Inhibition and mechanism of AEAMP9,a novel compound formula of alcoholic extract of anticancer medicinal plants,on lung cancer A549 cells
AO Zhiguang,JIANG Yuhong,YANG Lei,LI Dong,WANG Jun,WANG Siyi,MAO Canquan
(Sichuan Engineering Research Center for Biomimetic Synthesis of Natural Drugs,School of Life Science and Engineering,Southwest Jiaotong University,Chengdu 610031,China)
Abstract In view of the high morbidity and mortality of lung cancer,lung cancer was concerned and human non-small cell lung cancer A549 cells were used as the target in this study. Here,after screening 124 kinds of medicinal plants in A549 lung cancer cells and the determination of their anticancer orders,an efficient and novel compound formula of AEAMP9 was developed and its underlying molecular mechanism was investigated. AEAMP9 was prepared from the alcoholic extracts of six anticancer medicinal plants including Hedyotis diffusa Willd. (Bai Hua She She Cao),Phellodendron amurense Rupr. (Huang Bai),Glycyrrhiza uralensis Fisch. (Gan Cao),Platycladus orientalis (L.) Franco (Ce Bai Ye),Chrysanthemum indicum L. (Ye Ju Hua) and Notopterygium incisum Ting ex H. T. Chang (Qiang Huo). It can effectively inhibit the viability and clone formation of cancer cells,provoke apoptosis and block migration in A549 cells. Mechanistically,AEAMP9 significantly decreased the expressions of ABCG2,FoxM1,Vimentin,MMP14 and the ratio of Bcl-2/Bax,consequently promoted the activation of apoptosis associated genes including p53,caspase 3,caspase 8 and caspase 9. Network pharmacology and transcriptome RNA-seq revealed that many genes were significantly and differentially expressed in A549 cells after treatment with AEAMP9. TNF,IL-17 and other important signal pathways related to tumor,inflammation and apoptosis were closely regulated. EGR1,IL-6,ALK and VEGF may be the key regulatory targets for the interaction of AEAMP9 and lung adenocarcinoma. The components of quercetin,kaempferol and luteolin in AEAMP9 may play an important role in antitumor activity. Safely,AEAMP9 had no obvious toxic effects on SPF Kunming mice. Furthermore,AEAMP9 can significantly inhibit the tumor growth in mouse A549 xenograft models.
Keywords AEAMP9; cell apoptosis; cancer; medicinal plant; network pharmacology
1 Introduction
Cancer is the second cause of morbidity and mortality worldwide,while lung cancer remains the leading cause of cancer death[1]. Tumorigenesis in humans is a multistep process and related to both interior and external factors[2-4]. The classical and current main methods for cancer therapy are still surgery,chemotherapy,radiotherapy,as well as the adjuvant biotherapy and the combination of them. Though there is a rapid progress in such areas including the PD-1/PD-L1 targeted immune checking point inhibition and Chimeric Antigen Receptor T-Cell (CAR-T) immunotherapy[5],the inefficiency in some cancers and cases,drug resistance,metastasis and relapse is still the main bottlenecks for current cancer therapy and survival[6-7]. Hence,it is still urgent to develop novel drugs or therapeutic methods to improve the prognosis of cancer patients.
Medicinal plants are the abundant and treasure sources for drug development. More than 50% of current anticancer drugs are of natural origin[8]. In China,there is thousands of years’ history to effectively treat various diseases by traditional Chinese medicine (TCM),while the main materials are just the medicinal plants. Many TCM components/plants/formulas had been reported to be active in anticancer therapy such as Taxol,Camptothecin,Berberine,HedyotisdiffusaWilld. as well as Bo-Er-Ningcapsule[9-10]. The multiple anticancer mechanisms of TCMs had been reviewed in many reports[11-12]. However,current studies focused mostly on the finding and identification of active components from medicinal plants and their mechanism investigation,rare works were on the simultaneous evaluation of the effects of medicinal plants in large numbers and scales. Therefore,efficacy-driven approaches for the finding of medicinal plants and novel compound formulas against cancers were greatly needed.
In view of the seriousness of cancer and the high morbidity and mortality of lung cancer especially in China,lung cancer was concerned and human non-small cell lung cancer A549 cells were used as the target in this study. In the work,we established a new method for developing anticancer formula based on medicinal plants. We started with a screening of 124 kinds of alcoholic extracts of medicinal plants(AEAMP)against lung cancer A549 cells,and then AEAMP9 was optimally designed and established. The efficiency and possible molecular mechanisms of AEAMP9 for the inhibition of A549 cells were further studied bothinvitroandinvivo.
2 Materials and methods
2.1 Chemicals,reagents and animals
Medicinal plants were bought from Chengdu traditional Chinese medicine market (Chengdu,P. R. China) and morphologically identified. Three medicinal plants includingGlycyrrhizauralensisFisch. (Gan Cao),Platycladusorientalis(L.) Franco (Ce Bai Ye) andNotopterygiumincisumTing ex H. T. Chang (Qiang Huo) were further molecularly identified by ITS2 DNA barcode. Trypsin,Dulbecco’s modified Eagle’s medium (DMEM) and fetal bovine serum (FBS) were obtained from Gibco (CA,USA). Bicinchoninic acid (BCA) Bradford assay kit was purchased from Beyotime (Haimen,China). MTT and caspase activity assay kits were obtained from KeyGEN BioTECH (Nanjing,P. R. China). Mouse monoclonal anti-FoxM1,anti-p53 (1∶500,Santa Cruz Biotechnology,USA); Rabbit polyclonal anti-MMP14,anti-Bcl-2 (1∶500,Elabscience,Wuhan,China),Rabbit monoclonal anti-GAPDH(1∶10 000,Proteintech,Wuhan,China). The PrimeScriptTMRT reagent Kit and SYBR®Premix Ex TaqTMII Kit were purchased from TaKaRa (Dalian,P. R. China). Tea oil was kindly provided by Yunnan Puer Danzhou Pharmaceutical Com. Ltd. (Ninger,P. R. China). Mice were purchased from Chengdu Dashuo laboratory animal Com.(Chengdu,P. R. China).
2.2 Preparation of alcoholic extract of medicinal plants
A total of 124 kinds of air-dried medicinal plants were milled and soaked in 75% alcohol respectively at room temperature with stirring once a week. After one month’s extraction,the supernatants were filtered through a Millipore 0.45 μm pore size membrane and used for this study. The formulas of AEAMP from no.1 to 10 (designated as AEAMP1-10) were the mixtures of different ratio of alcoholic extracts of medicinal plants includingHedyotisdiffusaWilld.(Bai Hua She She Cao),PhellodendronamurenseRupr. (Huang Bai),GlycyrrhizauralensisFisch.,Platycladusorientalis(L.) Franco,ChrysanthemumindicumL. (Ye Ju Hua),NotopterygiumincisumTing ex H. T. Chang,ZingiberofficinaleRoscoe (Sheng Jiang),SapindusmukorossiGaertn. (Wu Huan Zi),CurcumalongaLinn.(Jiang Huang) andFructuscamptothecaeacuminatae(Xi Shu Guo),which were the ten medicinal plants ranked ahead in their inhibitory effects for A549 cancer cells. For the preparation of the freeze-dried powder of AEAMP9,the filtered supernatants were firstly evaporated using a rotary evaporator (Yamato Sci.,Tokyo,Japan) at 60 ℃ for 4-6 h and then freeze-dried using a lyophilizer (FD-1A-80,Yuming instrument co. LTD,Shanghai,P.R. China) for 24 h at -50 ℃,-0.01 MP a until constant weight. The freeze-dried powder without alcohol was designated as AEAMP-NO and also used for the quantification of AEAMP9.
2.3 Cell strains and culture
Human non-small cell lung cancer (NSCLC) A549 cells,human hepatocellular carcinoma HepG2 cells,human breast cancer MDA-MB-231 cells and mouse melanoma B16 cells were obtained from Sichuan University,Chengdu,China. Bone mesenchymal stem cells(BMSCs) were isolated from a rat in our lab according to the standard protocol. The cells were maintained in DMEM medium (Gibco,USA) supplemented with 10% FBS (Hyclone,USA) and antibiotics (100.0 U/mL of penicillin and 100.0 mg/mL of streptomycin,Hyclone,USA). All the cells were cultured at 37 ℃ in a humidified atmosphere of 5.0% CO2.
2.4 MTT assay
Either A549,HepG2,MDA-MB-231,B16 or BMSC cells were seeded in a 96-well plate (5.0×103cells/well) and incubated overnight. After reaching 80% confluency,they were treated with the alcoholic extracts of medicinal plants. When the cell culture reached the endpoint,10.0 μL of MTT solution (5.0 mg/mL) was added to each well and the cells were incubated for another 4 h. Later,110.0 μL of DMSO was added to each well and incubated for 10 min to dissolve the formazan crystals. The absorbance was measured at 490 nm by using a microplate reader (Synergy H1,Biotek,USA). The percentage of cell viability versus concentration of the medicinal plants was plotted.
2.5 Colony formation assay
A549 cells were seeded in a 60 mm culture dish (500.0 cells/dish) and incubated overnight for 24 h. They were then treated with 0,250 and 375 μg/mL of AEAMP9 respectively. The cells were replenished every three days with fresh medium with or without AEAMP9. After incubation for 12 days,the cells were subsequently treated with PBS,4.0% paraformaldehyde and finally stained with Wright-Giemsa stain (Solarbio,Beijing,China). Pictures of the cells were taken using a digital camera,only colonies with >50 cells were counted.
2.6 Wound healing assay
A549 cells were seeded in a 6-well plate,once reached 80.0% confluency,a scratch wound was made using a pipette tip,the wells were then gently washed with PBS to remove the detached cells. Thereafter,the cells were treated with 0,250 and 375 μg/mL of AEAMP9 respectively and incubated. The distance of migration was measured under an inverted microscope after 0,12,24 and 36 h.
2.7 Transwell migration assay
A549 cells pretreated with 0,250 and 375 μg/mL of AEAMP9 respectively for 24 h were collected and re-suspended in FBS-free DMEM medium,then the cells were seeded onto the upper chamber (8.0 μm pore size,Corning,the Netherlands) at a density of 5.0×105cells/mL. 800 μL DMEM medium with 10.0% FBS was added in the lower chamber. After incubation for 24 h,the migrated cells on the bottom membrane were fixed with 4% polyoxymethylene and stained with 0.1% crystal violet. Non-migrating cells were gently removed and then rinsed three times with PBS. The cells across the membrane were captured using a light microscope (BX53,Olympus,Japan). Five different fields were selected randomly for calculation under the microscope at 400× magnification.
2.8 Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted using the TRIzol reagent (Invitrogen,USA). The content and quality of RNA were measured by 1.0% agarose gel electrophoresis. 2.0 μg RNA was converted to cDNA with a Revert PrimeScript RT Enzyme Kit (TaKaRa,Japan). qRT-PCR was performed using SYBR Green Master Mix according to the manufacturer’s instructions in a Light cycler96 instrument (Roche Diagnostics,Switzerland). The PCR cycling conditions were 94 ℃ for 10 min,followed by 40 cycles of 94 ℃ for 30 s,60 ℃ for 40 s and 72 ℃ for 20 s. Melting curve analysis was used to evaluate the primer specificity of the amplification. The relative mRNA expression of the genes was calculated by normalizing to the internal control ofGAPDHgene. Primers used for all the qRT-PCR assays were shown in Table 1.
2.9 Acridine orange/ethidine bromide (AO/EB) staining
A549 cells grown on coverslips were treated with 0,250 and 375 μg/mL of AEAMP9 respectively for 24 h. After washing three times with PBS,the mixed AO/EB solutions (Solarbio,Beijing,China) were added to overlay the coverslips and incubated for 5 min in dark. Cell morphologies were examined under a fluorescence microscope(IX71,Olympus,Japan).
2.10 Caspase activity assay
A549 cells were seeded in a 6-well plate and exposed to 0,250 and 375 μg/mL of AEAMP9 respectively for 12 h. Cell lysates were prepared by incubating 2.0×106cells/mL in 200.0 μL cold lysis buffer (25.0 mmol/L Tris-HCl,pH 7.5,20.0 mmol/L MgCl2,150.0 mmol/L NaCl,1.0% Triton X-100,25.0 μg/mL leupeptin,and 25.0 μg/mL aprotinin) for 40 min on ice. The lysates were centrifuged at 10 000 r/min for 1.0 min,the supernatants were collected and the protein concentrations were determined by Bradford assay (Beyotime,Haimen,P.R. China). The activities of caspase-3,caspase-8 and caspase-9 were determined using the caspase activity kits (KeyGEN,Nanjing,P.R. China). Briefly,the cellular extracts were incubated in a 96-well plate with 5.0 μL caspase-3,caspase-8,caspase-9 substrates for 4 h at 37 ℃ respectively,A405 was read on amicroplate reader (Synergy H1,Biotek,USA).

Table 1 Primers used in qRT-PCR
2.11 Flow cytometry analysis
A549 cells were seeded in a 6-well plate and exposed to 0,250 and 375 μg/mL of AEAMP9 respectively for 12 and 24 h. Annexin V-FITC and Propidium Iodide (PI) solution (KeyGEN BioTECH,Nanjing,P. R. China) were used to stain the cells in the dark for 15 min. The samples were analyzed by flow cytometry (Biosciences AccuriC6,BD Biosciences,USA). The percentages of apoptotic cells of each group were analyzed with Flow J software.
2.12 Western Blot
Cells were harvested and lysed in RIPA buffer (Beyotime,Shanghai,P.R. China). The lysates were centrifuged and the concentration of the total proteins was quantified by an enhanced BCA kit. Subsequently,protein samples (40.0 μg per gel lane) were separated by 12.0% SDS-PAGE and then transferred to a nitrocellulose membrane (Solarbio,Beijing,P. R. China). After blocking with 5.0% skim milk for 2 h at room temperature,the membrane was incubated with the primary antibodies overnight at 4 ℃. Later,the membranes were washed with TBST and then incubated with the Horse-radish peroxidase (HRP)-conjugated anti-rabbit or anti-mouse IgG secondary antibody for 2 h at room temperature. GAPDH was used as the protein loading control. Lastly,the immunoreactive bands were visualized using enhanced chemiluminescence (ECL) reagents (4A Biotech,Beijing,P. R. China) on X-ray films. Quantitative analysis was conducted by using the Image J software.
2.13 Network pharmacology study
TCMSP (http:∥tcmspw.com/tcmsp.php),Batman (http:∥bionet.ncpsb.org.cn/batman-tcm/) and YaTCM (http:∥cadd.pharmacy.nankai.edu.cn/yatcm/home) databases were used to retrieve the potential compounds contained in AEAMP9. The active compounds in AEAMP9 were selected according to the generally used criteria of oral availability (OB)≥30%,drug-likeness (DL)≥0.18 in TCMSP and YaTCM databases and Score≥20 in BATMAN database. The protein targets associated with those active components were collected and standardized in Uniprot protein database (http:∥www.uniprot.org). The disease targets corresponding to lung adenocarcinoma were retrieved from GEO database (https:∥www.ncbi.nlm.nih.gov/gds/?term=,Chip number: GSE118370). The intersection between AEAMP9 and disease targets was established to identify the potential targets of AEAMP9 for lung adenocarcinoma. The compound-target network was built by Cytoscape 3.7.2 (http:∥www.cytoscape.org/) to reflect the complex relationships between active compounds and potential targets. R language was used for enrichment analysis of potential targets. Cytoscape 3.7.2 was also used to visualize Kyoto Encyclopedia of Genes and Genomes (KEGG) regulation network.
2.14 RNA-seq and data analysis
Total RNA samples were extracted from A549 cells treated with or without AEAMP9.RNA (1.0 μg) was used as an input for transcriptome sequencing. Sequencing libraries were generated using NEBNext®UltraTM RNA Library Prep Kit for Illumina®(NEB,USA). The library was sequenced on an Illumina Novaseq platform and 150 bp paired-end reads were generated. Differential expression analysis of two samples was performed using the DESeq2 R package (1.16.1). The resulting P-values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with an adjustedP-value <0.05 were assigned as differentially expressed. The calculation of unigene expression was based on the FPKM method. We named genes of significantly differential expression when the log2 ratio of FPKM higher than 1. Gene Ontology (GO),KEGG pathways and Disease Ontology (DO) enrichment analysis of differentially expressed genes were implemented by the cluster Profiler R package,in which gene length bias was corrected.
2.15 Acute toxicity study
After adaptation to the environment for one week,the five-week-old specific pathogen free (SPF) Kunming mice with 24 males and 24 females were randomly divided into 6 groups (n=8): the blank control,20.0% alcohol,50.0% alcohol,20.0% AEAMP9,50.0% AEAMP9 and AEAMP9-NO group (AEAMP9 without alcohol,20.0 mg/mL). All the solutions were diluted in 0.9% concentration of NaCl. The mice were fasted for 12 h before the gavage (20.0 mL/kg body weight) and allowed to drink water freely. After 9 days of continuous observation,the mice were sacrificed. Blood routine,aspartate aminotransferase (AST) and alanine aminotransferase (ALT) were measured. Important organs including heart,liver,spleen,and kidney were removed and weighed. Organ index was calculated as the following equation: the organ index=organ weight/mice body weight ×100%.
2.16 Tumor xenograft model study
Five-week-old male SPF balb/c nude mice were adapted to the environment for one week and then inoculated with A549 cells (1.0×107/mouse) in the right axilla of the mouse in 200.0 μL DMEM medium. 18 days later,all the twenty-eight nude mice bearing tumors were randomly divided into four groups (n=7): 75.0% alcohol control,AEAMP9 treatment,invitroandinvivocombination treatment of AEAMP9 (co-AEAMP9) and cis-platinum (CDDP,Beijing Solarbio Science & Technology Co.,Ltd,P. R. China) positive control. For 75.0% alcohol or AEAMP9 groups,75.0% alcohol or 2.0% AEAMP9 were mixed with tea oil in a 2∶1 ratio respectively,and then approximately 200 μL of the mixture was gently rubbed on the epiderm of the tumor for 3 min and three times a day. For theCDDPgroup,cis-platinum (25.0 mg/kg) was injected beside the tumor every three days. For the co-AEAMP9 group,in addition to the topical treatment of AEAMP9,the mice were free to water which contained 10×diluted of 2.0% AEAMP9.After 14 days’ breeding,all the mice were sacrificed,tumors and bloods were collected and preserved. The tumor growth inhibition rate (IR)% on day 14 was used to quantify the antitumor effect. (IR)% = (mean tumor weight in the control group-mean tumor weight in the experimental group)/mean tumor weight in the control group ×100%.
Both the mouse acute toxicity and tumor xenograft studies were in accordance with the regulations for animal use and care of Southwest Jiaotong University.
2.17 Immunohistochemistry
The isolated tumor tissues werexed with 4% paraformaldehyde and embedded in paraffin. The slides of tumor tissue sections were firstly deparafinized and rehydrated in graded alcohols followed by endogenous peroxidase quenching,antigen retrieval and nonspecific binding site blocking. Then,the sections were incubated with the primary antibodies (Ki67,cleaved-Caspase3 and FoxM1) at 4 ℃ overnight. After washed with PBS for three times,the slides were incubated with Polymer Helper and poly anti-rabbit IgG conjugated with peroxidase. Finally,they were developed by diaminobenzidine (DAB),stained with hematoxylin and fixed with neutral balata. The sections were observed under a light microscope (BA400Digital,MOTIC,Xiamen,P.R. China).
2.18 Elisa assay
The serums from the bloods of mice were prepared by centrifuging the coagulated blood at 3 000 r/min (HERMLE,Germany) for 15 min and stored at -20 ℃ until use. Mouse tumor necrosis factor-α (TNF-α) and interleukin-6 (IL-6) were determined using the Elisa kit (Jiancheng Bioengineering Institute,Nanjing,P.R. China) and measured on microplate reader Synergy H1/H1MF (BioTek,Winooski,Vermont,USA).
2.19 Statistical analysis
SPSS19.0 and Microsoft Excel software were used for statistical analyses. Data were represented as the mean ± standard deviation. Student’st-test and one-way Anova were used to determine the statistical significance.Pvalue<0.05,<0.01 and <0.001 was considered statistically significant,extremely significant and the most significant. All experiments were conducted at least in three triplicates otherwise indicated.
3 Results and discussions
3.1 Medicinal plants screening and formula optimization
Alcoholic extracts of 124 kinds of medicinal plants at the concentration of 2.0% were at first screened against the A549 cells by MTT assay,showing that 22 kinds of medicinal plants had strong inhibitory effects (≥80%). It is unexpected that some medicinal plants reported to have active components in anticancer activities such asCatharanthusroseus(L.)G. Don (Chang Chun Hua),CephalotaxusfortuneHooker(Shan Jian Shan) displayed lower inhibition rates in this screening. Then,by reducing the concentration to 1.5% and 1.0%,we still found 13 medicinal plants including S1,S4,S6,S7,S31,S33,S39,S41,S44,S63,S74,S89,and S110 with a high suppression rate. Ten medicinal plants with strong inhibitions and high safety were then selected for designing the formula of AEAMP1-10. AEAMP9 exhibited the strongest inhibition rate among all the formulas to A549 cells [Figure 1(a)] though the rest formulas were also excellent. Therefore,AEAMP9 was selected for further study. AEAMP9 was a compound formula composed of six medicinal plants includingHedyotisdiffusaWilld.,PhellodendronamurenseRupr.,GlycyrrhizauralensisFisch.,Platycladusorientalis(L.) Franco.,ChrysanthemumindicumL. andNotopterygiumincisumTing ex H. T. Chang. Among them,HedyotisdiffusaWilld. is commonly used in the therapy of various types of cancer[13-14].PhellodendronamurenseRupris has been found widely in Asia and rich in berberine and palmatine alkaloids[15-16].GlycyrrhizauralensisFisch is always contained in TCM prescription to counteract the side effects of other components. The leaves ofPlatycladusorientalishave broad spectrum of pharmacological activities including anti-inflammatory,antimicrobial and anticancer activities[17].ChrysanthemumindicumL. is mainly used in Korea and China,it can exert anti-inflammatory effect by targeting AKT1 and AKT2 in the NF-κB signaling pathway in macrophage-mediated inflammatory responses[18]. Lastly,NotopterygiumincisumTing ex H.T. Chang has various therapeutic effects such as analgesic,anti-inflammatory,anticancer and antioxidants effects[19]. In China in 2020,the national medical products administration (NMPA) issued the new “Provisions for Drug Registration”,in which the compound traditional Chinese medicine was listed as 1.1 in the category of innovative traditional Chinese medicine,while previously ranked as 1.6 in the old edition,which to some extents indicated the more attentions of China’s government for compound TCM.

(a) Inhibitions of AEAMP1-10 (1.0%) to A549 cells for 24 h. (** P<0.01,*** P<0.001 compared with that of the AEAMP1). (b) Inhibitions of AEAMP9 at various concentrations and times for A549 cells. (c)-(d) Morphological changes and inhibition rates of AEAMP9 at different concentrations to 6 kinds of cell lines for 12 h (100×,magnification). (* P<0.05,** P<0.01,*** P<0.001 compared with that of the A549). (e) Inhibition of AEAMP9 treatment on the colony formation of A549 cells.Figure 1 The inhibition of the viability and clone formation of cancer cells by AEAMP9 图1 AEAMP9抑制肿瘤细胞的活力和克隆形成
By evaporation and vacuum freeze-drying,the crude dry weight of AEAMP9 was determined with an average concentration of 51.3 mg/L. The optimized concentration and time of AEAMP9 for the treatment of A549 cells were at 375 μg/mL and 12 h respectively [Figure 1(b)]. The effects of AEAMP9 were also tested in other cancer cells. The inhibition rate of AEAMP9 for A549,B16 and HepG2 cells reached to 98.1%,92.4% and 94.5%,respectively [Figure 1(c) and 1(d)]. It revealed the wide spectrum of anticancer effects for AEMAP9. More excitingly,AEAMP9 was found to be toxic to BMSC cells with the inhibition rate of 80.7%. Since it is widely accepted that the refractory cancers were related to tumor/cancer stem cells (TSCs/CSCs),the toxicity to BMSC cells indicates the possible inhibition of AEAMP9 for TSCs or CSCs[20-21]. The cytotoxicity of AEAMP9 to cancer cells were further supported by colony formation assay,concentrations of AEAMP9 at 250 and 375 μg/mL could completely prevent the colony formation of A549 cells compared to that of the negative control [Figure 1(e)].
3.2 Migration inhibition for A549 cells
The migration of A549 cells treated respectively with the concentrations of AEAMP9 at 250 and 375 μg/mL were at first determined by scratch wound healing and transwell migration assay. The migration inhibition of A549 cells was dependent on the concentrations of AEAMP9 and treating time [Figure 2(a)-(d)]. Moreover,as shown in Figure 2(e),the relative mRNA expressions ofMMP2,MMP9andMMP14in A549 cells treated with AEAMP9 (375 μg/mL) were significantly down-regulated with a decrease of 51.3%,52.7% and 77.1% compared with the control,respectively (P<0.01),suggesting the involvement of matrix metalloproteinases (MMPs) in migration inhibition of A549 cells. MMPs are a class of zinc-dependent endopeptidase superfamily,the members of MMP2,MMP9 and MMP14 are closely related to the invasion and metastasis of cancers[22].

(a)-(b) Wound healing and relative migration rate of A549 cells treated with AEAMP9 (100×,magnification). (c)-(d) Images of the crossing cells and the calculation of the migration of A549 cells (100×,magnification). (e) qRT-PCR assay of the mRNA expression of MMP2,MMP9 and MMP14 in A549 cells after treatment with AEAMP9 for 12 h. * P<0.05,** P<0.01,*** P<0.001 compared with that of the control.Figure 2 The migration of A549 cells inhibited by AEAMP9图2 AEAMP9抑制A549细胞的迁移
3.3 Effect of AEAMP9 on apoptosis induction for A549 cells
Cell apoptosis can be determined in many ways including AO/EB double fluorescence staining,enzymatic caspase activity assay and apoptosis associated gene detection[23]. We could find large numbers of apoptotic and dead cells with colors of yellow-green or orange after treatment with AEAMP9 for 12 h [Figure 3(a)]. The proportion of apoptotic cells increased from 8.8% to 23.2% and 7.8% to 31.8% respectively after treated with AEAMP9 for 12 h and 24 h assayed by FCM,indicating the apoptosis inhibition of A549 cells was also in a dose and time dependent manner [Figure 3(c)]. The enzymatic activities of caspase 3,caspase 8 and caspase 9 were notably up-regulated in A549 cells when treated with AEAMP9 [Figure 3(b)],which were consistent with our previous apoptosis results. Unexpectedly,the increased activity folds of caspase 3,caspase 8 and caspase 9 at concentration of 375 μg/mL of AEMAP9 were all lower than those of the lower dose of 250 μg/mL. It may be caused by the fact that cell death other than apoptosis may also play an important role in AEAMP9 induced cell death as shown in Figure 3(a).
Further apoptosis supports come from the expressions of apoptosis-associated genes both at the transcription and translation levels. There was a significant increase in the expression of pro-apoptotic genes ofbax,p53,caspase8,caspase9(212.3%,85.6%,117.5% and 44.6% respectively,P<0.05) while the expressions ofbcl-2andcaspase3were dropped by 50.5% and 39.1% (P<0.05) as determined by qRT-PCR [Figure 3(d)]. Moreover,Western Blot showed that the expressions of p53 and Bax were significantly increased while the anti-apoptotic Bcl-2 was decreased,compared to that of the control [Figure 3(e) and 3(f)].
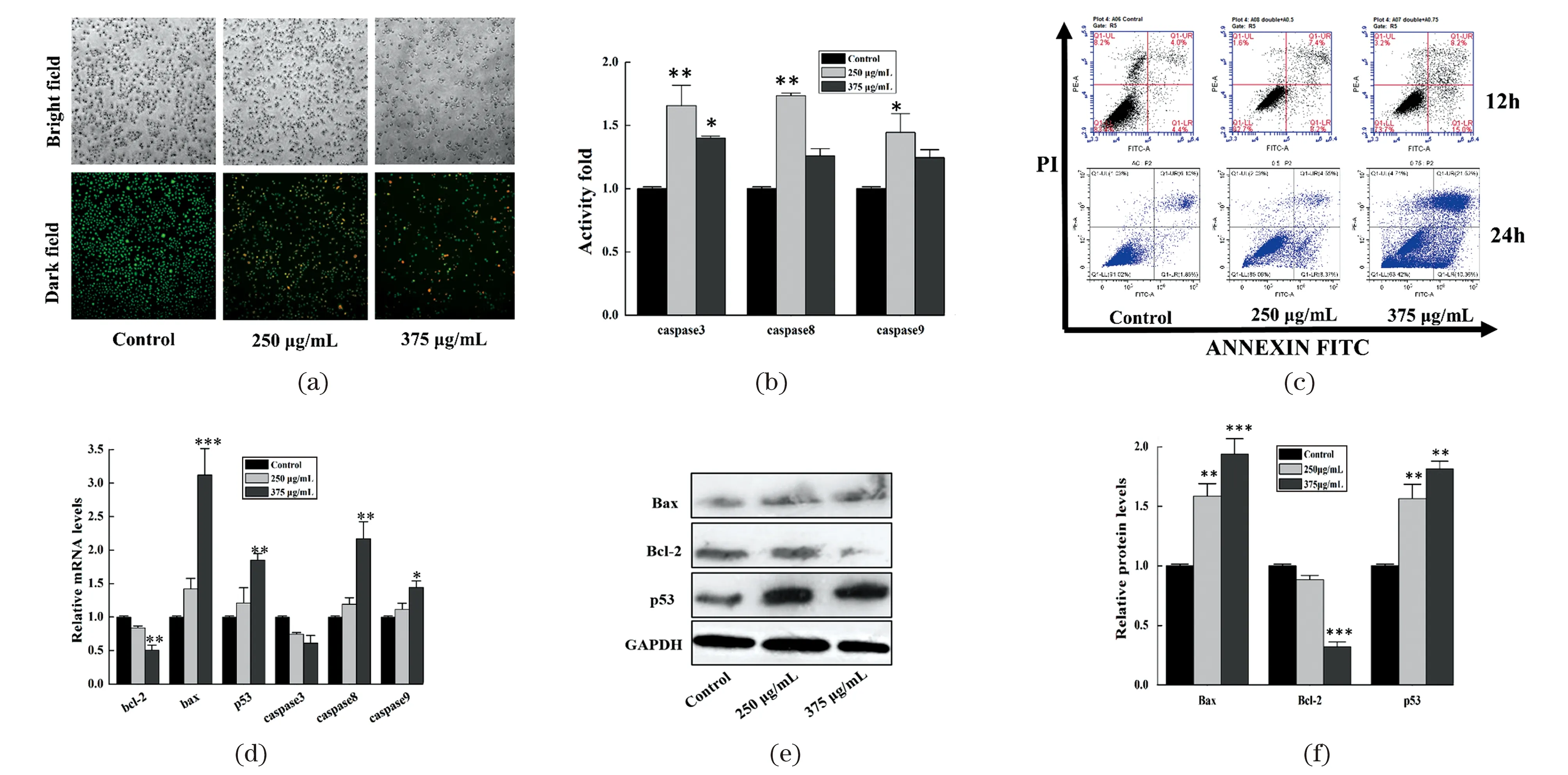
(a) A549 cells treated with AEAMP9 for 12 h were observed by AO/EB double staining (100×,magnification). (b) The enzymatic activities of caspase3,caspase8,and caspase9 were detected in A549 cells treated with AEAMP9 for 12 h. (c) A549 cells treated with AEAMP9 were detected by flow cytometry assay. (d) qRT-PCR assay of the relative mRNA expression of apoptosis-associated genes in A549 cells after treatment with AEAMP9 for 12 h,GAPDH was used as the internal control. (e) The protein expressions of Bax,Bcl-2 and p53 were detected after treatment with AEAMP9 for 24 h by Western Blot,GAPDH was used as loading control. (f) The mean density of Western Blot was determined by Image J software. * P<0.05,** P<0.01,*** P<0.001 compared with the control.Figure 3 The apoptosis of A549 cells induced by AEAMP9图3 AEAMP9诱导A549细胞凋亡
3.4 Drug resistance-associated genes inhibition in A549 cells
To explore the participations of AEAMP9 in regulation of drug resistance in A549 cells,we performed qRT-PCR and Western Blot to quantify the expression levels of drug resistance-associated genes including ABCG2,FOXM1 and MDR1. ABCG2 and MDR1 are both important drug resistant genes and implicated in cancers[24],FoxM1 is a key transcription factor involved in broad areas of cancer including drug resistance and regarded as the Achilles’s heel of cancer[25]. From Figure 4(a)-(b),we found that the mRNA expressions ofABCG2,FOXM1andMDR1were significantly down-regulated (P<0.05) after treatment with AEAMP9,the expressions ofFOXM1,ABCG2andMDR1were decreased by -86.5%,-49.6% and -67.3% when treated with the concentration of AEAMP9 at 375 μg/mL,respectively. The results were further convinced by the Western Blot assays,in which the protein expressions of FoxM1 and ABCG2 were significantly down-regulated by -30.1% and -54.2%,respectively [Figure 4(c)-(d)]. Our work was also consistent with the previous reports that the up-regulation of p53 could down-regulate the expression of FoxM1[26-27]. It should be emphasized that the inhibitions of AEAMP9 for the colony formation,migration,apoptosis and drug resistance of A549 cells were also verified in our study of a mouse cell line (data not shown).

(a) The purity and quantity of total RNA extracted from A549 cells assessed by 1.0% agarose gel electrophoresis. (b) The relative mRNA expression of ABCG2,FOXM1 and MDR1 genes in A549 cells detected after treatment with AEAMP9 for 12 h by qRT-PCR,GAPDH used as the internal control. (c)-(d) The protein expression of FoxM1 and ABCG2 in A549 cells detected after treatment with AEAMP9 for 24 h by Western Blot,GAPDH used as loading control. * P<0.05,** P<0.01,*** P<0.001 compared with that of the control.Figure 4 AEAMP9 down-regulating the expression of drugresistance-associated genes in A549 cells图4 AEAMP9下调A549细胞耐药相关基因的表达
3.5 Network pharmacology analysis for potentiality of AEAMP9
Network pharmacology was an important method to study the compound TCM formulas and predict their potential targets for specific diseases[28-30]. Here,a total of 168 compounds and 240 potential targets from the six medicinal plants of AEAMP9 were retrieved from TCMSP,YaTCM and BATMAN databases after removing the redundancy. Six chemicals including thalifendine (MOL006422),quercetin (MOL000098),beta-sitosterol (MOL000358),phellopterin (MOL002644),sitosterol (MOL000359) and stigmasterol (MOL000449) were contained in more than one medicinal plants. A total of 771 disease-related targets were retrieved from GEO database. 26 key target genes of AEAMP9 in lung adenocarcinoma were obtained. The AEAMP9 compound-target network covered 39 nods (13 compounds in AEAMP9 and 26 potential compound targets) and 44 edges. Quercetin (MOL000098),kaempferol (MOL000422),and luteolin (MOL000006) were found to function on more than 7 targets,respectively. Hence,they were supposed to be the crucial compounds in AEAMP9 for their wide involvement in the network.
The topological features of the original protein protein interaction (PPI) network were also analyzed. The subnetworks were obtained according to the criteria of degree centrality (DC≥60) and betweenness centrality (BC≥600) respectively. The subnetworks include 163 nodes and 28 593 edges. Proteins including FOS,JUN,CAV1,CDK4,CDC20,CDK1,EGFR,TP53,TOP2A,PCNA were ranked first,indicating being the major targets for AEAMP9. GO function enrichment analysis totally obtained 346 items,including 232 Biological processes (BP),25 Cell components (CC) and 89 Molecular functions (MF). By KEGG enrichment analysis,a total of 42 signal pathways were obtained,the top 10 pathways include IL-17,TNF,AMPK and EGFR tyrosine kinase inhibitor resistance etc. Finally,Cytoscape was used to construct the KEGG regulatory network,the latter was consisted of 45 nodes (20 signaling pathways,25 target genes) and 90 edges. It can be found that tumor and drug resistance associated signaling pathways were predominately engaged and JUN,IL-6,MMP1,FOS,VEGFA,EGR1 were among the vital targets in the regulatory network.
3.6 RNA-seq analysis for the effect of AEAMP9 on gene expression in A549
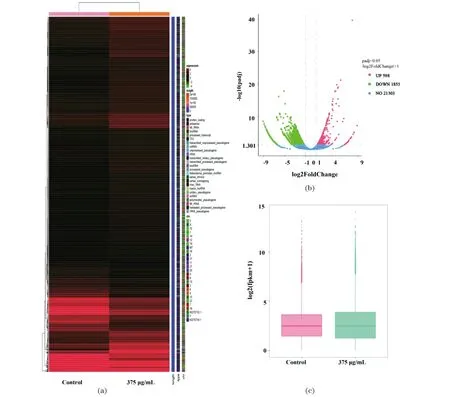
RNA-seq is commonly the most popular next-generation sequencing technique for the analysis of genome-wide RNA expression at the transcriptome level[31]. In this work,a total of 10 883 genes were found to be differentially expressed in A549 cells treated with AEAMP9 at the concentration of 375 μg/mL [Figure 5(a)]. From the volcano map,we can find that 598 genes in A549 cells were significantly up-regulated while 1 853 genes were significantly down-regulated with AEAMP9 treatment [Figure 5(b)]. FPKM values of most of the genes were down-regulated by 10-50 folds and up-regulated by 3-20 folds respectively in treatment group compared with that of the control [Figure 5(c)]. GO function enrichment analysis showed that most of the genes were significantly enriched in biological processes such as the down-regulation of embryonic morphogenesis,reproductive system development,extracellular matrix and up-regulation of transcriptional activator activity,response to lipopolysaccharide and bacteria [Figure 5(d)]. In the KEGG enrichment analysis,multiple signaling pathways such as TNF,IL-17,NF-kB,MAPK and EGFR related to inflammation and cancer were significantly enriched. Among them,TNF and IL-17 were both found in our KEGG analyses from network pharmacology and RNA-seq [Figure 5(e)]. Furthermore,from DO enrichment analysis,the genes relating to respiratory system cancer and lung cancer were significantly enriched [Figure 5(f)],the later further supported our study as A549 is a NSCLC cell line.
To verify the authenticity of our RNA-seq analysis,we used qRT-PCR to detect the top 30 up-regulated and down-regulated genes associated with cancer and found in RNA-seq. As shown in Figure 6,the expression patterns of the majority of these genes were consistent with the results from those of RNA-seq. The relative mRNA expressions ofEGR1,SUMO4,SOCS1,KLF6,TNFAIP3,SDC4,BIRC3,NGR1 andIL-6were significantly up-regulated [Figure 6(c)],while the relative mRNA expressions ofALK,UNTM2A,AXIN2,POU5F1,RNF43,SOX2,FoxA1,BCL11B,MMP12andLRP6 were significantly down-regulated and in a time-dependent manner [Figure 6(d)]. Important genes such asIL-6,EGR1andVEGFwere overlapped in both our network pharmacology and RNA-seq study. In addition,among all the genes verified,EGR1andALKwere the most differentially expressed genes,up-regulated by 785.5% and down-regulated by 646.8%,respectively.EGR1,a member of the immediate early gene of the early growth response (EGRs) family,is an important tumor suppressor gene.EGR1 can reverse the malignant phenotype of HT1080 fibrosarcomacells[32],it was also revealed thatEGR1 can not only inhibit the proliferation of osteosarcoma cells,but also regulate cell cycle and apoptosis-related genes[33].ALKis a member of the receptor tyrosine kinase family,EmL4-ALK is highly tumorigenic and the expression of the EmL4-ALK fusion protein occurs in approximately 3%-5% of the non-small cell lung cancer[34].
Continued Figure 5 (续图5)

(a) Heat map and cluster patterns of 2 451 significant differentially expressed genes between AEAMP9 treated sample and the control. Genes were clustered together with similar expression profiles using hierarchical clustering. (b) Volcano plot for differentially expressed genes. (c) Box plot showed differentially expressed gene profiles between the control and treatment groups. (d) GO terms of the significant up-regulated and down-regulated differentially expressed genes. (e)-(f) Top 20 enriched KEGG/DO pathways of the differentially expressed genes. The bigger size of the circle,the more expressed genes annotated to the terms.Figure 5 Transcriptome sequencing analysis in A549 cells treated with AEAMP9图5 AEAMP9处理A549细胞的RNA转录组测序结果分析

(a)-(b) Top 15 up-regulated and down-regulated cancer associated genes analyzed by RNA sequencing,respectively. (c)-(d) qRT-PCR verification of the up-regulated and down-regulated genes respectively. GAPDH was used as a control. * P<0.05,** P<0.01,*** P<0.001 compared with the control.Figure 6 qRT-PCR verification of the significantly expressed genesfound in transcriptome sequencing in A549 cells treated with AEAMP9图6 qRT-PCR验证AEAMP9处理A549细胞后转录组测序差异基因表达变化
3.7 Single dose acute toxicity of AEAMP9
During the experiment,no mouse died in all the groups,AEAMP9 treated groups showed no significant differences to those of the alcoholic control group no matter the organic index,routine blood test,and even the hepatic activity indexes of AST and ALT [Figure 7(b)]. This is reasonable,as the formula of AEAMP9 was well designed and contained no reported toxic medicinal plants,which are all safely used in traditional Chinese medicine and included in Chinese Pharmacopoeia (Chinese pharmacopoeia commission,2020). In particular,while there is a significant decrease in the body weight of negative control groups of 50% and 20% alcohol treatment,all the AEAMP9 treated groups showed no significance as compared with the blank control [Figure 7(a)]. We speculate that some constituents in AEAMP9 may promote the weight gain in mice and they are worthy further study.
3.8 Effect of AEAMP9 on nude mice xenograft tumor growth suppression
The tumor suppressive effect of AEAMP9 was evaluatedinvivoby using the nude mouse A549 xenograft model. At the endpoint of the experiment,the average tumor inhibitory rate (IR) of AEAMP9,co-AEAMP9 andCDDPgroup reached to 47.3%,68.2% and 85.7% compared with those of the alcohol group (P<0.05),respectively [Figure 8(a)].

(a) Body weight changes of SPF Kunming mice of the divided six groups after treatment for 9 days. (b) The contents of ALT and AST in the serum of each group. Normal saline (NS,0.9% NaCl) was used as the blank control. *** P<0.001 compared with the control.Figure 7 Effects of AEAMP9 treatment on the changes of body weight and liver transaminases in mouse acute toxicity test图7 AEAMP9处理对小鼠体重及肝脏转氨酶的急性毒性影响
We need to emphasize that our AEAMP9 treatment was just topical application. Topical application of drugs has many advantages at least including avoiding adverse effects,increasing drug concentration at the focus and also possible reducing the drug resistance. The efficiency of AEAMP9 treatment would be more improved if the mice were treated more times and the concentration of AEAMP9 was higher. The results were also supported by immunohistochemistry,in which the expressions of Ki67 and FoxM1 were significantly down-regulated while the cleaved caspase3 increased,indicating the propagation inhibition,apoptosis progression and the participation of FoxM1 in AEAMP9-induced tumor suppression [Figure 8(d)-(g)]. Consistently,TNF-α and IL-6 in serums were significantly decreased in the treatment groups as compared to negative control [P<0.05,Figure 8(b)-(c)]. Ki67 is a key proliferating factor,and TNF-α and IL-6 are pro-inflammatory cytokines which are all the important diagnosis markers and molecular targets for cancer[35-36]. The expressions of cleaved-caspase3 and FoxM1invivowere also consistent with those of ourinvitrostudy.
In conclusion,we established an efficacy-driven approach to find drugs against important targets based on large numbers of the alcoholic extracts of medicinal plants,the target can not only be the cancer cells but also extend to important proteins,pathogenic microbes,etc. The determined efficiency of the medicinal plants for cancer cells would aid the drug discovery as well the clinic application in the prescription of the TCM therapy. Also,we had developed a new formula of AEAMP9 which had strong inhibitory effects for A549 cancer cells as well as broad spectrum of cancer cells including B16,HepG2 and BMSC cells. Important bottlenecks for cancer therapy including tumor migration and drug resistance were particularly addressed in our study of AEAMP9. Lastly,the possible mechanisms of AEAMP9 for its inhibitory effects on A549 cancer cells were investigated. AEAMP9 can induce the apoptosis of A549 cancer cells by activation of caspase and p53 signal pathway,it can inhibit the migration of A549 cancer cells by down-regulating MMPs expression. In addition,AEAMP9 may overcome the drug resistance by down-regulating the expression ofFOXM1,MDRandABCG2genes. Nevertheless,there are limitations to this study. Further works are still needed,such as the deep deciphering of the material bases for AEAMP9 by modern chemical analysis methods,the in-depth antitumor mechanism study,and the possible development of the products for anticancer therapy.

