Bioinformatics and in vitro study reveal the roles of microRNA-346 in high glucose-induced human retinal pigment epithelial cell damage
Peng Li, Li Wang, Qing Liu, Zhao-Jiang Du
1Department of Ophthalmology, Xijing 986 Hospital Department, Fourth Military Medical University, Xi’an 710054, Shaanxi Province, China
2Ophthalmology Teaching and Research Section of Institute of Medical Technology, Xi’an Medical College, Xi’an 710032,Shaanxi Province, China
3Department of Ophthalmology, Xi’an Central Hospital, Xi’an 710001, Shaanxi Province, China
Abstract
● KEYWORDS: miRNA; miRNA-346; ARPE-19;bioinformatics analysis
INTRODUCTION
Diabetes is a public health issue with an estimated 415 million patients globally in 2015 and is projected to affect 642 million people by 2040[1].Diabetic retinopathy(DR), a major microvascular complication of diabetes,normally characterized by abnormal retinal microcirculation,affects a third of the diabetic patients and is the main cause of irreversible blindness in adults[2].It is associated with a poor life quality and increases the risk of other complications and mortality[3-5].Inflammatory processes play important roles in the pathogenesis of DR, which has been enforced by the clinical application of dexamethasone[6].There are three strategies to prevent blindness induced by DR.While primary prevention requires the prevention or delay of its onset,secondary prevention is to delay the progression of DR.Lastly,non-invasive laser photocoagulation and invasive ocular surgery are the treatment options.However, anti-vascular endothelial growth factor (VEGF) is also increasingly used to treat vision-damaging DR[7].Therefore, detailed studies of its molecular mechanisms are essential to provide a theoretical framework for research on DR treatment.
As we know, blood-retinal barriers (BRB) is a physiologic barrier that maintain the structural and functional integrity of the retinal tissues[8].The BRB composed of inner and outer components.The inner BRB is formed mainly by the tight junctions of retinal endothelial cells, and outer BRB is formed by the tight junctions of retinal pigment epithelial(RPE) cells[9].Alterations of both inner and outer BRB play an important role in the development of retinal diseases[8].RPE cells were previously believed unrelated to DR.However,increasing evidence suggests that all types of retinal cell are affected by diabetes[10], including the degeneration or dysfunction of the RPE[11-13].In diabetic retina, cellular changes in the RPE occur in the early stage of DR, Samuelset al[11]used three mouse models to compare the time course of RPE involvement in type 1 and type 2 diabetes, and all mouse models showed altered RPE function accompanied with the onset of hyperglycemia.Recent studies have emphasized the importance of RPE in DR[8,14].For example, RPE contribute to the development of DR probably by promoting the retinal vascular alterations in the early stages of the retinopathy[10].Additionally, there have been a substantial of studies focusing on the inner retina in DR, and studies on outer retina is still limited.
Previous studies have shown that the occurrence of DR is related to abnormal epigenetic regulation[15-17].In eukaryotic organisms, miRNA, an important component of epigenetics,is a highly conserved class of non-coding RNAs, which are about twenty-two nucleotides long.Endogenous miRNAs can regulate gene expression at the transcriptional level through specific interactions with target gene sequences and participate in many biological processes such as cell proliferation,differentiation and apoptosis[17-18].miRNAs are associated with DR microvascularization, and differentially expressed miRNAs (DEMs) in DR have been identified[19].miR-15b has been reported to inhibit angiogenesis by targeting VEGF in proliferative DR[20].miR-451a mediates the proliferation and migration of RPE cells in proliferative DR by regulating mitochondrial function[21].Previous studies have suggested that miRNAs could be potential biomarkers for DR therapeutic strategies[22-23].For instance, diabetes could induce the high expression of miR-21 in retina, and inhibition of miR-21 attenuated retinal neovascularization and inflammation,indicating that miR-21 might be a therapeutic target in DR[24].Α previous study identified 8 dysregulated miRNΑs and their key targets usinginsilicomethod and were successfully confirmed usingin vivomethod[25], suggesting the feasibility of identifying miRNAs using bioinformatics methods.Therefore, we screened key dysregulated miRNAs in DR using bioinformatics methods based on the miRNA expression profiling in the Gene Expression Omnibus (GEO) database.
The target genes of dysregulated miRNAs were predicted,followed by functional enrichment analysis and proteinprotein interaction (PPI) analysis to further study the role of dysregulated miRNAs in DR.In addition, we investigated the effect of miR-346 in high glucose (HG)-treated human retinal pigment epithelial cells.
MATERIALS AND METHODS
Data AccessThe microarray dataset of GSE52233 was downloaded from the GEO database (http://www.ncbi.nlm.nih.gov/geo/), which contained the miRNA expression data of agematched human autopsy normal (n=6) and DR (n=3) central corneas.The sequencing platform was GPL8786 [miRNA-1]Affymetrix Multispecies miRNA-1 Array.All methods were carried out in accordance with relevant guidelines and regulations.
Differential Expression AnalysisThe miRNA data from downloaded CEL files were processed using the R package of Affymetrix (Version 1.50.0, http://www.bioconductor.org/packages/release/bioc/html/affy.html)[26].The procedure included background correction and normalization.Additionally, expression calculations were performed using the robust multi-array average (RMA) method.The annotation file of the platform was used to annotate the probes for the miRNA chip, and the probes of non-human miRNAs were removed[27-28].The empirical Bayes linear model in the limma package was used to analyze DEMs between DR and control(normal).The criteria for differential expression were set as follows:P<0.05, |log2FC|>0.585 (fold change >1.5 or fold change <1.5).
Prediction of Target Genes of DEMsThe target mRNAs of the DEMs (top five upregulated and top five downregulated DEMs) were predicted using miRWalk 2.0[29-30](http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/).The miRNAmRNA pairs were included only when they appeared in all of the following twelve databases: miRWalk, microT v4,miRanda, miRBridge, miRDB, miRMap, miRNAMap, Pictar2,PITA, RNA22, RNAhybrid, and Targetscan.Cytoscape software (Version 3.4.0), which were used to visualize the miRNA-mRNA regulation network, and the CytoNCA[31]plugin (Version 2.1.6, http://apps.cytoscape.org/apps/cytonca) was used to determine the connectivity degree of each node by analyzing the topological properties.
Function Enrichment AnalysisTo explore the functions of the DEMs, function enrichment analysis was performed for the target mRNAs of the DEMs.The hypergeometric distribution test of ClusterProfiler in R package was used to analyze the biological processes from Gene Ontology[32]and Kyoto Encyclopedia of Genes and Genomes (KEGG)[33]pathways.Statistical significance was set atP<0.05.
PPI Network AnalysisSTRING database[34](Version: 10.0,http://www.string-db.org/) was used to analyze the interactions among the target mRNΑs of the DEMs with PPI score ≥0.4.Cytoscape software (Version 3.4.0) was used to visualize the PPI network.The CytoNCA[31]plug-in was used to analyze the topological properties of each node in the PPI network.
Cell CultureThe human retinal pigment epithelial cell line ARPE-19 was purchased from the Shanghai Cell Bank of the Chinese Academy of Sciences, China.The cells were cultured at 37℃ in a humidified incubator with 5% carbon dioxide and were maintained in basic DMEM (Catalog No.8117225,Gibco, Grand Island, NY, USA) supplemented with 10%fetal bovine serum (Catalog No.10099-141, Gibco, Grand Island, NY, USA) and 1% penicillin-streptomycin (Catalog No.BS734, Sangon biotech, Shanghai, China).
For HG treatment, ΑRPE-19 cells were treated with different concentrations of glucose (7.5, 17.5, 25, 50, 100, and 200 mmol/L; Catalog No.14431-43-7, Sinopharm, China)for 24, 48, and 72h.Cell viability was assessed, and 25 mmol/L and 200 mmol/L HG treatment for 48h were used in the following experiments HG treatment (7.5 mmol/L) was used as a control.
Real-time Polymerase Chain ReactionThe TRIzol reagent kit (Catalog No.15596018, Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) was used to isolate total RNA according to the manufacturer’s instructions.Total RNΑ (1 μg per sample) was used for the reverse transcription reaction with the PrimerScript reverse transcription reagent kit (Catalog No.RR036A, Takara, Shiga, Japan) according to the manufacturer’s protocol.After reverse transcription, the Power SYBR Green PCR MasterMix (Applied Biosystems,Foster City, CA, USA) was used for real-time polymerase chain reaction (RT-PCR), which was performed in an ABI 7500 thermocycler (Applied Biosystems, Foster City, CA,USA) following the manufacturer’s instructions.Each sample contained three replicates.The data were analyzed and calculated according to the cycle threshold values.
TransfectionARPE-19 cells were seeded onto 6-well plates at a density of 3×105cells/well.After overnight incubation,the hsa-miR-346 inhibitor or negative control (NC) sequence was transfected into cells using Lipofectamine 2000 reagent(Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA).After 24, 48, and 72h, the cells were harvested for subsequent experiments.
Cell Viability DetectionA cell counting Kit (CCK-8,Catalog No.C0039, Beyotime, Shanghai, China) based on WST-8 was used to detect cell viability and cytotoxicity.After overnight incubation, approximately 4×103ARPE-19 cells seeded in a 96-well plate were subsequently treated with different concentrations of glucose (7.5 mmol/L for the blank group,25 mmol/L for the HG group).After 24, 48, and 72h of cell culture, the medium in the wells was discarded and 100 μL CCK-8 (5 mg/mL) was added to each well to a final concentration of 10% and thoroughly mixed for an additional 2h of incubation.The absorbance of formazan dye in each well was measured using a microplate reader (Thermo Multiscan MK3; Thermo Fisher Scientific, Waltham, MΑ, USΑ) at 450 nm.
Flow Cytometry AnalysisFluorescence-activated cell sorting(FΑCS) was performed to detect apoptosis, cell cycle profiling,and ROS activity.For apoptosis detection, four different groups were formed: 1) ARPE-19-blank (ARPE-19 cells without treatment); 2) ARPE-19+HG (ARPE-19 cells treated with 25 mmol/L HG); 3) ARPE-19-NC+HG, (ARPE-19 cells treated with hsa-miR-346 NC and 25 mmol/L HG); 4) ARPE-19-inhibitor+HG (ARPE-19 cells treated with hsa-miR-346 inhibitor and 25 mmol/L HG).Cells were then harvested and stained with FITC-Annexin V and propidium iodide (PI,36 μg/mL; Catalog No.P4170, MilliporeSigma, St.Louis,MO, USΑ) with RNase (10 μg/mL; Catalog No.RNΑSEΑRO, MilliporeSigma, St.Louis, MO, USΑ) at 25℃ for 15min.Cell apoptosis was measured as the percentage of FITC+ and PI-cell populations using FACS.
For cell cycle detection, cells were harvested and stained with PI (36 μg/mL; Catalog No.P4170, MilliporeSigma, St.Louis,MO, USΑ) with RNase (10 μg/mL; Catalog No.RNΑSEΑRO, MilliporeSigma, St.Louis, MO, USΑ) at 25℃ for 15min.The cell cycle was analyzed by PI staining data from FACS.
For ROS detection, the ROS assay kit (Catalog No.S0033,Beyotime, Shanghai, China) was used.In details, ARPE-19 cells transfected with miRNA NC and miRNA inhibitor were pre-incubated for 48h.DCFH-DA was then added to four different groups of cells to a final concentration of 10 mmol/L and incubated at 37℃ for 30min in the dark.The cells were subsequently washed with PBS and adjusted to a concentration of 1×106cells/mL for FACS analysis under the condition that the excitation and emission wavelengths were 488 and 525 nm,respectively.Data were analyzed using ModFitLT software(Verity Software House, Topsham, ME, USA).Representative data from three independent experiments with similar trends are shown.
Dual-luciferase Reporter AssayIn order to verify the direct interaction between miR-346 and AGO2, the wild type or mutant 3’-UTR fragments of Argonaute 2 (AGO2) containing the binding sites were amplified and cloned into the pmirGLO reporter vectors (Promega Corp., Madison, WI, USA).Then,miR-346 mimic or mimic NC and pmirGLO AGO2 wild type (WT)/ mutant (MUT) were then transfected into 293T cells using Lipofectamine 2000 Reagent (Invitrogen).After 48h of transfection, luciferase activity of reporter vectors were determined, and the binding intensity between miR-346 and AGO2 was reflected by the firefly luciferase activity by normalizing against Renilla luciferase activity.

Figure 1 Results of differential expression analysis A: Volcano plot of DEMs.Red and blue nodes represent upregulated and downregulated miRNAs, respectively.B: Clustering heatmap for DEMs.Each row represents one DEM, and each column represents one sample.The top five upregulated and top five downregulated miRNAs are labeled.DEMs: Differentially expressed miRNAs.
Statistical AnalysisAll data are presented as the mean±standard deviation (SD).Thet-test and two-way ANOVA followed by Tukey’s post-hoc test were used for statistical analysis.If theP-value was less than 0.05, the difference was considered significant.
RESULTS
miRNAs Differentially Expressed in DRIn total, 39 miRNAs were found to be differentially expressed between the control and DR groups (P<0.05, absolute fold change >1.5), of which 19 were upregulated and 20 downregulated (Figure 1A).The clustering heatmap of DEMs showed different miRNA expression patterns in the DR and control groups, and the two groups could be clustered by differential miRNΑs (Figure 1B).Potential Function of DEMsTo investigate the potential target mRNAs of the key DEMs in DR, we predicted the miRNA-mRNA interaction pairs for the top 10 DEMs.A total of 119 mRNAs were predicted to be targets for seven DEMs,involving 125 miRNA-mRNA pairs (Figure 2).The seven DEMs included four upregulated miRNAs (e.g., miR-346) and three downregulated ones (e.g., miR-509-3P).There were 15 targets was found for miR-346, such as AGO2, RDX and MASP1.
To further investigate the functions and signaling pathways related to these seven DEMs, function enrichment analysis was performed for the target mRNAs (Figure 3).These targeted mRNA were involved in different biological processes and pathways,e.g., the target mRNAs of miR-346 were enriched in the biological processes of protein localization to the cell periphery, protein localization to the plasma membrane, and regulation of intracellular transport.They were also enriched in the KEGG pathways, including SNARE interactions in vesicular transport, retinoic acid-inducible gene I (RIG-I)-like receptor signaling pathway, and synaptic vesicle cycle.The target mRNAs of miR-34b-3P were enriched in the mTOR signaling pathway and regulation of miRNA metabolic processes, and in the negative regulation of T-helper 17 type immune response and activated T cell proliferation.
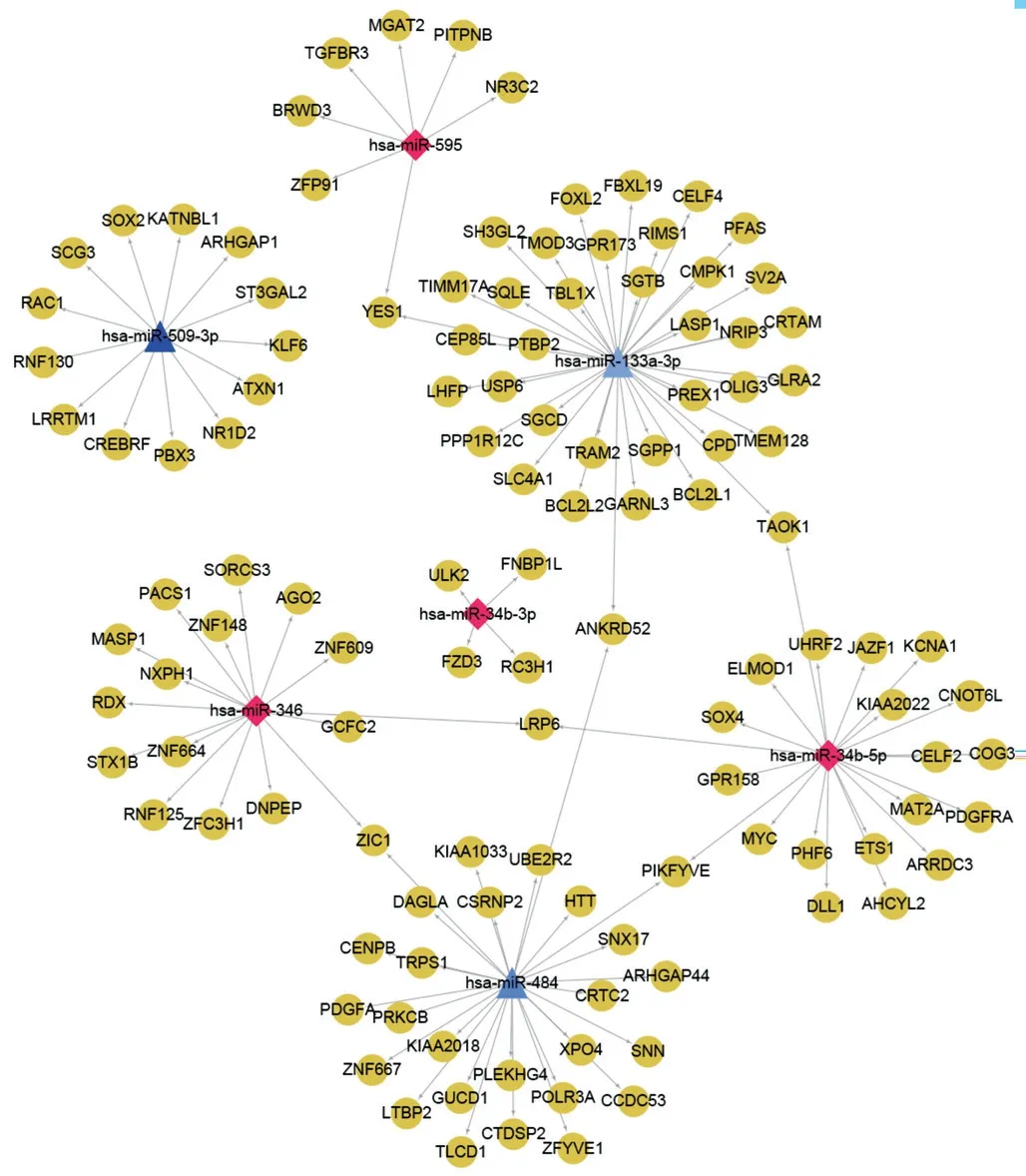
Figure 2 miRNA-mRNA interaction network Blue triangles represent down-regulated miRNAs, red diamonds represent up-regulated miRNAs, yellow circles represent target genes.The color darkness of the blue triangles and red diamonds represents the relative value of logFC; the darker the color is, the bigger the |logFC| value is.
PPI NetworkTo further determine whether there was crosstalk among the target mRNAs, we constructed a PPI network, which contained 67 nodes and 72 interactions(Figure 4).According to the degree ranking of the PPI network topological properties, MYC (degree=18), RAC1 (degree=11),and SOX2 (degree=6) were hub nodes with a higher degree in the PPI network.

Figure 3 Results of function enrichment analysis for target genes of key DEMs A: Bubble diagram shows the significantly enriched biological processes in GO annotation.B: Bubble diagram shows the significantly enriched KEGG pathway.The horizontal axis labels are miRNA IDs with the number of enriched genes in parentheses; the vertical axis labels are the names of KEGG pathways.The dot size GeneRatio is the gene proportion; the larger the gene proportion, the greater the proportion of the enriched genes.The colors from purple to green represents the relative P value; a lower P value indicates higher significance.DEMs: Differentially expressed miRNAs; KEGG: Kyoto Encyclopedia of Genes and Genomes.
Expression of miRNAs in Human Retinal Pigment Epithelial Cell Treated with HGExpression of three downregulated and up-regulated miRNAs were validated in human retinal pigment epithelial ARPE-19 cells after HG treatment(Figure 5).Consistent with the results in bioinformatics analysis, expression of miR-509-3p, miR-484 and miR-133a were significantly reduced in 200 mmol/L HG treated ARPE-19 cells, and the decreased expression of miR-509-3p and miR-133a in HG treated ARPE-19 cells seemed to be dose-dependent.Expression of miR-297 was significantly increased in 200 mmol/L HG treated ARPE-19 cells.Additionally,expression of miR-424 and miR-346 were elevated in 25 mmol/L HG treated ARPE-19 cells compared to that in control cells,and its expression continuously elevated in a dose-dependent manner up to treatment with 200 mmol/L HG.We mainly focused on the two miRNAs (miR-424 and miR-346) whose expression was significantly up-regulated with the increase of high glucose concentration.Previous studies have reported the involvement of miR-424 in retina-associated diseases[35-36],while miR-346 has not been investigated yet.Therefore,miR-346 was selected for further investigations.
Role of miRNA-346 in HG-induced Cell Proliferation Inhibition and ApoptosisExpression of miR-346 was markedly reduced in ARPE-19 cells after transfecting with miR-346 inhibitor (Figure 6A).Results of the CCK-8 assay showed that cell proliferation was substantially inhibited after 25 mmol/L HG treatment at both 48 and 72h (1.22 fold at 48h,1.29 fold at 72h,P<0.01), while such inhibition was partly reversed by inhibiting miR-346 expression (1.12 fold at 48h,1.09 fold at 72h,P<0.01; Figure 6B).

Figure 5 Expression of miRNAs validated in HG-treated ARPE-19 cells Expression of three upregulated miRNAs and three downregulated miRNAs in HG-treated ARPE-19 cells determined by qPCR.aP<0.05; bP<0.01, compared to control.qPCR: Quantitative polymerase chain reaction; HG: High glucose.

Figure 6 The effect of miR-346 on ARPE-19 cell proliferation A: Expression of miR-346 was verified to be decreased after transfecting miR-346 inhibitor.B: Cell proliferation of ARPE-19 cells determined by CCK-8 assay after HG treatment and miR-346 inhibition.bP<0.01, compared to control, ARPE-19-blank group; cP<0.05, dP<0.01, compared to NC+HG group.NC: Negative control; HG: High glucose; CCK: Cell Counting Kit.
As per cell cycle detection results, the proportion of cells in the G0/G1 phase (1.16 fold,P<0.01) and G2M phase (1.51 fold,P<0.01) increased considerably, but there was a substantial decrease in the proportion of cells in S phase (2.69 fold,P<0.01) after 25 mmol/L HG treatment.As expected, HGinduced changes in the cell cycle could be partly reversed by inhibiting miR-346 expression (1.20 fold at G0/G1 phase, 2.24 fold at S phase,P<0.05; Figure7A).In addition, HG treatment induced ARPE-19 cell apoptosis (2.36 fold,P<0.01), which was partly inhibited by inhibiting miR-346 expression (1.37 fold,P<0.01; Figure 7B).These results suggest that miR-346 expression is involved in HG-induced damage in human retinal pigment epithelial cells.
Role of miRNA-346 in HG-induced Oxidative StressHG treatment led to increased levels of reactive oxygen species(ROS, 1.64 fold,P<0.01), which substantially decreased after inhibiting the expression of miR-346 (1.23 fold,P<0.01,Figure 8), suggesting that miR-346 inhibition could reduce oxidative stress induced by HG.
AGO2 a Target of miR-346In order to verify the targets of miR-346, the binding probability between miR-346 and its targets were predicted.Among the 15 targets shown in the above miRNA-mRNA interaction network, AGO2 showed highest binding probability to miR-346.Expression of AGO2 was significantly increased after inhibiting miR-346 (Figure 9Α),indicating that there might be a negative regulatory between miR-346 and AGO2.Dual-luciferase reporter assay was further performed to validate the targeting relationship between miR-346 and AGO2.Luciferase activity of AGO2-WT was markedly reduced after transfection of miR-346 mimics, while luciferase activity of AGO2-MUT showed no significant changes after transfection of miR-346 mimics (Figure 9B).These results suggested that AGO2 was a target of miR-346.
DISCUSSION

Figure 7 The effect of miR-346 on cell cycle and apoptosis of ARPE-19 cells A: Cell cycle detection by FACS; B: Cell apoptosis detection by FACS.aP<0.05; bP<0.01, compared to ARPE-19-blank group; cP<0.001, compared to NC+HG group.HG: High glucose; FACS: Fluorescence-activated cell sorting; NC: Negative control.

Figure 8 The effect of miR-346 on HG-induced oxidative stress ROS detection by FACS.aP<0.001, compared to blank group; bP<0.001,compared to NC+HG group.HG: High glucose; ROS: Reactive oxygen species; FACS: Fluorescence-activated cell sorting; NC: Negative control.
DR develops at different levels in more than 40% of diabetic patients, and in about 4% of them, progresses to the proliferative type[37], which seriously impairs visual function.The development of proliferative DR seriously damages visual function.Although surgical treatment can restore the anatomical structure of the retina, visual function remains impaired[38-39].Therefore, searching for biomarkers with higher sensitivity, specificity, and stability may provide potential targets for the treatment of DR.An increasing number of studies have demonstrated the important role of miRNAs in DR[40-41].For example, Bao and Cao[42]suggested that the expression of miR-138-5p decreased in DR, and that it mediated the proliferation of retinal capillary pericytes and endothelial cells by regulating NOVA1.In this study, a total of 39 DEMs were screened for DR.Decreased expression of miR-509-3p, miR-484 and miR-133a, while elevated expression of miR-297, miR-424 and miR-346 were verified in HG treated ARPE-19 cells.The detailed roles of these miRNAs have not been reported in DR.Noticeably,miR-484 was reported to participate in apoptosis of the retinal ganglion cells following retinal ischemia reperfusion injury[43].MiR-133a-3p was found to locate at the outer nuclear layer in the damaged retina[44], and miR-424-5p was found to be upregulated in hypoxia-induced high-altitude retinopathy cell model[35].Further investigations focusing on the involvements of these miRNAs in DR should be carried out.

Figure 9 AGO2 was a target of miR-346 A: Expression of AGO2 after miR-346 inhibition; B: Dual-luciferase reporter assay for determining the interactions betwwen miR-346 and AGO2.aP<0.01, compared to control; bP<0.01, compared to inhibitor NC; cP<0.001, compared to WT-mimic NC.AGO2: Argonaute 2; WT: Wild type; NC: Negative control.
In this study, expression of miR-346 was found to increase in central corneas of DR patients, and its increased expression was verified in HG-treated ΑRPE-19 cells in a dose-dependent manner, suggesting the involvement of miR-346 in DR.However, there was no study to report the roles of miR-346 in DR.Here, we found that the target gene of miR-346 was mainly enriched in the regulation of intracellular transport,(RIG-I)-like receptor signaling pathway, and synaptic vesicle cycle.A study by Miaoet al[45]found that aberrantly hypermethylated genes that were enriched in the synaptic vesicle cycle and visual perception were involved in the pathophysiology of proliferative DR.(RIG-I)-like receptors play an important role in cytoplasmic RNA sensing, where viral RNAs are recognized and innate immune system and inflammation in cells is induced[46].The regulation of these signals is especially crucial to decrease inflammation in the eyes and other immune-tolerant organs[47].We speculated that miR-346 was involved in DR progression, probably by targeting genes associated with these signaling pathways.We further verified the targets of miR-346, and AGO2 was confirmed to be directly regulated by miR-346 in dualluciferase reporter assay.In HG-treated ARPE-19 cells,expression of ΑGO2 was significantly increased after inhibiting miR-346.Reportedly, either inhibiting or overexpressing ΑGO2 in mouse retina leaded to significantly changed retinal morphological and severely damaged retinal function[48].This finding also proven that miR-346 may be involved in the occurrence and development of DR to a certain extent.
RPE is a monolayer of pigment epithelium cells which comprises the outer blood-retinal barrier, and represents an important site for retinopathy[49].The damage of HG to RPE cells has been considered as an important event in DR, and the damage was implicated with apoptosis, inflammation, and oxidation[10].Especially, oxidative stress has been demonstrated to be an important contributor for the etiopathogenesis of DR[50].ARPE-19 cells are sensitive to oxidative damage,while abundant oxidative metabolites are produced in cells after HG stimulation[51-52].The accumulation of oxidative metabolites may result in irreversible cytotoxic damage to ARPE-19 cells[53].In addition, increased free radicals may trigger apoptosis of ARPE cells by damaging mitochondrial DNA[54].In order to confirm the involvement of miR-346 in the pathogenesis of DR, functional experiments were performed in HG-treated ARPE-19 cells.We found that miR-346 inhibition could alleviate HG-induced decreased cell viability and increased cell apoptosis.In addition, inhibition of miR-346 could also stabilize the increased rate of the G0/G1 phase and the decreased rate of the S phase after HG treatment.Most importantly, the increased level of ROS caused by HG could be reversed by inhibiting miR-346.These findings verified the involvement of miR-346 in the pathogenesis of DR.
To our knowledge, this was the first study to investigate the role of miR-346 in DR.However, this was just a preliminary study, and remained some limitations.The bioinformatics analysis revealed that the targeted genes of miR-346 enriched in several pathways reported to be involved in DR.Whether miR-346 affected these pathways had not been investigated in thein vitroexperiments.This study suggested that miR-346 mediated the basic biological processes in HG-induced damage of retinal pigment epithelial cell, such as cell proliferation,apoptosis and oxidative stress.However, the underlying specific regulation mechanism should be further investigated.For example, the signaling pathways that control cell cycle and apoptosis needed to be further explored.Inflammatory processes have been suggested to play important roles in the pathogenesis of DR, and whether miR-346 regulates the expression of inflammatory markers in DR should be further investigated.Additionally, many ocular cells are affected in DR.For example, apoptosis of neural and vascular cells in the retina had been demonstrated to be a contributing mechanism of DR[55].This study indicated that miR-346 mediated the cell proliferation, apoptosis and oxidative stress in HG-induced damage of RPE cell.It was necessary to investigated whether miR-346 mediated these processes of other ocular cells.Only based on this, the roles of miR-346 could be extended to DR in general.In conclusion, as per the current bioinformatic and experimental results, miR-346 is highly expressed in DR and plays an important role in HG-induced damage in human RPE cells.Therefore, miR-346 may play a key role in DR progression.However, in future, more rigorous, prospective,large sample sized experimental studies and long-term clinical trials are required to support this finding.
ACKNOWLEDGEMENTS
Foundations:Supported by the Social Development Project of Shaanxi Provincial Department of Science and Technology(No.2020SF-167); Supporting Fund Project of Shaanxi Provincial Department of Science and Technology Agency Project (No.2022SF-502); Xi’an Medical University 2022 Annual Scientific Research Capacity Improvement Plan Project (No.2022NLTS104).
Conflicts of Interest:Li P,None;Wang L,None;Liu Q,None;Du ZJ,None.
 International Journal of Ophthalmology2023年11期
International Journal of Ophthalmology2023年11期
- International Journal of Ophthalmology的其它文章
- Quantitative analysis of optic disc changes in school-age children with ametropia based on artificial intelligence
- Association analysis of Bcll with benign lymphoepithelial lesions of the lacrimal gland and glucocorticoids resistance
- In vitro protective effect of recombinant prominin-1 combined with microRNA-29b on N-methyl-D-aspartateinduced excitotoxicity in retinal ganglion cells
- Therapeutic effect of folic acid combined with decitabine on diabetic mice
- Comparison of visual performance with iTrace analyzer following femtosecond laser-assisted cataract surgery with bilateral implantation of two different trifocal intraocular lenses
- Comparison of three fundus inspection methods during phacoemulsification in diabetic white cataract
