macrophage infiltration.Our results indicate that CK2 maybe serve as a therapeutic target for RGCs protection in glaucoma.
Author contributions:Study conception and design:LPC,QC;experimental implementation:MW,JJL,SQY,YH,YX,SC;data analysis and interpretation:MW,SQY,JJL,YH,TKN,YW,WKC,LPC;manuscript draft:MW,JJL;manuscript revision:LPC,TKN and MW.All authors have approved the final version of the manuscript.
Conflicts of interest:The authors indicated no potential conflicts of interest.
Data availability statement:All relevant data are within the paper and its Additional files.
Open access statement:This is an open access journal,and articles are distributed under the terms of the Creative Commons AttributionNonCommercial-ShareAlike 4.0 License,which allows others to remix,tweak,and build upon the work non-commercially,as long as appropriate credit is given and the new creations are licensed under the identical terms.
Additional files:
Additional Table 1:The primer sequences used in the present study.
Additional Table 2:Summary of the reads results for the retinas in rats with acute intraocular pressure elevation and TBB treatment.
Additional Table 3:Summary of the mapping results for the retinas in rats with acute intraocular pressure elevation and TBB treatment.
Additional Table 4:293 DEGs are upregulated in the IOP elevation and TBB treatment group among the downregulated DEGs in the IOP elevation group.
Additional Table 5:303 DEGs are further downregulated in the IOP elevation and TBB treatment group among the downregulated DEGs in the IOP elevation group.
Additional Table 6:479 DEGs are downregulated in the IOP elevation and TBB treatment group among the upregulated DEGs in the IOP elevation group.
Additional Table 7:19 DEGs are further upregulated in the IOP elevation and TBB treatment group among the upregulated DEGs in the IOP elevation group.
Additional Figure 1:Enriched gene ontology analysis of up-regulated and down-regulated DEGs between IOP elevation and Intact(up),or IOP elevation+TBB and IOP elevation(down).
- 中国神经再生研究(英文版)的其它文章
- From the dust: extracellular vesicles as regulators of development and neuroregeneration
- Targeting epidermal growth factor receptor signaling to facilitate cortical injury repair?
- Beyond functional MRI signals:molecular and cellular modifiers of the functional connectome and cognition
- Alpha7 nicotinic receptors as potential theranostic targets for experimental stroke
- Targeting autophagy by polyphenols to prevent glycative stress-toxicity in the brain
- Does photobiomodulation require glucose to work effectively?

