Reviving and characterizing three species of dinof lagellate cysts dormant for about 70 years in the East China Sea:Biecheleria brevisulcata, Biecheleriopsis adriatica, and Scrippsiella donghaienis*
Zhangxi HU , Xiaoying SONG , Jinxiu WANG , Zhe TAO , Yuanyuan SUN ,Yuhang LI , Yuyang LIU , Yunyan DENG ,3,6, Lixia SHANG ,3,6, Zhaoyang CHAI ,3,6,Yingzhong TANG ,3,6,**
1 College of Fisheries, Guangdong Ocean University, Zhanjiang 524088, China
2 CAS Key Laboratory of Marine Ecology and Environmental Sciences, Institute of Oceanology, Chinese Academy of Sciences,Qingdao 266071, China
3 Laboratory of Marine Ecology and Environmental Science, Pilot National Laboratory for Marine Science and Technology(Qingdao), Qingdao 266237, China
4 CAS Key Laboratory of Experimental Marine Biology, Institute of Oceanology, Chinese Academy of Sciences, Qingdao 266071,China
5 Laboratory of Marine Organism Taxonomy and Phylogeny, Institute of Oceanology, Chinese Academy of Sciences, Qingdao 266071, China
6 Center for Ocean Mega-Science, Chinese Academy of Sciences, Qingdao 266071, China
7 University of Chinese Academy of Sciences, Beijing 100049, China
Abstract Many marine dinof lagellates can form resting cysts as a part of their life cycle, and the cysts could be buried in sediment and remained viable for as long as over 150 years. However, only a very limited number of cyst species have been revived from long-buried sediments and investigated in regard to a possible shift in the intra-specif ic genetic structure of a species detected from the historical record at a particular location. Here, we report a successful germination of three species of resting cysts that were sampled from the depth dated back to 1941±18 AD from a 44-cm sediment core from the East China Sea.Seven isolates were established from germination of single cyst isolation or multi-cyst germinations. LSU rRNA gene or ITS sequences of these strains were obtained, then they were identif ied to be Biecheleria brevisulcata (f ive strains), Biecheleriopsis adriatica (one strain), and Scrippsiella donghaienis (one strain)in terms of morphology and rRNA gene sequence. Biecheleria brevisulcata strain 1, Bps. adriatica strain 21, and S. donghaienis strain 23 were examined in detail with light microscope (LM) and scanning electron microscope (SEM), and analyzed with high performance liquid chromatography (HPLC) for their pigment compositions, and genetic diversity. We also conf irmed the presence of a resting cyst of Bps. adriatica in the f ield for the f irst time. The LSU rRNA gene-based genetic distances of Bps. adriatica from that obtained from water sample, single-cell PCR sequencing for the cysts isolated from the surface sediment of the same sea area and that reported from other regions during the recent years, and ITS-based genetic distances of S. donghaienis from that obtained from cysts isolated from the surface sediment of the same location and that reported from other regions during the recent years indicated that the intra-specif ic genetic structure of each species in the sampling area may have shifted during the last 70 years. Our work conf irms that B.brevisulcata, Bps. adriatica, and S. donghaienis, all described as new species around 2010, have inhabited the East China Sea for about 70 years. The present work reports for the f irst time the revival of dinof lagellate resting cysts long-buried in the coastal sediments of China, which facilitates further study on the historical occurrences of other harmful dinof lagellates and their relevance to the regional climate and environmental changes in China.
Keyword: core sediment; dinof lagellate resting cyst; germination; Biecheleria brevisulcata; Biecheleriopsis adriatica; Scrippsiella donghaienis
1 INTRODUCTION
Dinof lagellates constitute one of the main groups of marine phytoplankton in terms of their important contribution to the primary production, and comprise about 2 400 species belonging to 259 genera (Gómez,2012), which is still growing as more new taxa are to be described (Gómez et al., 2015; Takahashi et al.,2015, 2017, 2019; Boutrup et al., 2017; Luo et al.,2018; Hu et al., 2020b, 2021; Ok et al., 2021; Gu et al., 2022). The appearance of dinof lagellates dates back more than 400 million years in the fossil record,and this group of protists has evolved diverse features to adapt to their environments, including diverse morphologies, multiple-membrane cell walls, diff erent modes of nutrition, pigments, toxins, asexual and sexual reproduction, and other characters (Steidinger and Meave del Castillo, 2018). Among these features,the ability of about 10% of dinof lagellate species to produce resting cysts as a part of their life cycle is a vital one (Head, 1996; Bravo and Figueroa, 2014;Tang et al., 2016, 2021). It is now widely accepted that resting cysts play important roles in the biology and ecology of dinof lagellates (Anderson and Wall,1978; Dale, 2001; Bravo and Figueroa, 2014; Tang et al., 2016, 2021; Ellegaard and Ribeiro, 2018;Figueroa et al., 2018).
In the past, f ield studies on dinof lagellate cysts are generally focused on the surface sediments for mapping the distribution and abundance of cysts of important species, investigating species diversity of cyst assemblage in a region of concern, and conf irming the cyst presence for some species with particular importance (Luo et al., 2018; Limoges et al., 2020; Liu et al., 2020a, b, 2021; Mertens et al.,2020; Van Nieuwenhove et al., 2020; Hu et al., 2021,2022), whereas sediment cores representing a valuable archive of phytoplankton communities are usually used to reconstruct past environmental changes(Keafer et al., 1992; Dai et al., 2012; Ellegaard et al.,2013, 2020; Bringué et al., 2016; García-Moreiras et al., 2018; Kim et al., 2018; Price et al., 2018; de Freitas et al., 2020; Li et al., 2021; Siano et al., 2021).Cultures established from reviving dinof lagellate resting cysts collected from sediment cores have been used to evaluate the impact of environmental changes on the physiology, genetic structure, and diversity in various species (Ribeiro et al., 2011; Klouch et al.,2016; Lundholm et al., 2017; Kremp et al., 2018;Delebecq et al., 2020; Ellegaard et al., 2020; Girault et al., 2021). However, the numbers of dinof lagellate cysts remaining viable in long-buried sediments and the mechanisms behind it still need to be explored.Therefore, mining cyst records of dinof lagellate species buried in sediment cores is important in many aspects of the ecology of dinof lagellates and in reconstruction of the history of marine environmental changes.
Recently, we successfully established seven clonal cultures ofBiecheleriabrevisulcata,Biecheleriopsisadriatica, andScrippsielladonghaienisvia cyst germination from the depth dated back to 1941±18 AD of a sediment core collected from the East China Sea, and further characterized their morphologies,pigment compositions, and the genetic diversity in their LSU rRNA gene and ITS sequences.
2 MATERIAL AND METHOD
2.1 Sediment core and surface sediment sampling and dating
One sediment core (S06-2, 120.417°E, 26.122°N)was collected from the East China Sea in September 19, 2018 during the public cruise of R/VXiangYangHong18organized by the National Natural Science Foundation of China and the First Institute of Oceanography, Ministry of Natural Resources,China, and one surface sediment sample (0–2 cm;S01-1, 122.997°E, 31.000°N) was also collected from the East China Sea in September 17, 2019 during the public cruise of R/VXiangYangHong18.The sediment core was sliced into 2-cm layers (the top 20 cm) and 4-cm layers (20 cm to the bottom)by caution. A total of 16 subsamples were collected for the210Pb and137Cs measurements. The detailed measurement and age determination was reported in Liu et al. (2021), and the standard error of age was produced in regression uncertainties.
2.2 Culture establishment
Cyst assemblage in the subsample (42–44 cm) of core sediment (S06-2) and surface sediment sample(S01-1) was concentrated using sodium polytungstate solution (SPT) (Bolch, 1997). For subsample of core sediment, single cysts were washed at least three times using sterile seawater (with a salinity of 31)enriched with f/2-Si medium (Guillard, 1975), and then micropipetted to a 24-well culture plate with each well containing 2.5-mL fresh medium and 2%antibiotic solution (a mixture of 10 000-IU penicillin and 10 000-μg/mL streptomycin; Solarbio, Beijing,China). The rest of cyst assemblage was transferred to a 6-well culture plate with each well containing 10-mL fresh medium and 2% antibiotic solution. The plates were incubated at 21 °C, 12-h꞉12-h light꞉dark cycle, and ~100 μmol photons/(m2·s). Cysts and new germlings were observed every day or every other day with an inverted microscope (IX73, Olympus,Japan) and photographed by a DP80 digital camera(Olympus, Japan). Five strains ofB.brevisulcata(S1,S2, S3, S4, and S5) were established from the cyst assemblage germination experiment, but the resting and empty cysts ofB.brevisulcatawere not observed.One strain ofBps.adriatica(S21) and one strain ofS.donghaienis(S23) were established from single cysts germination experiments, their resting and empty cysts were clearly recorded using an inverted microscope (IX73, Olympus, Japan) equipped with a DP80 digital camera (Olympus, Japan). All cultures were routinely maintained in the same condition mentioned above.
2.3 Light microscopic observation
Live cells ofB.brevisulcata(strain S1),Bps.adriatica(strain S21), andS.donghaienis(strain S23)were observed and photographed using a Zeiss Imager Z2 (Carl Zeiss, Gottingen, Germany) equipped with diff erential interference contrast (DIC), or an inverted microscope (IX73, Olympus, Japan) equipped with a digital camera (DP80, Olympus, Japan). For observation of thecal plates ofS.donghaienis, live cells were stained with Calcof luor White (Sigma-Aldrich, St. Louis, MO, USA) and examined using an epif luorescence microscope (BX53, Olympus,Japan) with a UV f ilter set (Fritz and Triemer,1985). Cells sizes ofB.brevisulcata,Bps.adriatica,andS.donghaienisfor 50 live cells at the midexponential growth phase were measured at ×400 (forB.brevisulcataandBps.adriatica), and ×200 (forS.donghaienis) magnif ication using a DP80 digital camera (Olympus, Tokyo, Japan).
2.4 SEM observation
For SEM observation, vegetative cells ofB.brevisulcataandBps.adriaticaat mid-exponential growth stage were f ixed with OsO4(2% f inal concentration), andS.donghaieniswith glutaraldehyde(2.5% f inal concentration) for 40–50 min. Fixed cells were gently f iltered onto 5-μm (B.brevisulcataandBps.adriatica) and 11-μm (S.donghaienis) pore size Millipore nylon membranes, dehydrated in an acetone series (10%, 30%, 50%, 70%, 90%, and three times in 100%, 15 min for each step), and critical pointdried with liquid CO2(EM CPD300, Leica, Austria).They were sputter-coated with platinum-palladium(EM ACE200, Leica, Austria), and observed using an S-3400N SEM (Hitachi, Japan).
2.5 Pigment analyses
Fifty milliliter of each culture ofB.brevisulcata(strain S1),Bps.adriatica(strain S21), andS.donghaienis(strain S23) in exponential growth(in cell densities of ca. 102 150, 95 900, and 980 cells/mL, respectively) were f iltered through 25-mm diameter glass f iber f ilter (Whatman, Maidstone,UK) and immediately frozen at -80 °C for later analyses. Pigments were analyzed on an Alliance HPLC (e2695, Waters, Milford, Massachusetts,USA) using a 100-μL sample injection according to Kong et al. (2012) and Hu et al. (2020a). Pigments were identif ied and quantif ied using Shimadzu Class-VP software and by comparing pigment spectra and retention times with those of 26 standard pigments(DHI Water and Environment, Hørsholm, Denmark;Kong et al., 2012; Hu et al., 2020a).
2.6 DNA extraction, PCR amplif ication, and rDNA-based phylogenetic analyses
Genomic DNA ofB.brevisulcata,Bps.adriatica,andS.donghaieniswere extracted using a plant DNA extraction kit (Tiangen, Beijing, China) according to the manufacturer’s protocol. ForB.brevisulcata,Bps.adriatica, andS.donghaienis, about 1 400 bp of LSU rDNA were amplif ied using primers of D1R(forward, 5ʹ-ACCCGCTGAATTTAAGCATA-3ʹ)(Scholin et al., 1994) and 28-1483R (reverse,5ʹ-GCTACTACCACCAAGATCTGC-3ʹ) (Daugbjerg et al., 2000), and forS.donghaienis, about 660 bp of ITS was amplif ied using primers of ITS1 (forward,5ʹ-TCCGTAGGTGAACCTGCGG-3ʹ) and ITS4(reverse, 5ʹ-GCATATCAATAAGCGGAGGA-3ʹ)(White et al., 1990). Polymerase chain reaction (PCR)reactions were conducted using a PCR Master Cycler nexus gradient (Eppendorf, Hamburg, Germany), and performed with a f inal volume of 25 μL, containing 9.5-μL ddH2O, 12.5-μL 2×Taq PCR MasterMix,1 μL of each PCR primer (10 mmol/L), and 1 μL of the DNA template. The following cycling conditions were used: an initial denaturation at 94 °C for 5 min, 35 cycles at 94 °C for 20 s, 55 °C for 30 s,and 72 °C for 2 min, and a f inal elongation step of 10 min at 72 °C. The PCR products were conf irmed using 1% agarose gel electrophoresis and visualized with ultraviolet light. Targeted bands were purif ied using an agarose gel DNA fragment recovery kit(GENEray Biotechnology, Shanghai, China), ligated with pMD18-T cloning vector (TaKaRa, Tokyo,Japan), and then sequenced (Sangon, Shanghai,China). Sequences were deposited in GenBank with accession numbers OL355144–OL355148 (LSU,B.brevisulcata), OL355142 (LSU,Bps.adriatica),OL314541 (LSU,S.donghaienis), and OL314542(ITS,S.donghaienis).
For surface sediment sample (S01-1), the single cysts were individually micropipetted onto a glass slide, then photographed with an inverted microscope(IX73, Olympus, Japan) equipped with a DP80 digital camera (Olympus, Japan). Subsequently, individual cyst was micropipetted and transferred onto a sterile slide, then broken by another coverslip. The crushed cyst and the coverslip pieces were transferred into a 250-μL centrifuge tube, as the template to amplify about 1 400 bp of the LSU rDNA using the primer set, D1R(forward, 5ʹ-ACCCGCTGAATTTAAGCATA-3ʹ)(Scholin et al., 1994) and 28-1483R (reverse,5ʹ-GCTACTACCACCAAGATCTGC-3ʹ) (Daugbjerg et al., 2000). The following procedure was according to Shang et al. (2019).
For phylogenetic analyses of the LSU rDNA regions ofB.brevisulcataandBps.adriatica, and LSU and ITS rDNA regions ofS.donghaienis,newly obtained LSU rDNA and ITS sequences were incorporated into those of closely related species available in the GenBank and that of outgroup taxa were f irst aligned using MAFFT v7.475 (Katoh et al.,2002) online program (http://maff t.cbrc.jp/alignment/server/) with default settings, and alignments were manually checked with BioEdit v7.2.5 (Hall, 1999).The f inal alignments of the LSU and ITS rDNA sequences ofS.donghaienisconsisted of 97 and 41 taxa and contained 750 and 600 positions (including gaps introduced from alignment), and the sequences ofCryptoperidiniopsisbrodyi(DQ991374) andPentapharsodiniumdalei(JX262496) were used as the outgroup, respectively. LSU rDNA sequences ofBps.adriaticaandB.brevisulcataconsisted of 66 taxa with 1 590 positions (including gaps introduced from alignment), and the sequence ofAlexandriummargalef ii(AY154957) was used as outgroup. The program jModelTest 2.1.4 was used to select the most appropriate model of molecular evolution with Akaike information criterion (AICc) (Posada, 2008),models GTR+G+I and TrN+I+G were selected as the best-f it model for the LSU rDNA and ITS datasets ofS.donghaienis, and TrN+I+G for the LSU rDNA dataset ofBps.adriaticaandB.brevisulcata.Phylogenetic trees were constructed using Bayesian inference (BI) and maximum likelihood (ML)analyses. Bayesian inference (BI) was performed with MrBayes 3.2.6 (Ronquist and Huelsenbeck,2003) with the best-f itting substitution models(GTR+I+G for LSU rDNA dataset ofS.donghaienis,TrN+I+G for ITS dataset ofS.donghaienis, and TrN+I+G for LSU rDNA dataset ofBps.adriaticaandB.brevisulcata). Four independent Markov chain Monte Carlo simulations were run simultaneously for 5 000 000 generations and trees were sampled every 1 000 generations. The f irst 10% trees were discarded as burn-in. The convergence was judged based on the average standard deviation of split frequencies(all less than 0.01). The remaining trees were used to generate a consensus tree and calculate the posterior probabilities of all branches using a majority-rule consensus approach. Maximum likelihood (ML)analyses were conducted with raxmlGUI v1.3.1(Silvestro and Michalak, 2012; Stamatakis, 2014)using the models GTR+I+G (for LSU rDNA dataset ofS.donghaienis), GTR+G (for ITS dataset ofS.donghaienis; the model GTR+G ranked the third, and the score of this model was close to model TrN+I+G),and GTR+I+G (for LSU rDNA dataset ofBps.adriaticaandB.brevisulcata; the model GTR+I+G ranked the second, and the score of this model was close to model TrN+I+G). Node support was assessed with 1 000 bootstrap replicates. FigTree (v1.4.4) was used to view and edit trees for publication.
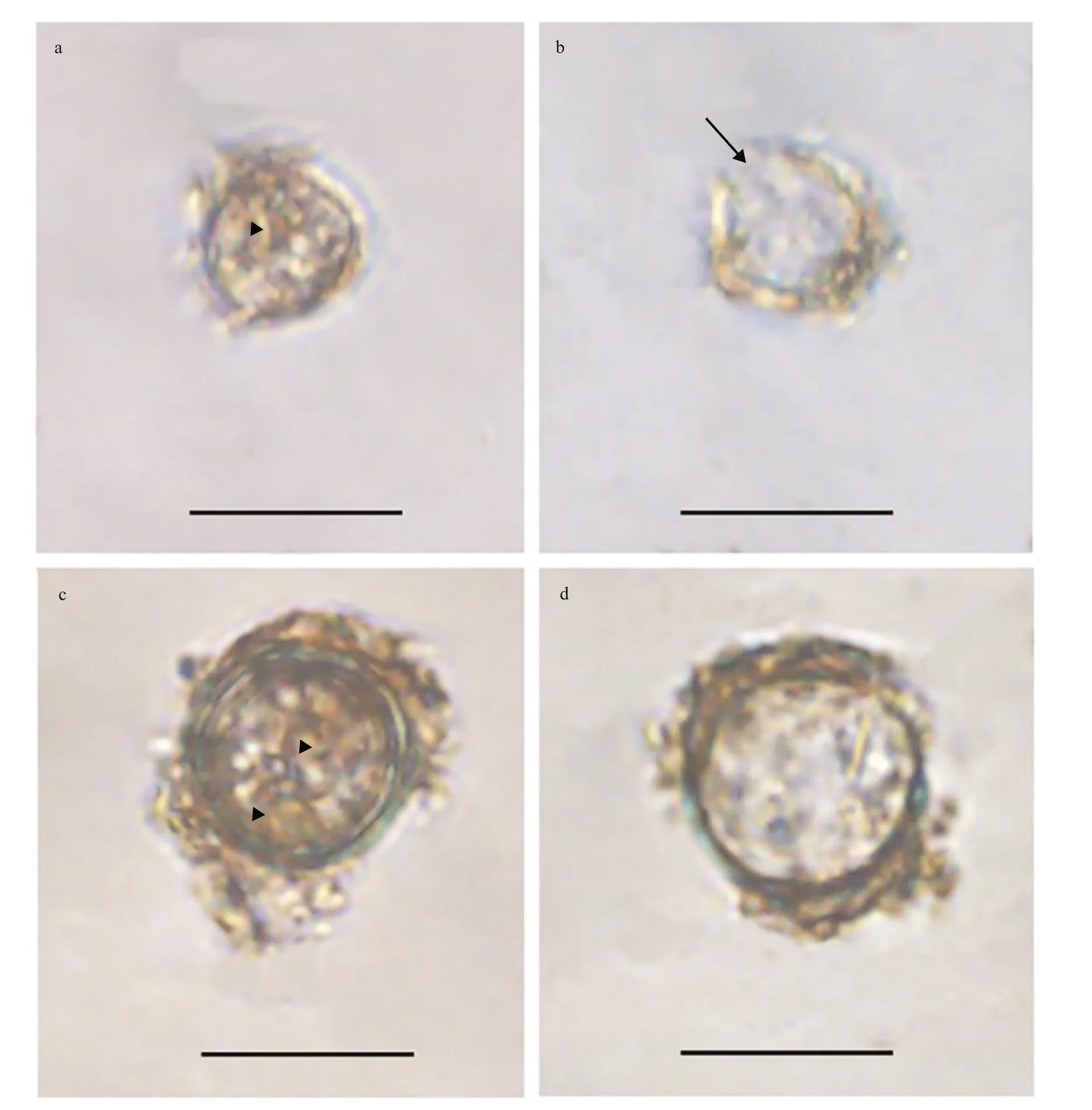
Fig.1 Light microscopy photographs of Bps. adriatica and S. donghaienis cysts
2.7 Genetic diversity analyses
The pairwise distances were computed among all sequences that were newly obtained in the present work forB.brevisulcata,Bps.adriatica, andS.donghaienisand that retrieved from the NCBI database for these three species together with other reference sequences. Sequences were aligned using the MAFFT v7.475 with the default settings (Katoh et al., 2002) (http://maff t.cbrc.jp/alignment/server/) and modif ied manually using BioEdit v7.2.5 (Hall, 1999).Pairwise evolutionary distances were then computed using Jukes and Cantor algorithm implemented in the MEGA X (Tamura et al., 2004; Kumar et al., 2018).
3 RESULT
3.1 Morphological observations of the resting cysts
Resting cyst ofBps.adriaticawas sub-spherical to spherical and light brown, with a diameter of ~7.3 μm,full of small granules and had 2–3 red accumulation bodies (Fig.1a). After four days’ incubation, the cyst was germinated. The cyst wall was thick and smooth,and the archeopyle was tremic (Fig.1b). Resting cyst ofS.donghaieniswas noncalcareous, spherical and brown, full of diff erent sizes of granules (Fig.1c). The diameter of the cyst was ~10.7 μm, and it contained several red accumulation bodies (Fig.1c). After four days’ incubation, the cyst was germinated. The cyst wall was thick, and the archeopyle was unclear(Fig.1d). As the f ive clonal cultures ofB.brevisulcatawere established from cyst assemblage germination,the resting and empty cysts of this species were not observed.
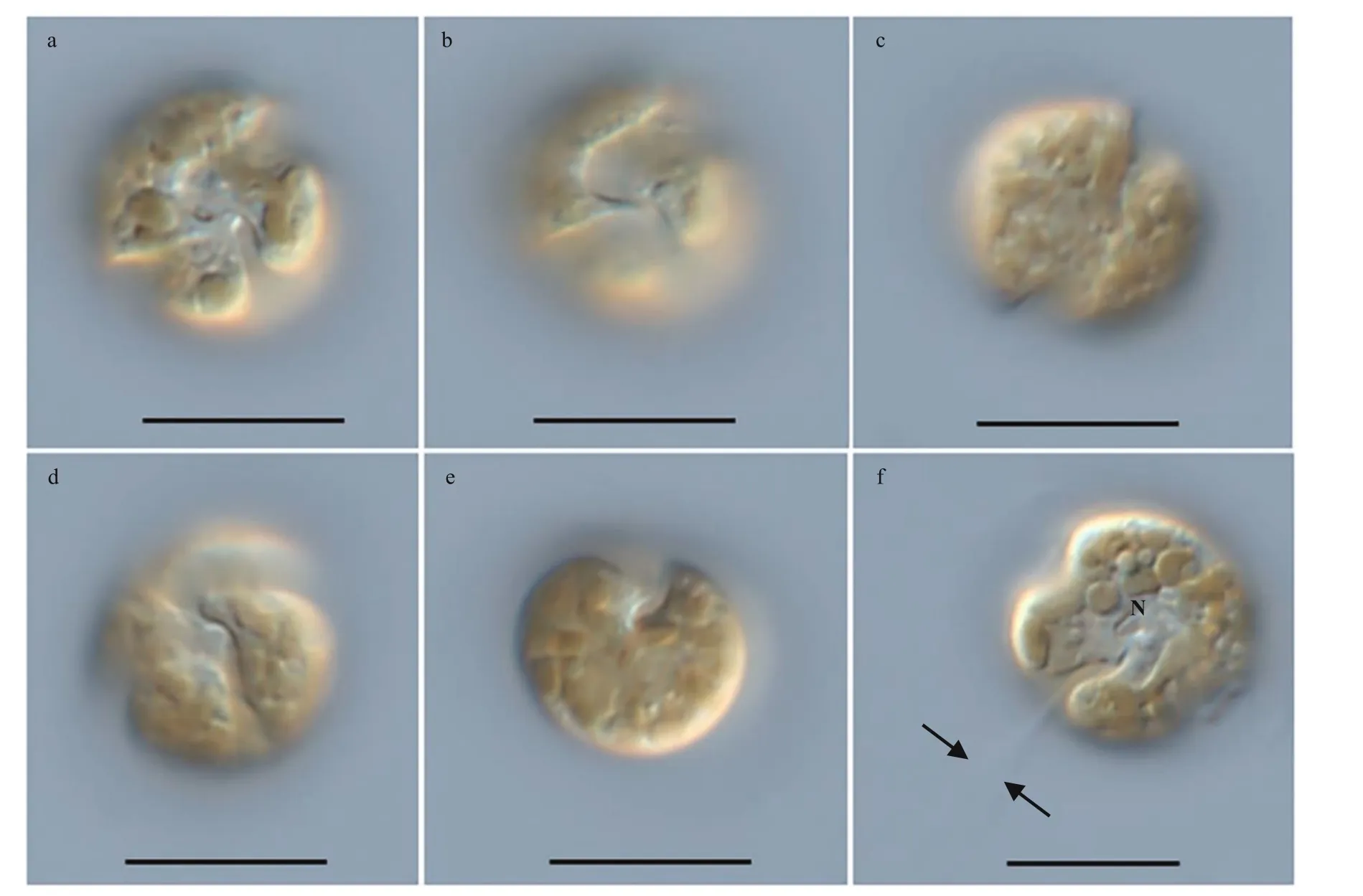
Fig.2 Light microscopy photographs of B. brevisulcata strain S1 germinated from resting cyst
3.2 Morphological observations for the vegetative cells of Biecheleria brevisulcata, Biecheleriopsis adriatica, and Scrippsiella donghaienis
The vegetative cells ofB.brevisulcatawere spherical to ellipsoidal, 7.2–9.2 μm long (average 8.2±0.6 μm;n=50) and 5.8–8.9 μm wide (average 7.3±0.7 μm;n=50) (Figs.2–3). The epicone was slightly wider and longer than the hypocone, and mushroom-shaped (Figs.2–3). The hypocone was bilobed (Figs.2–3). The descending cingulum was deep and wide, and displaced by 1.5 times its own width (Figs.2a–b, 3a–b). The sulcus was in the form of a sigmoid curve (Figs.2a, b, d, & 3a). The nucleus was round and located in the middle or slightly upper part of the hypocone (Fig.2f). Numerous yellowbrownish and reticulated or granulated chloroplasts were distributed peripherally (Fig.2). The arrangement of polygonal amphiesmal vesicles (AVs) was shown in Fig.3. Latitudinal rows of AVs were in four series on the epicone (E1–E4), three series on the hypocone(H1–H3), and three series in the cingular area (C1–C3; Fig.3b, d & e). A narrow elongate apical vesicle(EAV) was present in the apical area (Fig.3e–f), which was surrounded by f ive AVs (a small four-sided AV(X) and four elongated quadrangular AVs) and eight irregular pentagonal AVs (E1-1–E1-8; Fig.3e–f).
The vegetative cells ofBps.adriaticawas spherical to ellipsoid, 5.8–10.1 μm in length (average 7.8±1.2 μm;n=50) and 4.1–8.0 μm in width (average 6.0±0.9 μm;n=50). The epicone was rounded,and almost equal to the hypocone (Figs.4–5). The cingulum was deeper and wider than the sulcus(Figs.4–5). The cingulum was median and displaced by 1.5 times its own width (Figs.4–5). The sulcus was slightly sigmoid (Figs.4a–b, e–f, & 5a). Chloroplasts were yellowish-brown, and reticulated or granulated,which were distributed peripherally (Fig.4). The round refractive bodies were commonly observed(Fig.4). The nucleus was located in the center or slightly upper part of the hypocone (Fig.4c). Many pentagonal or hexagonal AVs were observed on the cell surface (Fig.5). Four epiconal, three cingular, and four hypoconal AV series were formed the latitudinal series (Fig.5a & c). The apical furrow was composed of an EAV and several surrounding AVs (Fig.5a–b, d,& f).
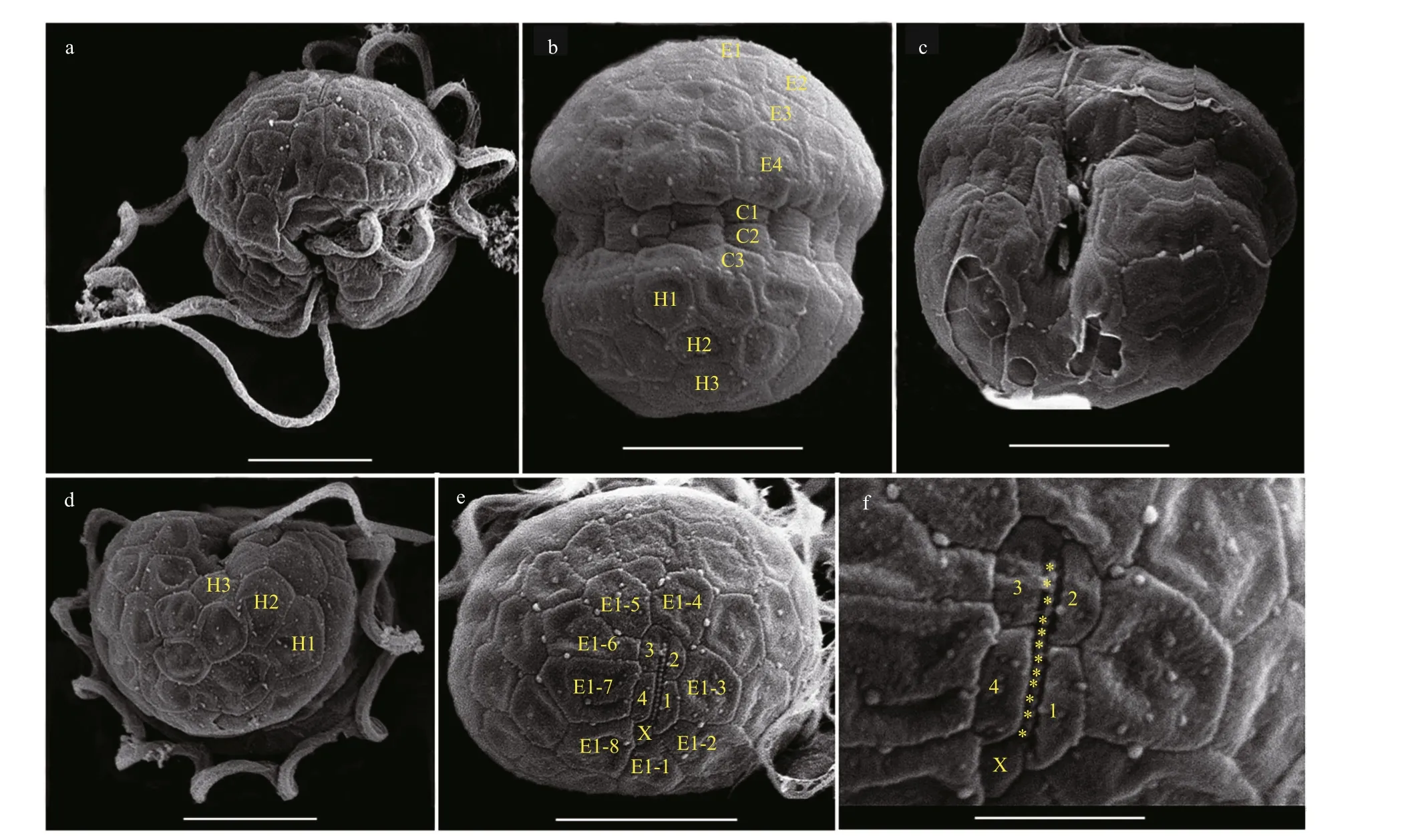
Fig.3 Scanning electron micrographs (SEM) of B. brevisulcata strain S1 germinated from resting cyst
The vegetative cells ofS.donghaieniswas 12.8–20.2 μm long (average 16.4±2.1 μm;n=50) and 9.6–16.0 μm wide (average 12.9±1.5 μm;n=50). The epitheca was conical and longer than the hypotheca,and the hypotheca was rounded and bilobed (Figs.6–7). The plate formula is Po, x, 4ʹ, 3a, 7″, 6c, 6s, 5‴, 2■(Fig.7). The cingulum was wide and deep (Fig.7a–c).The upper part of sulcus was narrower than its lower part (Figs.6a–b & 7a). The apical pore complex (APC)comprised a round apical pore plate and a long canal plate (Fig.7a & e–f). The nucleus was rounded and located centrally (Fig.6d). The yellowish-brown, and granulated chloroplasts were distributed peripherally(Fig.6d–e).
3.3 Pigment composition
Based on available standards, four photosynthetic pigments were identif ied inB. brevisulcata(strain S1),Bps.adriatica(strain S21), andS.donghaienis(strain 23), including one kind of chlorophyll (Chla)and three carotenoids (peridinin, diadinoxanthin, and diatoxanthin; Fig.8). Chl-acontents ofB.brevisulcata(strain S1),Bps.adriatica(strain S21), andS.donghaienis(S23) were 1.02, 0.25, and 10.89 pg/cell, respectively. Peridinin (3.51, 4.91, and 79.11 pg/cell) was the most abundant carotenoid for the three species, then diadinoxanthin, and diatoxanthin.There were 10, 11, and 11 unidentif ied small peaks(either new pigments or known pigments but without standards) forB.brevisulcatastrain S1,Bps.adriaticastrain S21, andS.donghaienisstrain S23.
3.4 Molecular phylogeny
Seven partial LSU rRNA gene sequences ofB.brevisulcata(strains S1, S2, S3, S4, and S5;OL355144–OL355148),Bps.adriatica(strain S21; OL355142), andS.donghaienis(strain 23;OL314541), and one ITS sequence ofS.donghaienis(strain 23; OL314542) were obtained from the clonal cultures. One partial LSU rRNA gene sequence ofBps.adriatica(ON350794) was obtained using single-cell PCR sequencing for the cyst from surface sediment sample of S01-1.
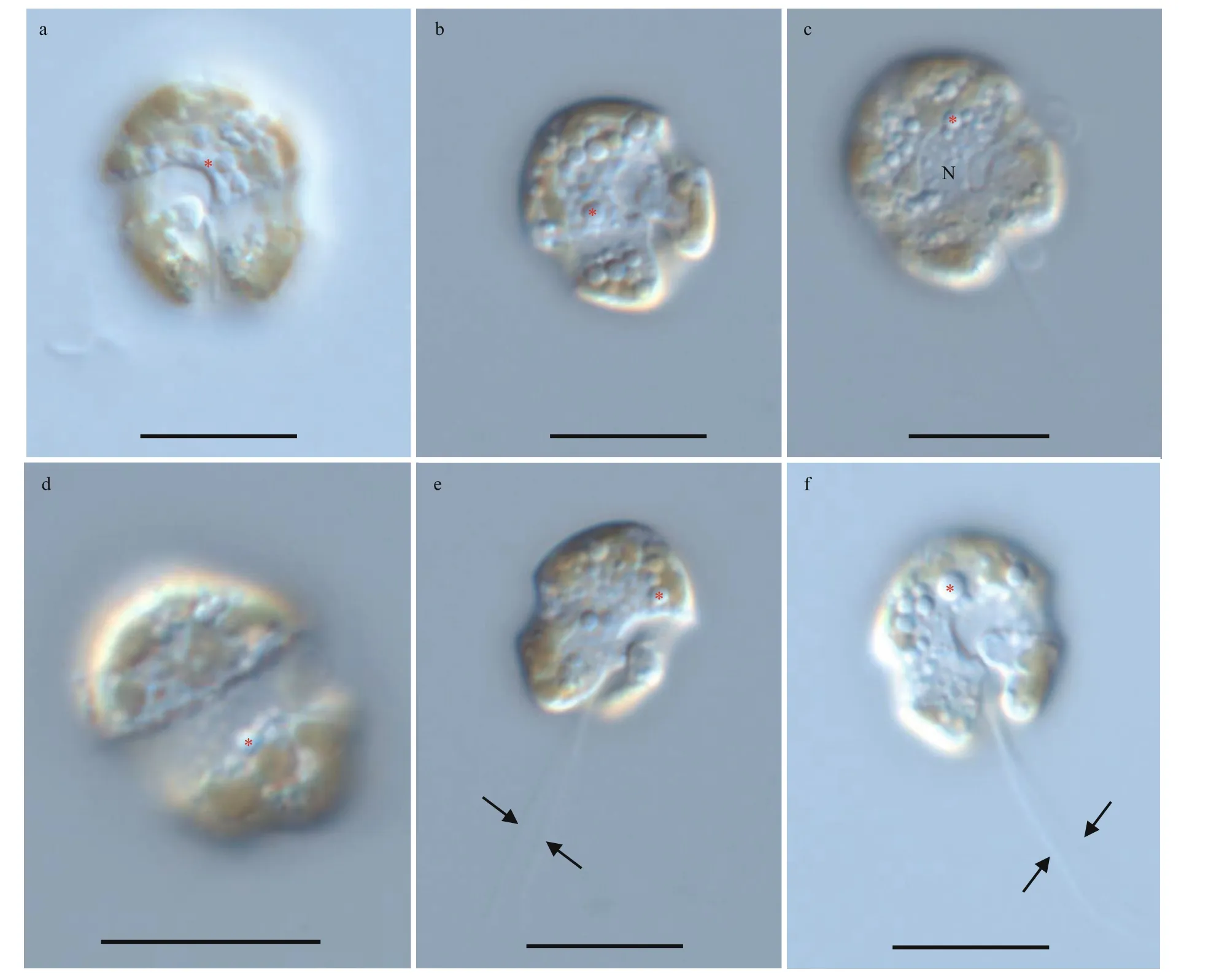
Fig.4 Light microscopy photographs of Bps. adriatica strain S21 germinated from resting cyst
The partial LSU rRNA gene sequence ofB.brevisulcatastrain S1 (1 374 bp; OL355144)was 99.93% (1 373 bp/1 374 bp) identical to the entity of the type material (AB858351), 99.88%(852 bp/853 bp) to 99.93% (1 359 bp/1 360 bp)identical to four entities (AB858352, AB858353,LC068842, and OL699922) deposited asB.brevisulcata, and 98.60% (845 bp/857 bp), 99.41%(509 bp/512 bp), 99.30% (853 bp/859 bp), and 99.32%(877 bp/883 bp) identical toB.pseudopalustris(syn.Woloszynskiapseudopalustris; AF260402),B.baltica(syn.Woloszynskia halophila sensuKremp et al.(2005); AY628430),B.cincta(syn.Woloszynskiacincta; FJ024705), andB.tirezensis(LT601379).
The partial LSU rRNA gene sequence ofBps.adriaticastrain S21 (1 425 bp; OL355142) was 99.69% (1 267 bp/1 271 bp) identical to the entity of the type material deposited at GenBank asGymnodiniumpygmaeumstrain K-0968, 99.24% (1 172 bp/1 181 bp)to 99.79% (1 422 bp/1 425 bp) identical to 12 entities(AB858354–AB858356, LC068843, LC413947–LC413950, LM992904–LM992906, and OL691545)deposited asBps.adriatica, 99.78% identical to 11 entities (KM603188–KM603198) deposited asBps. cf.adriatica, and 99.13% (794 bp/801 bp) to 99.30% (1 415 bp/1 425 bp) identical to four entities(JN558103–JN558105, KM603185) deposited asProtodiniumsimplex. Among all these entities in GenBank,Bps.adriaticaalso corresponded to the cultures established from vegetative cells isolated from the same area where the cultures from cysts were established (i.e., East China Sea; Luo et al., 2015).
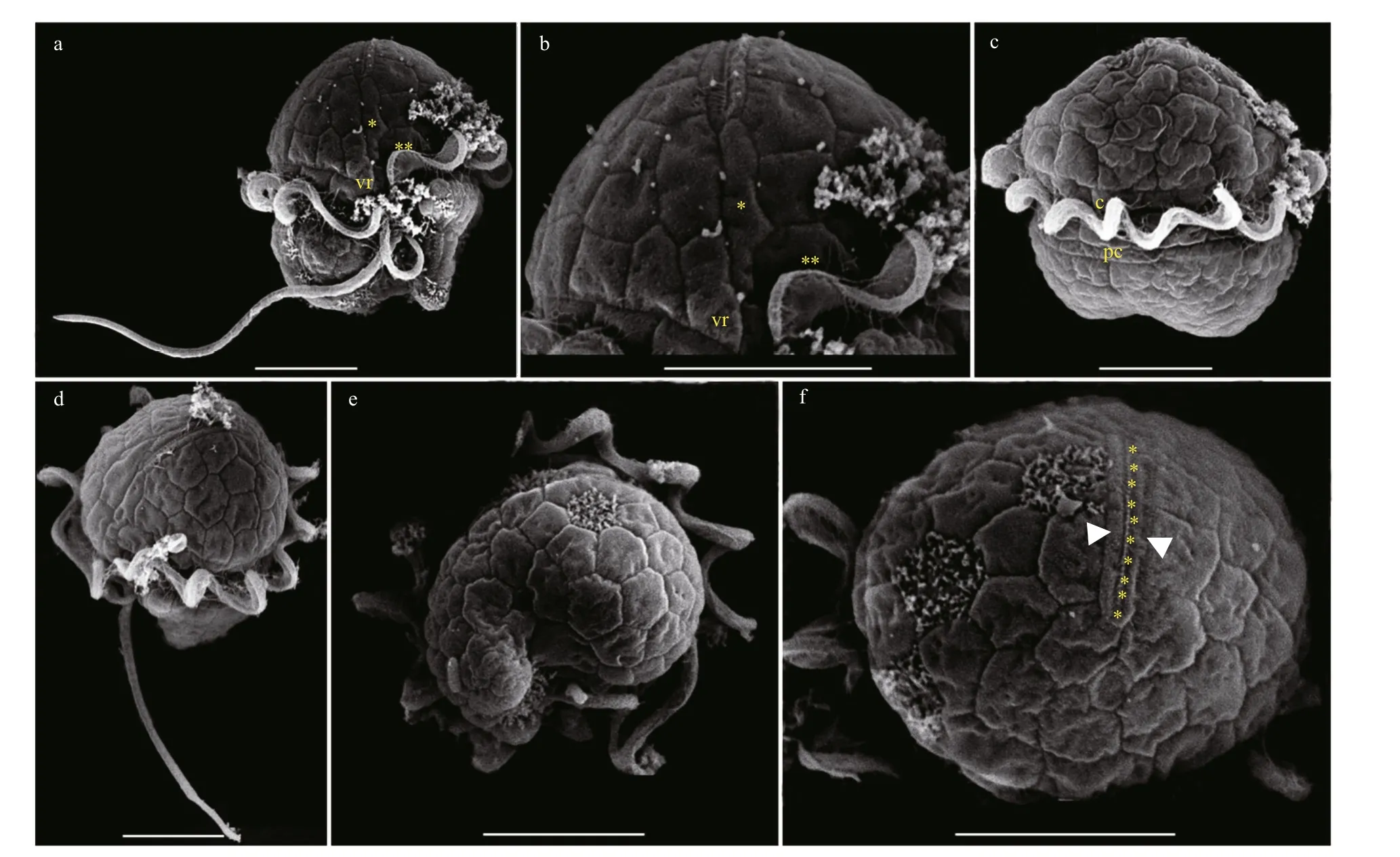
Fig.5 Scanning electron micrographs (SEM) of Bps. adriatica strain S21 germinated from resting cyst
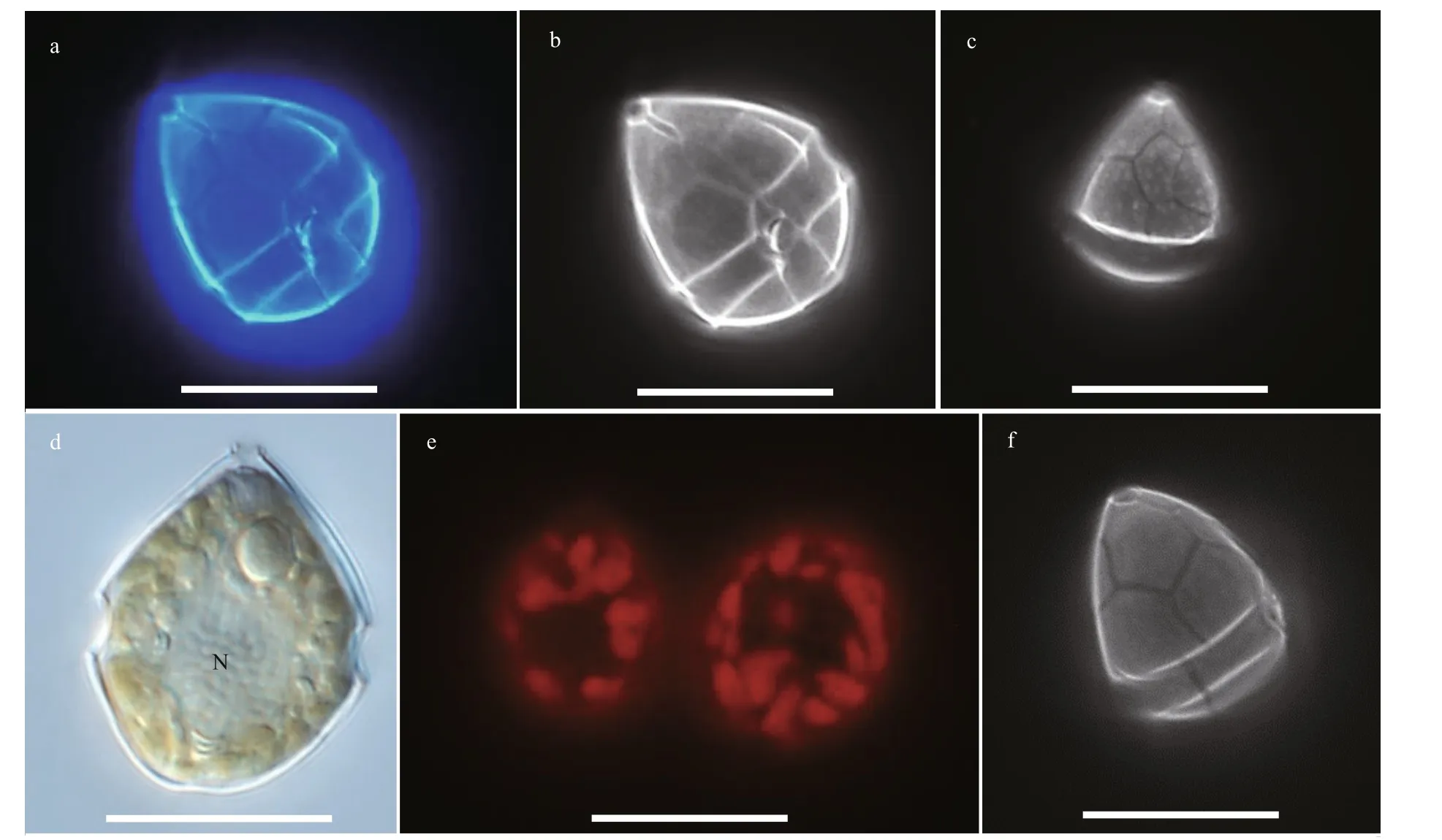
Fig.6 Light microscopy photographs of S. donghaienis strain S23 germinated from resting cyst
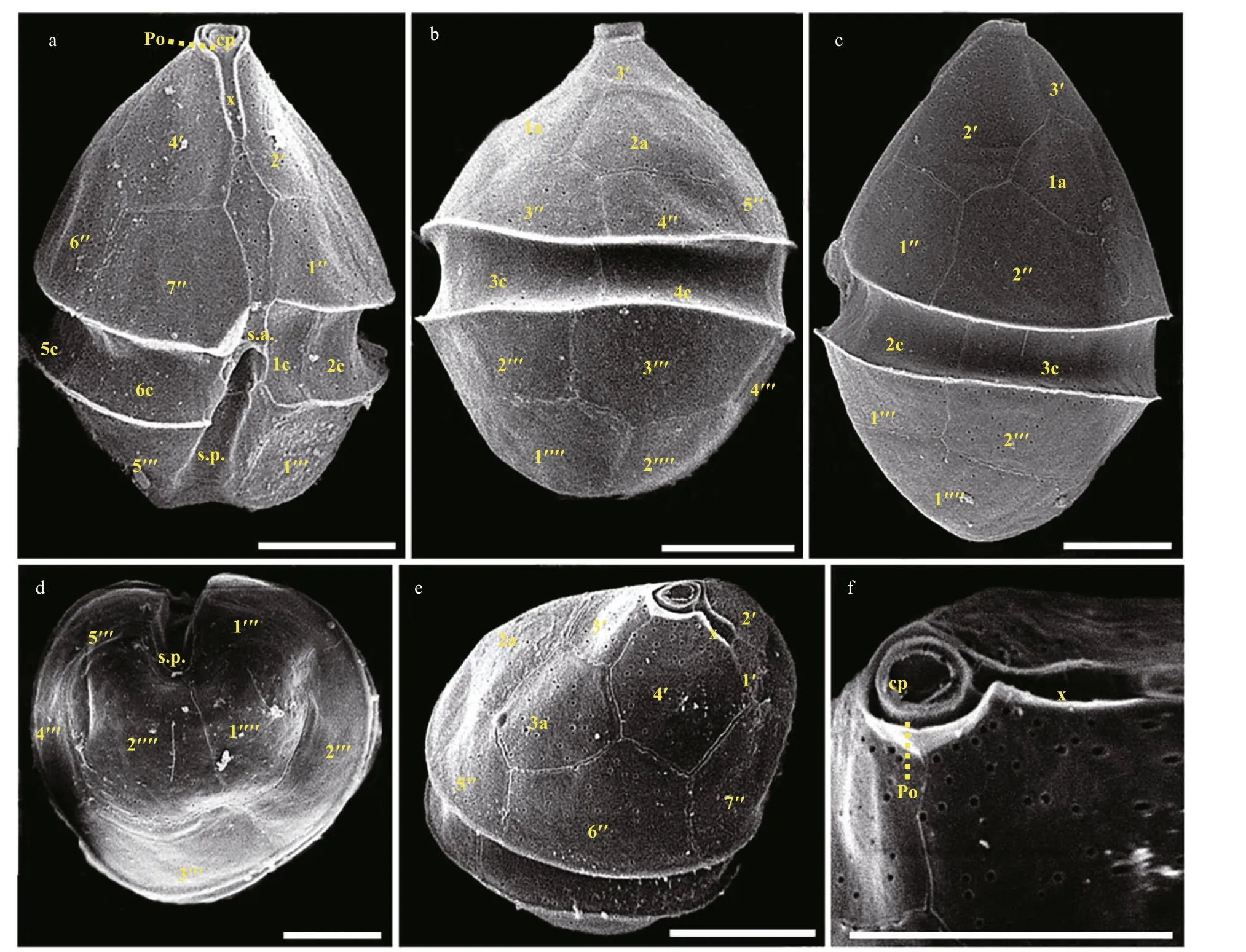
Fig.7 Scanning electron micrographs (SEM) of S. donghaienis strain S23 germinated from resting cyst
The partial LSU rRNA gene sequence ofS.donghaienisstrain 23 (1 433 bp; OL314541) was 98.51% (796 bp/808 bp) to 99.51% (812 bp/816 bp)identical to 69 entities deposited asS.donghaienisin GenBank. The ITS sequence ofS.donghaienisstrain 23 (663 bp; OL314542) was 93.80% (454 bp/484 bp)to 99.64% (558 bp/560 bp) identical to f ive entities(AY685008, HQ729492, HQ729502, JN982374, and MG914024) deposited asS.donghaienis, and 99.28%(550 bp/554 bp) to 99.68% (617 bp/619 bp) identical to seven entities (AY499533, AY676151, AY67615,AY788357, and FJ823594–FJ823596) deposited asScrippsiellasp. in GenBank.
Phylogenetic analyses ofB.brevisulcata,Bps.adriatica, andS.donghaienisusing maximum likelihood (ML) and Bayesian inference (BI)generated similar trees based on LSU rRNA gene and ITS sequences but diff ered at a few internal nodes(Figs.9–11). ForB.brevisulcata, our sequences and other sequences (AB858351–AB858351,LC068842) formed a coherent clade with strong support (0.71/100), which is sistering to the clade includingWoloszynskiahalophila(EF205019,AY628430),B.cincta(FJ024705),Woloszynskiapseudopalustris(AF260402), andGymnodiniumsp. (AY318248) with strong support (0.95/100;Fig.9). ForBps.adriatica, our sequence and other sequences (LC068843, LC413947, and LC413948),Bps. cf.adriatica(KM603188–KM603198), andG.corii(GU477610) formed a coherent clade with strong support (0.93/100; Fig.9), and formed a wellsupported sister clade (0.98/100; Fig.9) including the type material ofBps.adriatica(“G.pygmaeum”,EU857537), andG.corii(EU165298, AF318226).For the phylogenetic analysis ofS.donghaienisbased on LSU rRNA gene sequences, our sequence(OL31454), 69 sequences deposited asS.donghaienis,andScrippsiellasp. (AY685011) formed a coherent clade with maximal support (1/100; Fig.10),and formed sister groups with otherScrippsiellaspecies (Fig.10). For the phylogenetic analysis ofS.donghaienisbased on ITS sequences, our sequence (OL314542), four sequences deposited asS.donghaienis(HQ729492, HQ729502, JN982374,and MG914024), and seven sequences deposited asScrippsiellasp. (AY499533, AY676151, AY676155,AY788357, FJ823594–FJ823596) formed a coherent clade with medium support (0.51/88; Fig.11), and formed a sister group with one sequence deposited asS.donghaienis(AY685008) with maximal support(1/100; Fig.11).
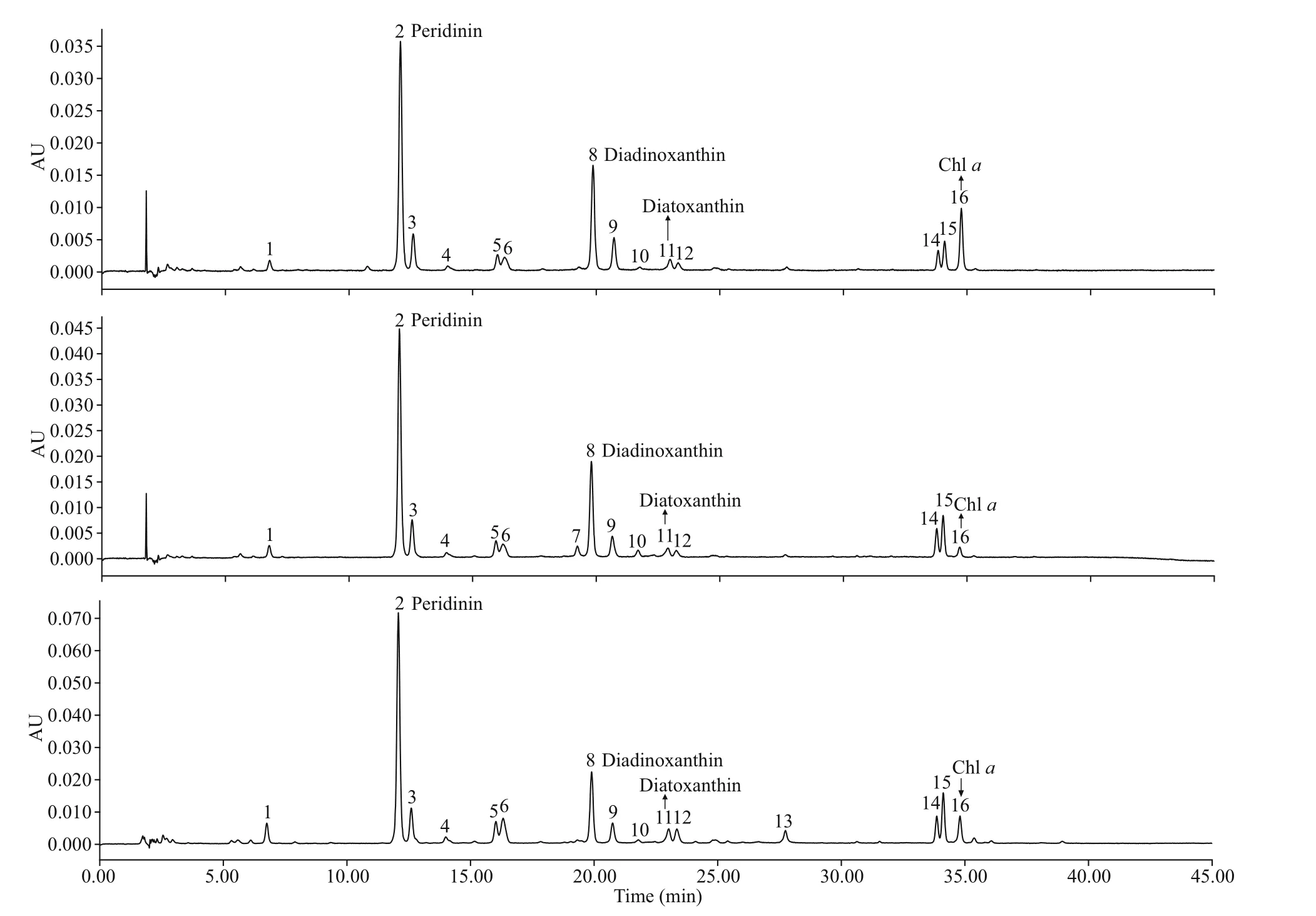
Fig.8 Absorption chromatograms (440 nm) of the pigment extracts of B . brevisulcata strain S1, Bps. adriatica strain S21, and S. donghaienis strain 23 from the East China Sea, China
3.5 The genetic diversity ref lected in partial rDNA and ITS sequences
The pairwise distances computed using Jukes-Cantor model showed that the sequence divergence among the LSU rRNA gene sequences ofBps.adriaticafrom the present study, type material(deposited in GenBank asG.pygmaeumwith the accession No. EU857537), and sequences deposited asBps.adriatica(AB858354–AB858356, LC068843,LC413947–LC413950, and LM992904–LM992906)ranged from 0.000 to 0.006, the distances between our sequence andBps. cf.adriatica(KM603188–KM603198) were 0.002, and among our sequence, and the sequences deposited asP.simplex(AF060901),Gymnodiniumsp. (EF205006),G.corii(AF318226,EU165298, GU477610, KT389967, KT389942),P.simplex(FJ024704, JN558103–JN558105,KM603185–KM603187) ranged from 0.000 to 0.007 (Supplementary Table S1). However, the distances between our sequence andB.brevisulcata(AB858351),Ansanellagranifera(HG529980),andA.margalef ii(AY154957) were 0.039, 0.029,and 0.244, respectively (Supplementary Table S1).Sequence diff erences among our strain (S21), the other stains from water samples or surface sediments formed a well-supported clade with ours (Fig.9), and the one we found in the surface sediment sample in the same sea area were found mainly at three stable positions (3 bp/1 438 bp (D1–D6 regions); Fig.12).
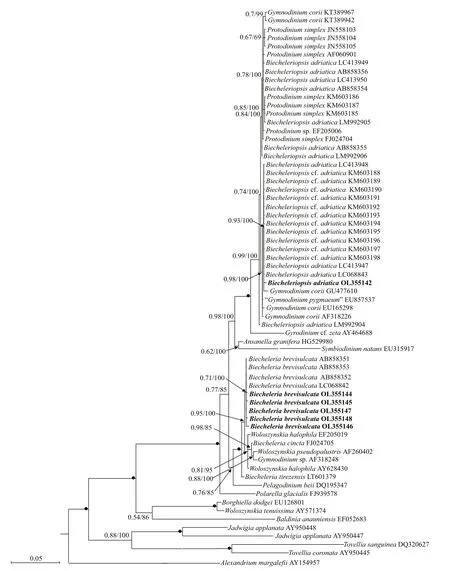
Fig.9 Molecular phylogeny of B. brevisulcata, Bps. adriatica, and diverse assemblage of other dinof lagellates inferred from the partial LSU rRNA gene sequences based on Bayesian inference (BI) with A. margalef ii (AY154957) as outgroup
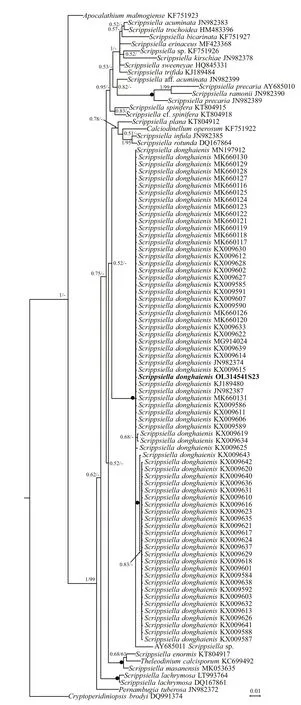
Fig.10 Molecular phylogeny of S. donghaienis and diverse assemblage of other dinof lagellates inferred from the partial LSU rRNA gene sequences based on Bayesian inference (BI) with C. brodyi (DQ991374) as outgroup
The genetic distance based on LSU rRNA gene ofB.brevisulcataobtained in the present study and previous works, and other related species were compared (Supplementary Table S2). The sequence divergence among the LSU rRNA gene sequences ofB.brevisulcataobtained in our work (OL355144–OL355148) and other strains previously deposited in GenBank (AB858351–AB858353, LC068842)ranged within 0.000–0.007 (Supplementary Table S2), the sequence divergence amongB.brevisulcata(OL355144–OL355148) and otherBiecheleriaspecies (B.baltica,B.cincta,B.pseudopalustris, andB.tirezensis) ranged 0.002–0.015 (Supplementary Table S2), but the sequence divergence amongB.brevisulcata(OL355144–OL355148) and other distant speciesPelagodiniumbeii(DQ195347),Polarellaglacialis(FJ939578), andA.margalef ii(AY154957) were 0.055–0.057, 0.077–0.079, and 0.808–0.875, respectively (Supplementary Table S2).
The genetic distance based on LSU rRNA gene and ITS ofS.donghaienisobtained in the present study and previous work, and other related species were compared(Supplementary Tables S3 & S4). The sequence divergence among the LSU rRNA gene sequence ofS.donghaienisobtained in our work (OL314541) and other strains previously deposited in GenBank ranged 0.000–0.009 (Supplementary Table S3), 0.013–0.139,and the sequence divergence amongS.donghaienis(OL314541) and otherScrippsiellaspecies (S.acuminate,S.erinaceus,S.sweeneyae,S.spinifera,S.plana,S.bicarinata,S.kirschiae,S.trif ida,S.infula,S.rotunda,S.lachrymose,S.enormis,S.masanensis,S.precaria, andS.ramonii) ranged 0.002–0.015(Supplementary Table S3). The pairwise distances computed using Jukes-Cantor model showed that the sequence divergence between the ITS sequences ofS.donghaienisobtained in the present study, and sequences deposited asS.donghaienis(AY685008,HQ729492, HQ729502, JN982374, and MG914024)ranged from 0.000 to 0.015 (Supplementary Table S4),the sequence divergence between the ITS sequences ofS.donghaienisobtained in the present study, and the otherScrippsiellaspecies ranged 0.175–0.360(Supplementary Table S4). Sequence diff erences between our strain (S23) and the other stains from water samples or surface sediments formed a wellsupported clade with ours (Fig.11) were found mainly at two stable positions (2 bp/599 bp; Fig.13).
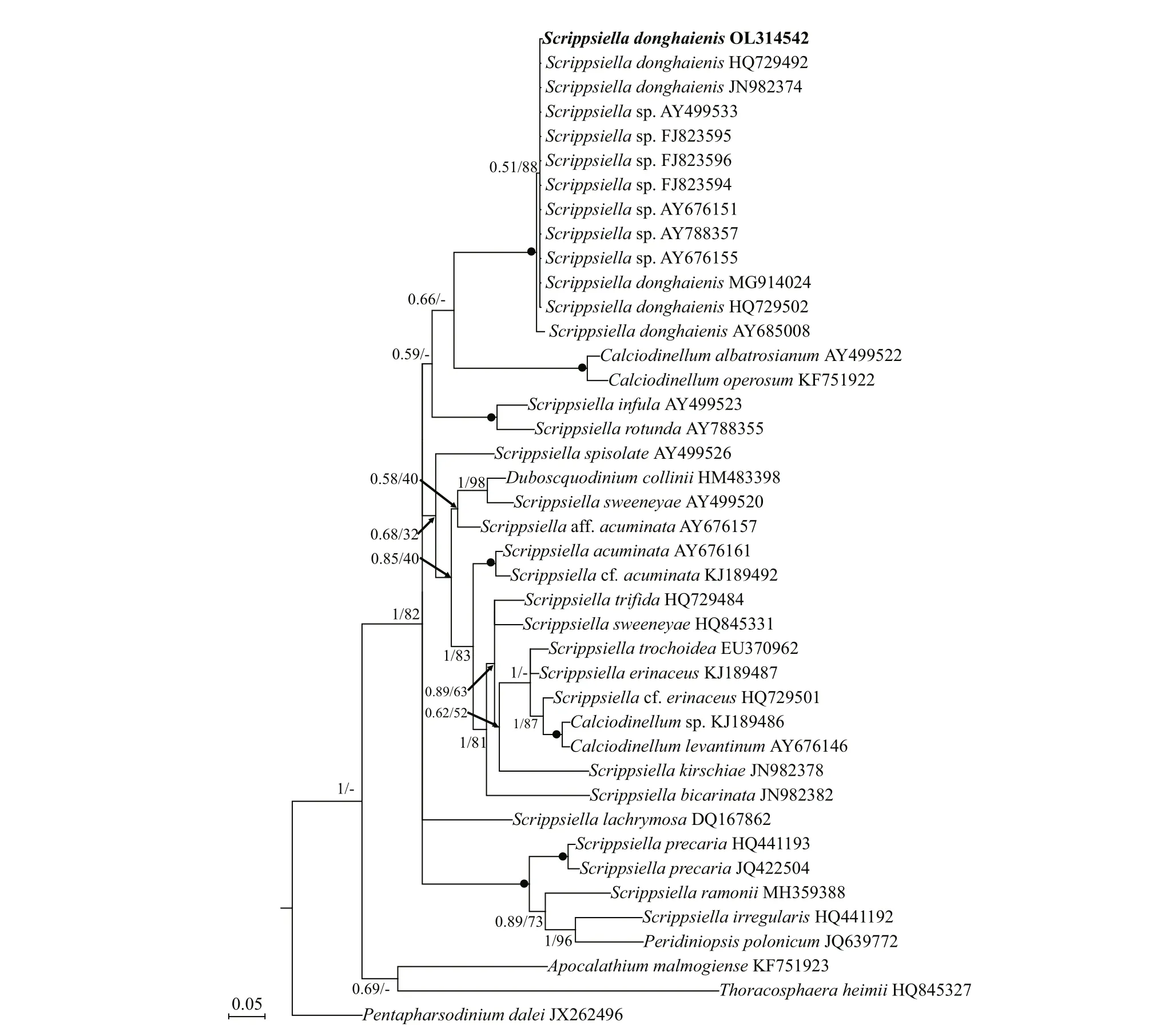
Fig.11 Molecular phylogeny of S. donghaienis and diverse assemblage of other dinof lagellates inferred from the internal transcribed spacer (ITS) region sequences based on Bayesian inference (BI) with P. dalei (DQ991374) as outgroup
4 DISCUSSION
4.1 Identif ication of Biecheleriopsis adriatica resting cyst presented in the f ield sediment

Fig.12 Sequence variation in the LSU rDNA sequences among Bps. adriatica strain 21, the one cyst of this species found in the surface sediment sample in the same sea area, and other strains of Bps. adriatica formed a well-supported clade with ours in Fig.9
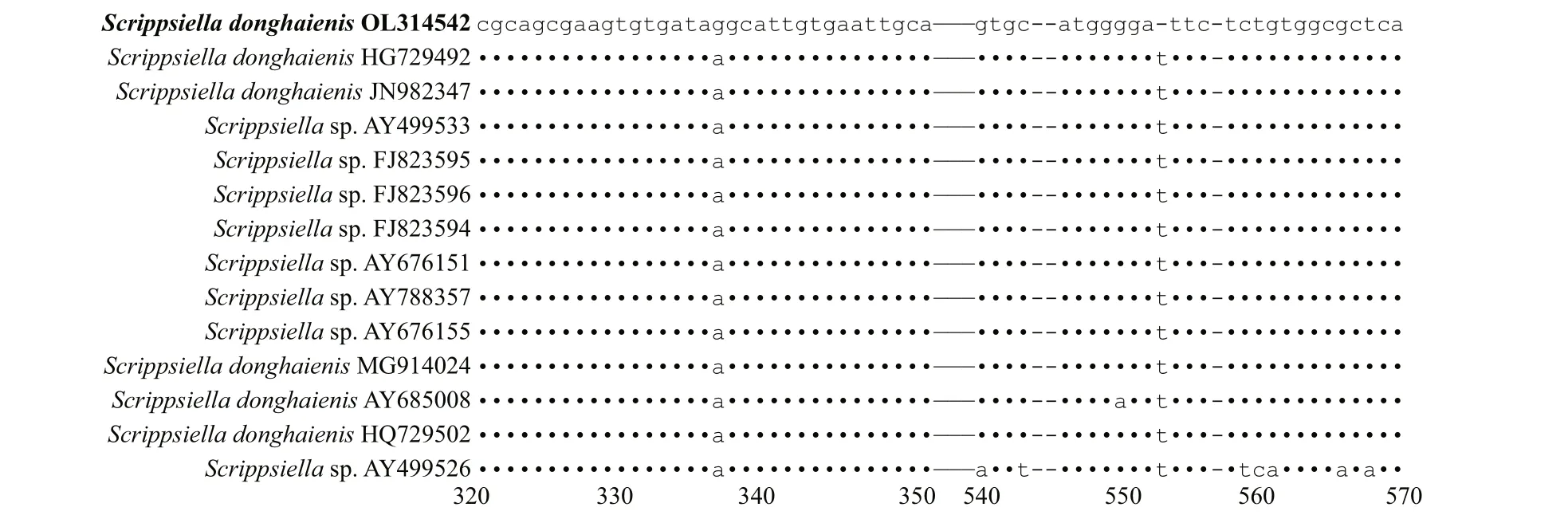
Fig.13 Sequence variation in the ITS sequences between S. donghaienis strain 23 and the other strains formed a well clade with ours in Fig.11
Biecheleriopsisadriaticawas described by Moestrup et al. (2009) who diff erentiated it from the species ofBiecheleriabased on the presence of a nuclear connector and a 51-bases long fragment of D2 domain of LSU rRNA gene. Moestrup et al. (2009)also found resting cysts in the culture ofBps.adriatica,described as having ellipsoidal to ovoid shapes and numerous spines, a size of 7–8-μm length and 5–6-μm width. Benico et al. (2019) found resting cyst-like cells in unialgal culture being morphologically diff erent from vegetative cell, and they called them as resting cyst-like cells, which were ~16 μm, spherical,covered by a transparent thick wall, and contained a red accumulation body. Kang et al. (2009), Wang et al. (2013), and Kang and Wang (2018) germinatedBps.adriaticafrom cyst assemblages (identif ied asG.coriiin Kang et al. (2009) and Wang et al. (2013))from Southern Chinese coastal sediments, but did not observe the genuine resting cyst of this organism.Luo et al. (2015) also germinatedBps. cf.adriatica(very possible conspecif ic with our strain S21) from the sediment collected from the Yellow Sea and the South China Sea, but also without observation on the morphology ofBps. cf.adriaticacyst. The abovementioned works onBps.adriaticahave conf irmed that it could form resting cyst, but none of them reported the morphology of cyst from the f ield. In this work, we germinatedBps.adriaticafrom a single cyst collected from the East China Sea, and the species was identif ied with morphological and molecular characterization allowing us to conf idently conf irm the cyst-motile stage relationship of this small-sized species. Resting cyst ofBps.adriaticawas spherical and sub-spherical, which is similar to the so-called“resting cyst-like cells” as observed by Benico et al.(2019), but diff erent from the cyst from culture having an ellipsoidal to ovoid morphology (Moestrup et al.,2009). The diameter ofBps.adriaticain our work(~7.3 μm) is very close to the cyst (7–8 μm long and 5–6 μm wide) observed by Moestrup et al. (2009), but much smaller than the resting cyst-like cells (~16 μm)observed by Benico et al. (2019). The sieves with the pore size larger than 20 μm were routinely used to concentrate cysts, and then followed by cyst assemblage germination, the cysts with smaller size(<20 μm) would be lost during processing, therefore,the eff orts in discovering more small sized cysts should focus on the cyst assemblage being smaller than 20 μm.
4.2 Viability of dinof lagellates cysts stored in longburied sediments
In the past, changes of species composition in sediments have been used to assess environmental changes including eutrophication, changes in salinity, or oxygen concentration (Dale et al., 1999;Ellegaard et al., 2013, 2020; Bringué et al., 2016;García-Moreiras et al., 2018; Li et al., 2021; Siano et al., 2021). The adaptive responses of dinof lagellate species might be inf luenced by anthropogenic activities or climate changes, and revealing the evolutionary processes in a species is very important to understanding the genetic structure of populations.However, the length of live resting cyst preserved in the sediment cores (e.g., decades to centuries) or the molecular information (rRNA genes or other gene markers) for the buried cysts or residual fragments have not been well explored before 2010. After that, the impacts of environmental changes on the physiology, genetic structure, and diversity in various dinof lagellate species (Ribeiro et al., 2011; Klouch et al., 2016; Lundholm et al., 2017; Kremp et al.,2018; Delebecq et al., 2020; Ellegaard et al., 2020;Girault et al., 2021) have been investigated. In this study, we successfully revivedB.brevisulcata,Bps.adriatica, andS.donghaienisfrom a sediment core dated back to 1941±18 AD from the East China Sea,which indicates that these cysts are viable for at least 70 years. Klouch et al. (2016) detected molecular signal ofS.donghaienisin a sediment core sample dated back to 1866±7 AD from the Bay of Brest,France, and successfully germinated this organism only from the layers of 2–17 cm corresponding to the years of 2010±1 AD to 1978±2 AD, much more recent than 1941±18 AD. In older sediments (70–100 years),Protoceratiumreticulatum,Lingulodiniumpolyedrum, andP.daleiwere successfully germinated(Lundholm et al., 2011; Ribeiro et al., 2011). Girault et al. (2021) successfully germinatedA.minutumandS.acuminatafrom the sediment corresponding to 1947±11 AD. Kremp et al. (2018) foundApocalathiummalmogienseis viable in the sediment layers of 106-year old. Recently, Delebecq et al. (2020) germinatedA.minutum,Heterocapsaminima,Margalef idiniumpolykrikoides,Protoperidiniumspp., andDiplopsalisgroup in sediments dated back to 117±21 years ago,andS.acuminatabeyond 156±27 years ago. Our work increases the diversity of species (B.brevisulcataandBps.adriatica) potentially revivable from more longburied sediments, which will promote studies in the f ield of resurrection ecology.
4.3 Genetic diversity among dinof lagellates revived from cysts stored in long-buried sediments and current condition
Genetic diversity is an important aspect of biodiversity, which is def ined as measurements that determine the changes of genetic variability within any level of a taxon but more often within a species or even population, e.g., allelic diversity or richness,mutational diversity, and eff ective population size(Hughes et al., 2008; Ebenezer et al., 2012; Ellegren and Galtier, 2016). Previous phylogeographic studies have revealed high levels of genetic diversity in a number of dinof lagellate species, e.g.,A.fundyense(Erdner et al., 2011),Amphidiniumspp. (Murray et al., 2012),Gambierdiscusspp. (Nishimura et al., 2013),Margalef idiniumfulvescens(Lin et al.,2020),Ostreopsisspp. (Lee and Park, 2020), andPseudocochlodiniumprofundisulcus(Hu et al., 2021).However, all above-mentioned works examined the genetic diversity among contemporary populations or even within a population rather than that between historical and contemporary populations as done in the present work, although the history here is less than a century (~70 years). From our phylogenetic tree, our f ive strains ofB.brevisulcatabranched together with the other strains of this species, but their evolution distance varied. Our strain ofBps.adriaticaand three strains ofBps.adriatica, eleven strains ofBps. cf.adriatica, and one strain ofG.coriiformed a coherent clade, and a sister group with the type material of this species, but diff ered at three stable positions. Our strain ofS.donghaienis, the other strains of this species, and one strain ofScrippsiellasp. formed a coherent clade. Within this clade, there were four branches, our strain grouped together with those historical and contemporary populations.We also compared the rRNA gene based genetic distances of the seven strains ofB.brevisulcata,Bps.adriatica, andS.donghaienisrevived from a long-buried sediment dated back to 1941±18 AD and those from water samples and surface sediments (the contemporary age), and found the genetic distance between our strain ofB.brevisulcataand other strains range from 0.002 to 0.006, but among other strains,the genetic distances are 0.000–0.005, most 0.001,and similar trends were also found inBps.adriatica,andS.donghaienis. We also found diff erences in three stable positions of LSU rRNA gene sequences between the population ofBps.adriaticawe found 70 years ago and the contemporary or present-day population (including the cyst of this species we found in the surface sediment and vegetative cells found by Luo et al. (2015) in the same sea area),and ITS sequences in two stable positions between the population ofS.donghaieniswe found 70 years ago and the contemporary or present-day population(including the cyst of this species found by Gu et al.(2008) in the surface sediment in the same sea area).It seemed these current populations are genetically diff erent from those that existed in the area 70 years ago, which suggests that there has been a shift in the populations ofBps.adriaticaandS.donghaienis.Due to a limited size of dataset for the present work,the possible historical succession of populations ofBps.adriatica,S.donghaienisand even other species in the area requires a more intensive and extensive investigation, as these possible shifts may be highly indicative of the environmental changes and anthropological activities that occurred in the area during the past.
5 DATA AVAILABILITY STATEMENT
All data generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
6 ACKNOWLEDGMENT
We are highly grateful of the two anonymous reviewers for their patience, critical comments, and generous suggestions, which helped greatly the improvement of the manuscript. Sediment samples’collections were supported by NSFC Open Research Cruise, funded by Shiptime Sharing Project of NSFC.We appreciate for the cruises conducted by R/VXiangYangHong18and staff from First Institute of Oceanography, Ministry of Natural Resources, China.
 Journal of Oceanology and Limnology2022年6期
Journal of Oceanology and Limnology2022年6期
- Journal of Oceanology and Limnology的其它文章
- Overview of harmful algal blooms (red tides) in Hong Kong during 1975–2021
- Information standardization for typical toxic and harmful algae in China’s coastal waters—a case study of Karenia mikimotoi*
- Biochemical composition of the brown tide causative species Aureococcus anophageff erens cultivated in diff erent nitrogen sources*
- Identif ication of paralytic shellf ish toxin-producing microalgae using machine learning and deep learning methods*
- Screening for lipophilic marine toxins and their potential producers in coastal waters of Weihai in autumn, 2020*
- First observation of domoic acid and its isomers in shellf ish samples from Shandong Province, China*
