Heterologous expression of the Haynaldia villosa pattern-recognition receptor CERK1-V in wheat increases resistance to three fungal diseases
Anqi Fn,Luyng Wei,Xu Zhng,Ji Liu,Li Sun,Jin Xio,Yji Wng,Hiyn Wng,Jin Hu,b,Rvi P.Singh,Zongkun Wng,*,Xiue Wng,*
a State Key Lab of Crop Genetics and Germplasm Enhancement,Cytogenetics Institute,Nanjing Agricultural University/JCIC-MCP,Nanjing 210095,Jiangsu,China
b Department of Plant Biology,Cornell University,Ithaca,NY 14853,USA
c International Maize and Wheat Improvement Center(CIMMYT),El Batan,Texcoco 56237,Estado de Mexico,Mexico
Keywords:Wheat Fungal disease LysM PRR Haynaldia villosa CERK1-V
ABSTRACT Wheat production is under continuous threat by various fungal pathogens.Identification of multipledisease resistance genes may lead to effective disease control via the development of cultivars with broad-spectrum resistance.Plant Lysin-motif(LysM)-type pattern-recognition receptors,which elicit innate immunity by recognizing fungal pathogen associated molecular patterns such as chitin,are potential candidates for such resistance.In this study,we cloned a LysM receptor-like kinase gene,CERK1-V,from the diploid wheat relative Haynaldia villosa.CERK1-V expression was induced by chitin and Blumeria graminis f.sp.tritici,the causal agent of wheat powdery mildew.Heterologous overexpression of CERK1-V in wheat inhibited the development of three fungal pathogens,thereby increased resistance to powdery mildew,yellow rust,and Fusarium head blight.CERK1-V physically interacted with the wheat LysM protein TaCEBiPs.CERK1-V/TaCEBiPs interaction promoted chitin recognition and activated chitin signal transduction in wheat.Transgenic plants with excessively high CERK1-V expression showed high resistance but abnormal plant growth,whereas plants with moderate expression level showed adequate resistance level with no marked impairment of plant growth.In transgenic lines,RNA-seq showed that gene expression involved in plant innate immunity was activated.Expression of genes involved in photosynthesis,ER stress and multiple phytohormone pathways was also activated.Optimized expression of CERK1-V in wheat can confer disease resistance without compromising growth or defense fitness.
1.Introduction
During their growth period,plants face the threats of harmful organisms or microorganisms,including insects,bacteria,and fungi.Plants employ a variety of immune receptors to sense pathogen infection or insect attack.A major class of plant immune receptors is that of membrane-anchored pattern-recognition receptors(PRRs).Once microbial conserved signatures,collectively referred as pathogen(or microbe)-associated molecular patterns(PAMPs or MAMPs)are detected,PRRs trigger PAMP-triggered immunity(PTI)[1].Flagellin and Ef-Tu are two well-characterized bacterial PAMPs.In Arabidopsis,flagellin is recognized by the leucine-rich repeat receptor-like kinase(LRR-RLK)AtFLS2[2],and Ef-Tu is recognized by AtEFR[3].Chitin,a common component of fungal cell walls,is the most well-studied fungal PAMP.During plant-fungus interaction,plant chitinases degrade the linear N-acetyl-Dglucosamine polymer of chitin into chitin oligosaccharides(with degree of polymerization[DP]6 to 8),which can be detected by plant PRRs and induce PTI[4].
In plants,fungal chitin is perceived by two types of PRRs:Lysinmotif(LysM)receptor-like proteins(LYPs)and kinases(LYKs).The Arabidopsis LYK cerk1 mutant completely lost the ability in responding to the chitin elicitor,thus the AtCERK1 was considered as the major signaling receptor[5].AtCERK1 did possess chitin binding ability via its second LysM domain[6],but isothermal titration calorimetry indicated that its chitin binding affinity was relatively low[7].Mutation in its chitin binding site(AtCERK1A138H)did not block chitin-induced phosphorylation of AtCERK1A138H[6].It is accordingly proposed that besides AtCERK1 there is another co-receptor playing a role in chitin binding.This proposal has been supported by the identification of the LYK AtLYK5,which showed about 200-fold higher chitin affinity than AtCERK1[7].AtLYK5 and its closest homolog AtLYK4 were required,and they formed a protein complex with AtCERK1 during chitin recognition in Arabidopsis.The rice LYK OsCERK1 was indispensable for rice chitin recognition:knockdown of OsCERK1 resulted in suppression of chitin-induced defense response[8].The rice LYP OsCEBiP is the first identified chitin receptor[9].OsCEBiP targets the plasma membrane through a glycosylphosphatidylinositol(GPI)anchor.Gene silencing or knockout of OsCEBiP impaired rice defense responses induced by chitin and the fungal pathogen Magnaporthe oryzae[10].In response to chitin treatment,OsCERK1 and OsCEBiP formed hetero-oligomers to coregulate chitin-elicited signal transduction[8].There are three homologs of CEBiP(AtLYM1,AtLYM2,and AtLYM3)in Arabidopsis,but their triple mutant did not show compromised chitin recognition[11].The rice LYPs OsLYP4 and OsLYP6,have been shown[12]to play dual roles,in both chitin and peptidoglycan(PGN)recognition.Silencing of OsLYP4 and OsLYP6 led to increased rice susceptibility to the bacterial pathogen Xanthomonas oryzae and the fungal pathogen M.oryzae.However,the LYP4 and LYP6 homologs in Arabidopsis,AtLYM1 and AtLYM3,only induced immunity mediated by bacteria or PGN,a PAMP of bacteria[13].These studies show that chitin-responsive PRRs employ different chitin receptor systems in Arabidopsis and rice.
Once chitin-LysM recognition has been established,chitin signal can be transmitted to initiate an endogenous immune response via various downstream pathways,including activation of Ca2+influx and mitogen-activated protein kinase(MAPK)cascades.MAPK cascades are the critical modules in chitin signal transduction.In Arabidopsis,a MAPK phospho-signaling cascade pathway,MAPKKK5-MAPKK4/MAPKK5-MAPK3/MAPK6,was induced by chitin.In the mapkkk5 mutant,the activation of MAPK3,MAPK4,and MAPK6 was strongly reduced[14].In rice,chitin signal transduction is accomplished by another MAPK cascade,consisting of MAPKKK11/18-MKK4-MPK3/6.MAPKKK18 could be phosphorylated by OsRLCK185,which is involved in the downstream signal transduction of OsCERK1[15].Silencing of MAPKKK18 and MAPKKK11 suppressed chitin-induced MAPK activation and reactive oxygen species(ROS)production.Chitin induces extracellular Ca2+influx,which forms potential calcium signatures in both cytoplasm and nucleus within a few minutes[16].Plants employ calcium-dependent protein kinases(CPKs)to decode specific calcium signatures.In Arabidopsis,four CPKs including CPK4,CPK5,CPK6,and CPK11 involved in PAMP induce early transcriptional reprogramming.Mutations of different cpks differed in their ability to impair flg22-induced gene expression[17].AtCPK1 is reported to play an essential role in early plant defense against fungal pathogen[18].AtCPK1 expression was rapidly induced by fungal elicitors.Overexpression of AtCPK1 led to salicylic acid(SA)accumulation and broad-spectrum resistance against two fungal pathogens,Fusarium oxysporum and Botrytis cinerea.In a recent study,CPKs regulated chitin-induced immunity by directly targeting PRRs[19].The Arabidopsis AtCPK5 phosphorylated the LYK AtCERK5,and the atcpk5 mutant showed impaired chitin-induced immune response.
Wheat production is threatened by many fungal diseases,such as powdery mildew(Pm)caused by Blumeria graminis f.sp.tritici(Bgt),yellow or stripe rust(Yr)caused by Puccinia striiformis f.sp.tritici(Pst)and Fusarium head blight(FHB)caused by Fusarium graminearum(Fg)[20].Development and deployment of cultivars carrying resistances against multiple diseases is the most environment-friendly and efficient approach.Bgt and Pst are biotrophic fungi,while Fg is a hemibiotrophic fungus.Nucleotidebinding domain leucine-rich repeat(NLR)-type Resistance(R)genes confer race-specific resistance against certain isolates carrying specific Avr proteins.Most resistance genes,especially typical NLR genes,confer only race-specific resistance or resistance to several races of single pathogen species and are readily overcome due to the emergence of new virulent races.Identification of genes that confer high level and broad-spectrum disease resistance has great potential in breeding varieties with durable resistance.Among cloned powdery mildew or rust resistance genes,most are typical NLRs[21].Four genes that individually provide partial resistance to multiple fungal pathogens have been cloned,they all are non-NLR type[22].Wheat Lr34/Yr18/Sr57/Pm38 encodes an ABC transporter[23],wheat Lr67/Yr46/Sr55/Pm46 encodes a hexose transporter[24],barley mlo encodes the only family of seventransmembrane(TM)proteins[25]and rice GH3-2 is regulator of resistance and growth[26].Lr34/Yr18/Sr57/Pm38[23]and Lr67/Yr46/Sr55/Pm46[24]confer partial resistance to three rust diseases and powdery mildew.Lr34 is suspected to have a role in exporting metabolites that affect fungal growth[23]and Lr67 acts as an inhibitor to reduce hexose uptake through heterodimerization with these functional transporters[24].The substrate of the ABC transporter and the mechanisms of multiple-disease resistance genes have remained unclear.Neither of the two cloned Fhb-resistance genes,Fhb1 and Fhb7,are NLR-type R genes[27-29].
Although wheat employs distinct downstream pathways in defending against biotrophic or hemibiotrophic pathogens,early detection and signal transduction of fungal chitin may be a conserved defense signal pathway.Such conservation might assist in identifying genes contributing to resistances against multiple fungal diseases.Haynaldia villosa(L.)Schur[syn Dasypyrum villosum(L.)P.Candargy],a diploid wheat wild relative,carries high resistance to multiple wheat fungal diseases,including powdery mildew,stem rust,yellow rust,and cereal eyespot[30-33].Identification and application of genes from such alien sources will broaden wheat genetic basis of disease resistance.
The objective of the present study was to clone H.villosa LysMtype PRRs and investigate their role in resistance against fungal diseases of wheat.By genome-wide characterization of the LysM type PRRs in H.villosa,we aim to identify the functional PRRs and elucidate their functions in fungal disease resistance.Our research will not only provide valuable genes for improving multiple disease resistance,but also help to reveal the mechanism underlying broad spectrum resistance.
2.Materials and methods
2.1.Materials
Haynaldia villosa(Accession No.91C43,genome VV)was introduced from Cambridge Botanical Garden,UK.A Triticum durum-H.villosa amphiploid and the T.aestivum-H.villosa addition line DA7V,and the wheat-H.villosa T6VS/6AL translocation lines 93R137 and Nannong 9918,were developed by the Cytogenetics Institute of Nanjing Agricultural University(CINAU).The wheat cultivars Sumai 3,Mingxian 169,Chinese Spring,Fielder,and Taishan 23 are maintained in CINAU,and Fielder was used as the recipient for genetic transformation.
2.2.Evaluation of disease resistance
Bgt isolate E26 was kindly provided by Yilin Zhou,Chinese Academy of Agriculture Sciences.The Bgt isolate mixture was collected from Nanjing,Jiangsu province,China.The pathogen was maintained on seedlings of the Pm susceptible variety Sumai 3 in a spore-proof greenhouse under a 14-h light/10-h dark(24/18 °C,70% humidity)regime.Pm resistance at the seedling stage was evaluated using detached leaves inoculated with isolate E26.Infection types(ITs)were graded as 0-4 following Liu et al.[34].Pm resistance at adult stage was evaluated by inoculation with Bgt isolate mixture.ITs were graded as 0-9 following Jakobson et al.[35].Infected leaf segments were bleached in 3:1 ethanol:acetic acid solution,followed by Coomassie blue(6 mg mL-1)staining,and were observed under an Olympus BX-60 microscope(Olympus,Tokyo,Japan).For each plant,the Bgt hyphal length of 20 germinated spores was measured with CellSens Entry(https://www.olympus-lifescience.com.cn/en/software/cellsens/)software,and three biological replicates were used.
Pst isolate CYR32 was provided by Zhensheng Kang(Northwest Agriculture and Forestry University,Yangling,Shanxi,China),and propagated on seedlings of the susceptible cultivar Mingxian 169(Mx169)in a growth chamber under a 16-h light/8-h dark(15/10°C,70%humidity)regime.Plants at the two-leaf stage were evaluated for Yr resistance against isolate CYR32 following Fu et al.[36],and ITs were graded 0-9 at 14 days after inoculation(dai).Yr resistance at adult stage was evaluated by natural infection of Pst mixture in the field.ITs were graded 0-9 following Krattinger et al.[23].For observation of Pst development,infected leaf segments were immersed in 1 mol L-1KOH,resuspended in 50 mmol L-1Tris-HCl(pH 7.5),and stained with fluorescein isothiocyanate isomer(FITC)-labeled wheat germ agglutinin(WGAFITC)and aniline blue.Pst hyphal length was measured under a fluorescence microscope(BX53F,Olympus).
Fg isolates Fg0609 and Chi-2 labeled with green fluorescent protein(GFP)were kindly provided by Xu Zhang(Jiangsu Academy of Agricultural Sciences,Nanjing,Jiangsu,China)and Yucai Liao(Huazhong Agriculture University,Wuhan,Hubei,China).FHB resistance was evaluated by a single-floret inoculation method using Fg0609.For each genotype,FHB severity was measured as the mean percentage of scabbed spikelets(PSS)of 30 individual spikes at 14 dai,and three biological replicates were tested.The GFP produced by Chi-2 was used to trace the infection and spreading of Fg under a macro-zoom fluorescence microscope(MVX10,Olympus).
2.3.Genome-wide identification of LysM gene family
LysM protein sequences were searched using the Pfam(https://www.sanger.ac.uk/software/Pfam)number of the LysM domain(Pf01476)with a HMMER(https://www.hmmer.org/)model against the genome of H.villosa(unpublished).LysM gene structures were visualized with TBtools(https://www.tbtools.com/).Domain structures of potential LysM proteins were identified with Pfam and Inter-ProScan(https://www.ebi.ac.uk/Inter-ProScan).
Phylogenetic analysis of LysM genes was performed with MEGA X(https://www.megasoftware.net).Full-length amino acid sequences of LysM proteins were selected for generating a bootstrap neighbor-joining phylogenetic tree.Bootstrap probabilities were calculated from 1000 replicates.
2.4.Cloning and expression profiling of CERK1s
cDNAs of H.villosa and Chinese Spring were isolated at 30 min after chitin treatment.Using the primer pair CERK1-FL-F and CERK1-FL-R(Table S8),CERK1-V was isolated from H.villosa,and TaCERK1-A,TaCERK1-B,and TaCERK1-D were isolated from Chinese Spring.PCR was performed at 95 °C for 5 min,followed by 30 cycles of 95°C for 15 s,56°C for 15 s and 72°C for 2 min,and then for 10 min at 72 °C.Phanta Max Super-Fidelity DNA polymerase(Vazyme,catalog number P505,Nanjing,Jiangsu,China)was used for amplification.
The qRT-PCR was used to measure CERK1-V expression in H.villosa.Samples of 0.1 g of leaf tissue at the two-leaf stage were collected at 0 and 24 h before and after inoculation with three pathogens(Bgt isolate E26,Pst isolate CYR32 and Fg isolate Fg0609),and 0.1 g of leaf tissue was collected before and at 1 h after treatment with three PAMPs[100μg mL-1chitin(Tokyo Chemical Industry,catalog number C2762,Tokyo,Japan),100μg mL-1PGN(Yuanye Bio-Technology,catalog number S11184,Shanghai,China)and 500 nmol L-1flg22(PhytoTech,catalog number P6622,Shawnee Mission,USA)].Total RNA was isolated with Trizol reagent(Invitrogen,catalog number 15596018,USA)according to the manufacturer’s instructions.First-strand cDNA was synthesized from 1μg total RNA using HiScript Q-RT SuperMix for qPCR(Vazyme,catalog number R223).The Tubulin gene was used as the internal control for normalization.qRT-PCR was performed with AceQ qPCR SYBR Green Master Mix(Vazyme,catalog number Q111)using PCR LightCycler 480(Roche,Switzerland).Each sample was assayed with three technical replicates,and the expression data for transcripts were fitted with ANOVA.The sequence information of primer pairs was listed in Table S8.
2.5.Subcellular localization
The expression vector CERK1-V-GFP was constructed by fusing CDS of CERK1-V to the N-terminal of GFP using the backbone vector pCambia 1305.1-GFP,and then transformed into the Agrobacterium strain GV3101 and infiltrated into the abaxial leaf surface of 4-to 6-week-old Nicotiana benthamiana.Images were captured under a LSM780(Carl Zeiss,Jena,Germany)confocal microscope at the GFP channel(488 nm).
2.6.Genetic transformation
An overexpression vector ProUbi::CERK1-V was constructed by inserting the open reading frame of CERK1-V into the pMWB110 vector using a Sac I cloning site.The resulting plasmid was transformed into wheat cultivar Fielder via Agrobacterium-mediated transformation[37].The specific primer pair CERK1-V-T-F/R(Table S8),which amplifies the first intron and part of the first and second exons of CERK1-V,was designed and used to screen for positive transgenic plants in the T0to T3generations.
2.7.Single-cell transient overexpression assay(TOA)
TOA was performed following Shirasu[38].The tested genes were inserted into vector pBI-220.The resulted plasmids were mixed with the reporter plasmid pWMB002 carrying theβglucuronidase(GUS)gene before particle-coating(at a molar ratio of 1:1;1μg).Bombarded leaves were transferred to 1%agar plates supplemented with 85μmol L-1benzimidazole,incubated at 22°C for 8 h,inoculated with a high density of fresh Bgt spores,and GUSstained to identify transformed cells at 48 hai.The haustorium indexes(HIs,percentage of GUS-staining cells with haustoria in the total GUS-staining cells attacked by Bgt)were measured in three independent experiments,each contributing at least 40 interactions.
2.8.RNA-seq and data analysis
RNA-seq was performed using Fielder at the three-leaf stage and the transgenic lines(OE-L and OE-H).Three biological replicates were used for RNA extraction and library construction.The library was sequenced with an Illumina HiSeq 4000(Illumina,San Diego,CA,USA).The RNA-seq reads were aligned to the wheat genome of the International Wheat Genome Sequencing Consortium with the RefSeq v2.0 annotation(https://wheat-urgi.versailles.inra.fr/Seq-Repository/Annotations).Differentially expressed genes(DEGs)were detected with PossionDis[39]as requested and thresholds were set as fold change≥2.00 and false discovery rate(FDR)≤0.001.
2.9.Chitin binding assay and Western blotting
Chitin magnet beads(New England BioLabs,catalog nu mber E8036S,UK)were co-incubated with purified maltose binding protein(MBP)-tagged recombinant proteins.The chitin magnet bead was washed five times with binding buffer[50 mmol L-1Tris-HCl,pH 7.0,100 mmol L-1NaCl,and 0.1 mmol L-1phenylmethanesulfonyl fluoride(PMSF)]and boiled with SDS-PAGE loading buffer.For binding competing assay,soluble chitin was added as competitors in the binding buffer.The presence of target proteins in the chitin binding fraction was determined by Western blotting.
Western blotting was performed following Liu et al.[12]using primary anti-CERK1(Agrisera,catalog number AS164037,1:1500 diluted,Sweden),anti-CPK1(Agrisera,catalog number AS194315,1:1500 diluted),anti-MBP(Abmart,catalog number 1:3000 diluted,Shanghai,China),and anti-phospho-p42/p44(Cell Signaling Technology,catalog number 9101,1:1000 diluted,USA).Secondary goat anti-mouse antibody or goat anti-rabbit antibody was conjugated to horseradish peroxidase and visualized using ECL solution(Vazyme,catalog number E412).Pictures were scanned and the image intensities were quantified with ImageJ(https://imagej.nih.gov/).
2.10.Yeast two-hybrid(Y2H)assay
TaCEBiPs and CERK1-V interaction was tested using the yeast HF7c strain.Using the primer pair CEBiPs-F and CEBiPs-R(Table S8),TaCEBiPs was isolated from cDNA of Chinese Spring.The extracellular(LysM)domains of the TaCEBiPs and CERK1-V were fused to the BD and AD domains,respectively.The resulting constructs and empty vectors were transformed into yeast as described by Xie et al.[40].The transformation mixtures were plated on yeast drop-down selection media(SD-LT)deprived of leucine(Leu)and tryptophan(Trp).Protein interactions were evaluated by observing the growth status of yeast cells on SDLTHA media after incubation at 30 °C for 3 days.
2.11.Measurement of programmed cell death(PCD),H2O2 accumulation,and SA content
Leaves were decolorized for 24 h in decolorizing solution(0.15%trichloroacetic acid dissolved in a mixture of ethanol and chloroform).The PCD was detected by staining the leaves with trypan blue,and the H2O2accumulation was measured by staining the leaves with diaminobezidine(DAB).H2O2concentrations of three biological replicates were calculated following Jiang et al.[41].For each genotype,SA concentrations of three biological replicates were calculated following Lee et al.[42].
2.12.Measurement of photosynthetic rate
Net photosynthesis rate was determined at the four-leaf stage following Gao et al.[43]using the red and blue light source chambers of a Li-Cor 6400(LI-COR,Lincoln,NE,USA).For each genotype,nine plants were used as a biological replicate,and three biological replicates were used.
3.Results
3.1.H.villosa CERK1-V is a chitin-responsive PRR
Twenty-four LysM proteins were identified in the genome of H.villosa.Based on composition of the functional domains,they could be classified as nine LYKs,five LYPs,and 10 extracellular Lysinmotif proteins(LysMes)or non-secretory Lysin-motif proteins(LysMns)(Fig.S1A).The LysM gene(s)involved in Pm resistance was preliminarily predicted by expression profiling using RNAseq and qRT-PCR.Upon infection by Bgt,gene Dv07G125800 was the most abundantly expressed among the identified 24 H.villosa LysM genes(LysMs),as shown by its expression increase to a peak of 86-fold at 24 hai(Fig.1A left panel,S1B left panel;Table S1).Dv07G125800 expression was also induced by infection of Pst(53-fold increase)or Fg(10-fold increase)(Fig.1A,left panel).LysMs expression was also induced by treatments with fungal PAMP chitin,and bacterial PAMPs,either flg22 or PGN(Fig.1A).Among 24 LysM genes,Dv07G125800 was the most significantly upregulated in response to chitin treatment(Fig.1A right panel).Dv07G125800 was expressed in all tested organs,and the highest expression was in roots(Fig.S1B right panel;Table S2).These findings suggested the involvement of Dv07G125800 in innate immunity against different plant pathogens.
Phylogenetic analysis revealed that Dv07G125800 was the closest homolog of rice OsCERK1 and Arabidopsis AtCERK1(Fig.1B).Dv07G125800 in H.villosa was designated as CERK1-V.The fulllength coding sequence(CDS)of CERK1-V was cloned from H.villosa at 30 min post-chitin treatment.CERK1-V encoded a protein consisting of 621 amino acids(Fig.S1D).Its full-length gDNA was 8677 bp,containing eight exons and seven introns(Fig.S1C).Similar as OsCERK1 and AtCERK1,CERK1-V contained one N-terminal signal peptide,three extracellular LysM domains,one transmembrane domain,and an intracellular Thr/Ser kinase domain(Fig.S1D).CERK1-V was rapidly accumulated in response to chitin treatment(Fig.1C)and localized on the plasma membrane(Fig.1D).
3.2.Heterologous overexpression of CERK1-V increased wheat resistance to three fungal diseases
CERK1-V was overexpressed in the recipient cultivar Fielder by genetic transformation.Five positive transgenic plants were identified,and all displayed increased Pm resistance in T0generation(Fig.S2A,B).Two lines representing independent transgenic events and showing significantly different transgene expression levels were selected for further analysis.OE-CERK1-V-T0-2(36-fold up-regulated expression,hereafter OE-L)and OE-CERK1-V-T0-5(303-fold up-regulated expression,hereafter OE-H)represented respectively lines having moderate and excessive higher transgene expression.Their derived T3lines were analyzed by qRT-PCR and Western blotting confirming the differences of transgene abundance at transcription and translation levels(Fig.S2C,D).Genomic qPCR indicated that OE-H and OE-L both carried two copies of the transgene CERK1-V(Fig.S2E).
Pm resistance at seedling stage was evaluated by inoculation of detached leaves with Bgt isolate E26.The mean ITs of Fielder,OE-L,and OE-H were 4,1,and 0,respectively(Fig.2A).The CERK1-V was located on chromosome 7 V of H.villosa.Wheat-H.villosa addition line DA7V showed increased seedling stage Pm resistance relative to Fielder,but its resistance level was lower than those of the two transgenic lines(Fig.2A).At adult stage,Pm resistance against Bgt mixture in field was evaluated.The mean ITs of Fielder,Sumai 3,OE-L,and OE-H were 7-8,8-9,2-3 and 0,respectively(Fig.S2F).These values indicated that OE-L and OE-H showed increased Pm resistance at both stages,with that of OE-H higher than that of OE-L.Bgt development was compared by Coomassie blue-stained Bgt-inoculated leaves.At 24 hai,Fielder and two transgenic lines showed no significant difference in Bgt spore germination and development.At 96 hai,significant differences in the number and length of developed hyphae were observed.OE-L and OE-H had fewer Bgt hyphae than Fielder,with OE-H showing many fewer than OE-L(Fig.2B,C).Thus,the inhibition of Bgt development in transgenic lines may have been due to higher expression of CERK1-V.
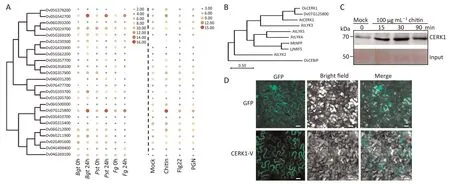
Fig.1.CERK1-V was predicted as a chitin-responsive PRR in H.villosa.(A)Phylogenetic and expression analysis of H.villosa LysM genes induced by fungal pathogens,including Bgt isolate E26,Pst isolate CYR32 and Fg isolate Fg0609(Left)and PAMPs,including 100μg mL-1 chitin,500 nmol L-1 flg22,and 100μg mL-1 PGN(right).Circles indicate the log2 fold change calculated by qRT-PCR,in which the values were normalized by the gene Dv05G378200.(B)Amino acid sequence-based phylogenetic analysis of Dv07G125800 with known LYK proteins.The rice LYP OsCEBiP(UniProtKB,Q8H8C7)was included as outgroup.AtCERK1(UniProtKB,A8R7E6),AtLRK2(UniProtKB,Q9SGI7),AtLYK3(UniProtKB,F4IB81),AtLYK4(UniProtKB,O64825)and AtLYK5(UniProtKB,O22808)in Arabidopsis thaliana;OsCERK1(UniProtKB,A0A0P0)in Oryza sativa;MtNFP(UniProtKB,Q0GXS4)in Medicago truncatula;LjNFP(UniProtKB,Q70KR1)in Lotus japonicus.(C)Detection of CERK1-V in leaves of H.villosa under 100μg mL-1 chitin treatment,the protein of CERK1-V was detected by anti-CERK1,Mock represents water treatment.(D)Subcellular localization of CERK1-V.Constructs using GFP alone and CERK1-V fused GFP were individually expressed in epidermal cells of Nicotiana benthamiana,visualized under confocal microscopy.Scale bars,20μm.
The role of CERK1-V and TaCERK1s in pre-haustorium Pm resistance was investigated by TOA.The HI of the negative control was 62.6%.In contrast,HIs of transiently overexpressed CERK1-V,TaCERK1-A,TaCERK1-B,and TaCERK1-D were 38.2% 46.8%,44.6%and 44.7%,respectively(Fig.S2G).Thus,transient-overexpression of CERK1s from H.villosa and wheat inhibited the haustorium formation,with heterologous overexpression of CERK1-V showing a stronger effect.
Yr resistance at seedling stage was evaluated by artificial inoculation with Pst isolate CYR32 in the growth chamber.At 14 dai,the mean ITs of Fielder,OE-L and OE-H were 7-8,3-4 and 0,respectively(Fig.2D).Yr resistance at adult stage was evaluated by natural infection with Pst mixture in field.The mean ITs of Fielder,OE-L and OE-H were 7-8,3-4 and 0,respectively(Fig.S2H).Thus,transgenic lines showed increased Yr resistance at both stages.Pst development was compared by staining CYR32 inoculated leaves with WGA-FITC.At 24 hai,they showed no significant difference in development of Pst urediniospores.At 96 hai,urediniospores germinated well and produced germ tubes(GTs)in Fielder;whereas Pst development in the transgenic lines stagnated,as shown by the reduced hyphal length and formation of few GTs(Fig.2E).The length and quantity of developed Pst hyphae in OE-H were shorter and lower than those in OE-L,indicating that higher CERK1-V expression/protein level also contributed to greater Yr resistance(Fig.2F).
The function of CERK1-V in resistance to FHB,a hemibiotrophic fungal disease,was also evaluated.At 14 dai of Fg0609,the percentage of symptomatic spikelets(PSS)was much higher in Fielder(28%)than two transgenic lines.The PSS of OE-H(7%)was less than that of OE-L(11%)(Fig.2G,H).The Fg infection was compared by observing the GFP signal distribution of the inoculated Fg isolate Chi-2.They showed no significant difference at 72 hai.At 144 hai,GFP signals were distributed in the whole spikelet in Fielder,but were limited in the inoculated florets in the transgenic lines.The signal intensity in OE-H was lower than that in OE-L,suggesting less fungal development in OE-H(Fig.2I).Thus,heterologous overexpression of CERK1-V increased wheat resistance against both biotrophic and hemibiotrophic fungal diseases,in an expression level-dependent manner.
3.3.CERK1-V promoted chitin binding by interacting with TaCEBiPs
To elucidate the molecular mechanism of multiple fungal disease resistance conferred by CERK1-V,gene expression of the two transgenic lines and Fielder was profiled by RNA-seq.Compared with Fielder,there were 6533 and 16,060 DEGs in OE-L and OE-H,respectively.In OE-L,5791 were upregulated and 742 were downregulated;while in OE-H,13,255 were upregulated and 2805 were downregulated(Fig.S3A),indicating higher change in gene expression in OE-H.Gene ontology(GO)and Kyoto encyclopedia of genes and genomes(KEGG)analysis showed that the most markedly enriched DEGs present in two transgenic lines included chitin sensing(response to chitin,polysaccharide/chitin binding,chitinase activity),signal transduction(plant-pathogen interaction,MAPK signaling pathway,calmodulin/calcium ion binding),phytohormone(phenylalanine metabolism,cinnamic acid biosynthesis, arachidonic acid metabolism), and photosynthesis-related biological process(photosynthesis,oxidative phosphorylation)(Fig.S3B).
Once sensing the fungal infection,plant chitinases degrade fungal chitin into chitin oligomers which can be recognized by PRRs.CERK1-V may function as such a PRR in sensing chitin and transduce defense signal pathways.How overexpressed CERK1-V regulated the expression of genes involved in chitin degradation and binding pathways was determined.From RNA-seq data,we found that 49 chitinase genes belonging to five basal classes(class I to class V)were enriched in transgenic lines.Class I,III and IV chitinase genes were all up-regulated,and very few class II or V chitinase genes were down-regulated(Fig.3A;Table S3).Overexpression of CERK1-V may have activated the expression of wheat chitinase genes,and increased resistance to the three fungal diseases.

Fig.2.Overexpression of CERK1-V in wheat increased resistance to three fungal pathogens.(A-C)Pm resistance of two transgenic lines and Fielder at seedling stage.(A)Disease symptom and infection type(IT)infected by Bgt isolate E26.ABV(T.durum-H.villosa amphiploid),DA7V(an addition line carrying a pair of 7 V chromosomes in a hexaploid wheat background)and Chinese Spring(CS)were used as controls.The ITs of Pm were measured in ten leaves of each line at 7 dai.Scale bars,1 cm.(B)Bgt development at 24 and 96 hai on leaves stained with Coomassie blue and observed under microscope.Scale bars,20μm.(C)Hyphal length of Bgt isolate E26 at 96 hai.Error bars represent standard deviation of three biological replicates.Different letters indicate significant difference by one-way ANOVA/Duncan(P<0.05).(D-F)Yr resistance of two transgenic lines and Fielder at seedling stage.(D)Disease symptoms and infection type(IT)of infection by Pst isolate CYR32.92R137 was used as resistant control,Mingxian(Mx)169 and Fielder were used as susceptible controls.The ITs of Yr were measured on 10 leaves of each line at 14 dai.Scale bars,1 cm.(E)Pst development at 24 and 96 hai on leaves stained with WGA and observed under microscope.Scale bars,25μm.(F)Hyphal length of CYR32 at 96 hai.Error bars represent standard deviation of three biological replicates.Different letters indicate significant difference by one-way ANOVA/Duncan(P<0.05).(G-I)FHB resistance of two transgenic lines and Fielder.(G)Disease symptom at 14 dai of Fg0609.Sumai 3 was used as resistant control,Taishan 23 and Fielder were used as susceptible controls.Scale bars,1 cm.(H)Percentage of symptomatic spikelets at 14 dai of Fg0609.Error bars represent standard deviation of three biological replicates.Different letters indicate significant difference by one-way ANOVA/Duncan(P<0.05).(I)Fg development at 72 and 144 hai of Chi-2.The GFP signal was observed under a macro-zoom fluorescence microscope system(Olympus MVX10).BF,bright field.Scale bars,1 mm.White arrows indicate inoculation points.
In monocotyledons,direct chitin binding depends on LYPs.The role of CERK1-V overexpression in regulating the expression of wheat endogenous LYPs,especially homologs of known chitin binding-associated LYPs(TaCEBiPs,TaLYP4s,and TaLYP6s)was also investigated.TaCEBiPs were the only significantly upregulated LYPs in transgenic lines(Figs.3B,S4;Table S3).TaCEBiPs could be pulled down by chitin in vitro,and TaCEBiPs decreased with the increase of soluble chitin(Fig.3C).Thus,TaCEBiPs did have the direct chitin binding activity,and the direct interaction of the extracellular domain of CERK1-V with the chitin-binding domain of TaCEBiPs was confirmed(Fig.3D).From the above findings,we propose that in transgenic wheat,CERK1-V promoted chitin binding by acting as the partner of TaCEBiPs.
3.4.Heterologous overexpression of CERK1-V activated expression of genes involved in both PTI and ETI
MAPK cascade and Ca2+are early transducing signals for PTI.RNA-seq revealed that 22 MAPKs were differentially expressed in transgenic lines,and 11(MAPKKK17,MAPKKK1,MAPKKK YODA,MAPKKK ANP1,MAPKK2,MAPKK3,MAPKK4,MAPK3,MAPK5,MAPK6,and MAPK12)were the most highly enriched(Fig.4A;Table S4).MAPK3 and MAPK6 are two conserved MAPKs in early stages of plant defense against fungal pathogens.In transgenic lines,phosphorylated TaMAPK3 and TaMAPK6 were clearly accumulated before and after Bgt inoculation(Fig.4B).Thus,overexpression of CERK1-V not only activated MAPK cascade gene expression,but also promoted protein phosphorylation.Five representative CPK genes(CPK1,CPK4,CPK5,CPK6,and CPK11)were significantly up-regulated in transgenic lines(Fig.4C;Table S4).CPK1 was the most highly up-regulated,and more abundant CPK1 protein accumulation was observed(Fig.4D).These results suggested that CERK1-V-mediated resistance was achieved by activating PTI,possibly via regulation of MAPK and CPK signaling pathways.

Fig.3.Overexpression of CERK1-V promoted wheat chitin recognition.(A)Relative expression of chitinase related genes in Fielder and transgenic wheat.I to V indicate chitinases of classes I to V.Expression values are fragments per kilobase per million(FPKM)obtained from RNA-seq and normalized to Fielder.Error bars represent standard deviation of three biological replicates.(B)Relative expression of TaCEBiPs,TaLYP4s,and TaLYP6s in Fielder and transgenic wheat.Expression levels were calculated by qRTPCR and normalized to Tubulin,and are presented as fold changes relative to Fielder.Letters indicate significant difference by one-way ANOVA/Duncan(P<0.05).Error bars represent standard deviation of three biological replicates.(C)The LysM domain of TaCEBiPs binds to chitin.Commercial chitin beads were used to pull down MBP-tagged recombinant LysM domain of TaCEBiPs(top).Soluble chitin competed with chitin beads.Commercial chitin beads were used to pull down MBP-tagged recombinant LysM domain of TaCEBiPs in the presence or absence of soluble chitin(0,20,50 and 100μg mL-1,bottom).(D)Y2H assay showing interaction between LysM domain of TaCEBiPs and LysM domain of CERK1-V.Combinations of plasmid encoding LysM domain of TaCEBiPs with plasmid encoding LysM domain of CERK1-V were co-transformed into yeast strain HF7c.Cells were plated on SD medium lacking Leu,Trp,histidine(His),and adenine(Ade)(-Leu/-Trp/-His/-Ade).
Previous studies[44,45]have indicated that the potentiation of PTI is indispensable for ETI during bacterial infection.We further investigated whether overexpression of CERK1-V activated the ETI pathway in wheat.In the reference genome of Chinese Spring(IWGSC RefSeq v2.0),2,151 NLR-type R genes were annotated in wheat.RNA-seq indicated that 282(13.11%)and 422(19.61%)wheat NLRs were up-regulated and that 10(0.46%)and 8(0.37%)wheat NLRs were down-regulated in OE-L and OE-H,respectively(Fig.S5A).The activation of such a large number of NLRs may be associated with the increased multiple-disease resistance of the transgenic lines.
SA and ROS burst are two typical hallmarks of PCD.Phenylalanine ammonia lyase(PAL)and isochorismate synthase(ICS)are two key pathways for SA biosynthesis.Trypan blue and DAB staining revealed that the transgenic line OE-H showed strong PCD and accumulated abundant ROS(Fig.5A).SA accumulation and transcription of SA responsive genes TaPR1 and TaPR2 were significantly increased in transgenic lines(Figs.5B-D,S6).In transgenic lines,gene expression associated with the PAL pathway was activated,while that related to ICS pathway remained at a level similar to that of Fielder(Fig.S5B;Table S5).We propose that CERK1-Vactivated SA biosynthesis depended mainly on the PAL pathway·H2O2accumulation was not significantly different in Fielder and OE-L.Although two ROS pathway-associated genes,TaNOX and TaGST,were up-regulated both in OE-L and OE-H(Figs.5F,G,S6),only OE-H showed significantly more H2O2(Fig.5E).Jasmonic acid(JA)and ethylene(ET)pathways are involved in plant resistance against necrotrophic and hemibiotrophic pathogens.Two marker genes for JA and ET pathways,TaCOI1 and TaERF,were significantly up-regulated in OE-L and OE-H.Their up-regulated expression levels were dependent on that of CERK1-V(Figs.5H,I,S6).
3.5.CERK1-V showed a tradeoff effect on disease resistance and plant growth
The disease resistance and plant growth of the transgenic lines were investigated in successive generations.One of the transgenic plants showing more than 100-fold transgene expression,OECERK1-V-T0-5,showed higher disease resistance and abnormal plant growth(Fig.S2A,B and F).Its derived T1,T2,and T3(OE-H)lines all showed severe growth defects,with yellowish leaf,few tillers,and reduced plant height,spike length,and thousand-kernel weight(Figs.6A-G,S7A-H).Another transgenic plant showing<100-fold transgene expression,OE-CERK1-V-T0-2,showed increased disease resistance and normal plant growth.Its derived T1,T2,and T3(OE-L)lines were similar as Fielder with respect to all tested agronomic traits(Figs.6A-G,S7A-H).Thus,CERK1-V influenced the balance between plant defense and growth,depending on its expression level.Excessive expression of CERK1-V could increase disease resistance at the cost of normal plant growth and development.

Fig.4.Overexpression of CERK1-V activated downstream signaling of chitin recognition.(A)Heat map showing the expression level of MAPK associated genes in Fielder and transgenic wheat.Expression values are FPKM obtained by RNA-seq.(B)Phosphorylated TaMAPK3 and TaMAPK6 were detected in Fielder and transgenic wheat after inoculation with Bgt isolate E26 using phospho-p42/p44 antibody.(C)Relative expression of TaCPK1,TaCPK4,TaCPK5,TaCPK6,and TaCPK11.Expression levels were calculated by qRT-PCR and normalized to Tubulin,and are presented as fold changes relative to Fielder.Letters indicate significant differences by one-way ANOVA/Duncan(P<0.05).Error bars represent standard deviation of three biological replicates.(D)The protein level of CPK1 was detected by anti-CPK1 in Fielder and transgenic wheat.
The photosynthetic rate in OE-H was significantly decreased(Fig.S8A).RNA-seq revealed that DEGs involved in photosynthesis were significantly enriched.Genes involved in light harvesting and antenna were significantly down-regulated in OE-H,while remained at similar levels in OE-L and Fielder(Fig.S8B,C;Table S6).We accordingly speculated that CERK1-V overexpression not only induced the changes of plant defense signal pathways,but also reprogrammed the gene expression involved in photosynthesis.These two effects may explain the tradeoff effect in OE-H on disease resistance and growth.However,it is feasible to obtain both increased resistance and normal plant growth by optimizing CERK1-V expression,just as in OE-L in this study.
Endoplasmic reticulum stress(ER stress)and phytohormones including auxin,gibberellins(GA),and brassinosteroids(BRs)regulate the balance of plant growth and immunity.Expression of markers or representative genes associated with the above pathways were measured.In OE-H,the expression of ER stressassociated genes was strongly induced,while expression of marker genes of auxin-responsive and GA pathway was suppressed.OE-L and Fielder showed similar expression patterns for the above genes(Fig.S8D-F;Table S7).The expression of marker genes associated with the BR pathway resulted in no marked change in any of the three materials(Fig.S8G;Table S7).We suggest that an excessively high level of CERK1-V expression activated ER stress,suppressed auxin and GA pathways,and finally resulted in the growth defect in OE-H.
4.Discussion
Because PAMPs,such as fungal chitin,are conserved modules,PRR-mediated PTI is usually shown as basal resistance but has potential value for increasing broad-spectrum resistance to multiple pathogen species.In rice,owing to its ability to recognize two PAMPs(fungal chitin and bacterial PGN),the OsLYP4 and OsLYP6 activated innate immunity against fungi and bacteria.Silencing of OsLYP4 or OsLYP6 led to compromised production of ROS in response to elicitor treatment,suggesting that OsLYP4/OsLYP6-mediated multi-resistance is associated with the ROS pathway[12].Following the recognition of PAMPs,defense-signaling proteins(such as RLKs,RLCKs,and MAPKs)are activated.In rice,OsMAPK15 negatively regulated PR gene expression,ROS and phytohormone(JA and SA)accumulation.Knockout of osmapk15 increased resistance to X.oryzae pv.oryzae(Xoo)and M.oryzae[46].In the present study,CERK1-V acted as a chitin receptor and activated the expression of genes related to MAPK cascade,CPKs,NLR genes,ROS,and several phytohormone signaling pathways(Fig.7).The observation that heterologous overexpression of CERK1-V activated multiple immunity pathways may explain increased resistances of the transgenic lines against both biotrophic pathogens Bgt,Pst,and hemibiotrophic pathogen Fg.
LysM PRRs recognize chitin oligosaccharides,and plants employ PRR pairs and protein components to transduce chitin-induced PTI signal.In rice,chitin binding and signal transduction are accomplished by LYP and LYK proteins,while in Arabidopsis,both processes are dependent on CERK1 and LYK5[7].In the present study,CERK1-V activated the gene expression of wheat chitinases and wheat LYP TaCEBiPs.Pull-down and Y2H assays supported direct interaction between CERK1-V and TaCEBiPs.We propose that activated wheat chitinase degrade chitin into oligosaccharides,which are recognized by CERK1-V/TaCEBiPs complex,which finally activates and transmits a chitin signal to induce PTI.In rice,the MAPK cascade consists of MAPKKK11/18-MKK4-MPK3/6 involved in chitin signal transduction,while in Arabidopsis,MAPKKK5-MAPKK4/MAPKK5-MAPK3/MAPK6 were reported[14,15]to play a major role.In the present study,although the up-regulated MAPKKK and MAPKK genes are different from those in rice and Arabidopsis,the phosphorylation of MAPK3 and MAPK6 suggested their conserved roles among plant species in chitin signal transduction.In Arabidopsis,CPK1,CPK4,CPK5,CPK6,and CPK11 are involved in PTI signal transduction[17,18].The observation that wheat CERK1-V also activated the expression of these CPKs supports the conservation of the calcium signal pathway in PRR-mediated defense in multiple plant species.

Plant chitinases are associated with resistance to multiple fungal diseases.PR3(chitinases from classes I,II,and IV),PR8(chitinases from class III),PR11(chitinases from class V),and PR4(proteins with low endo-chitinase activity)can inhibit fungal development by degrading fungal cell walls[47].LysMs or their complex recognize the degraded products(chitin oligosaccharides)and amplify PTI signaling.CERK1-V activated the SA and ROS pathways in wheat.We propose that SA-and ROS-induced PCD and activated downstream defense pathways are involved in CERK1-V mediated resistance against biotrophic pathogens.Exogenous application of SA or its analog benzothiadiazole did not increase FHB resistance of type I(Fg initial infection)or type II(Fg spreading)[48].However,expressing Arabidopsis SA receptor NPR1 in wheat did increase inheritable type II resistance[49].For a hemibiotrophic pathogen like Fg,SA or ROS-dependent PCD may suppress pathogen infection at the early biotrophic stage but accelerate pathogen growth and spread at the later necrotrophic stage.Arabidopsis knockout mutants of JA receptor and ET signaling genes coi1 and ein2,showed increased susceptibility to the necrotrophic pathogens Alternaria brassica and B.cinerea[50,51].Consistently,overexpression of Ethylene Response Factor 1(ERF1)increased resistance to B.cinerea[52].These indicate the positive roles of JA and ET in plant defense against a necrotrophic pathogen[53].Our observation that the expression of marker genes for JA and ET pathways was activated in CERK1-V transgenic wheat may explain the increased FHB resistance of the transgenic lines.
The activation of plant immune systems is accompanied by high energy consumption and metabolic resource reallocation.For this reason,over-activation of innate immunity usually impairs plant growth[54].Phytohormones play major roles in regulating the trade-off of plant immunity with growth.SA was required for the activation of immune response to biotrophic pathogens and SA accumulation triggered a series of immune-responsive signaling[55].In contrast to its positive role in immunity,SA negatively regulates plant growth in many ways,including inhibiting auxin perception[56],repressing GA signaling[57],and inducing ER stress[58].In the present study,excessively high expression of CERK1-V in wheat(OE-H)resulted in constitutive activation of ER stressassociated genes and suppression of IAA and GA signaling pathways(Fig.7).However,those genes in OE-L(with optimized expression of CERK1-V)displayed expression patterns similar as those in Fielder.The above findings may explain why the inhibition of plant growth and development was only observed in OE-H while not in OE-L.Optimizing CERK1-V expression may optimize both disease resistance and development.

Fig.6.Excessively high expression of CERK1-V caused abnormal growth of transgenic wheat.(A)Phenotype of Fielder and transgenic wheat at jointing(left)and heading stages(right).Scale bars,10 cm.(B,C)Statistical analysis of(B)plant height and(C)tiller numbers of Fielder and transgenic wheat.(D)Spike morphology at heading stage.(E)Statistical analysis of spike length.(F)Grain morphology at maturity.(G)Statistical analysis of thousand-kernel weight at maturity.Difference was tested by one-way ANOVA/Duncan(P<0.05).Error bars represent standard deviation of three biological replicates.
Due to the coevolution of pathogens and plants,PTI can be broken by adapted pathogens via delivering virulent effectors into plants.The Pseudomonas syringae AvrPto and AvrPtoB strongly suppressed Arabidopsis PTI signaling by interacting with two PRRs,FLS2 and EFR[59,60].Rice OsCEBiP and OsCERK1 form a receptor complex and function in chitin-triggered PTI defense signaling.The formation of a unique sandwich-type dimer of OsCEBiP acts in activation of chitin signaling[61,62].OsCEBiP homolog in barley,HvCEBiP,contributed to basal resistance to M.oryzae in barley[63].In wheat,TaCERK1 provided complete resistance to the mg3lysm(homolog of Ecp6)deletion mutant of nonpathogenic Mycosphaerella graminicola(syn.Zymoseptoria tritici),but conferred no resistance to the wild-type pathogen[64],suggesting that endogenous chitin receptors have been partially overcome by adapted pathogens in wheat.Thus,we propose the introduction of exogenous chitin receptors may provide a potential new way to increase wheat disease resistance.
Inter-species transformation allows the use of a large repertoire of PRRs or R genes with a range of substrate affinities.By heterologous expression of alien PRRs or R genes,it is possible to increase multiple disease resistances in cultivated crops.Transgenic plants heterologously expressing AtEFR in rice,wheat,and tomato showed increased resistance to several bacterial pathogens[65-67].Heterologous expression of rice XA21 in banana conferred complete resistance to X.campestris pv.musacearum[68].However,different species may exploit different resistance signaling components,such that the transfer of exogenous PRR alone cannot activate enough endogenous immunity in all cases.Expression of Arabidopsis EFR in rice provided only limited resistance to avirulent X.oryzae,while failing to increase resistance to virulent races[69].There are more than 350 Triticeae species in nature[70].The sequence and function of genes from subgenomes of various species are highly conserved.From the released genome sequence of Triticeae genomes,more than 1000 RLKs or RLPs have been identified.They are potentially valuable gene resources for improving PTI of cultivated wheat or other crops.In our study,overexpressing H.villosa CERK1-V in wheat fully activated wheat immune responses,leading to increased resistance to three fungal diseases.We propose that H.villosa and wheat employ conserved PTI signaling components in CERK1-mediated defense.Our results also provide a successful example for increasing wheat resistance by using PRRs from its wild relatives.

Fig.7.Proposed working model of the role of CERK1-V in regulating plant defense and growth.In response to infection by fungal pathogens,CERK1-V positively regulates the expression and physically interacts with the chitin-binding protein TaCEBiPs.Endogenous wheat chitinases degrade fungal cell wall chitins into chitin oligomers,which can be recognized by CERK1-V/TaCEBiPs complex.Overexpression of CERK1-V activates PTI by up-regulating the expression of chitinase genes,multiple MAPK cascadesespecially TaMAPK3 and TaMAPK6-and early defense-associated CPKs,including CPK1,CPK3,CPK4,CPK5,CPK6,and CPK11.Overexpression of CERK1-V also activates ETI by regulating expression of NLR genes and genes involved in the ROS pathway and phytohormones SA,JA,and ET.These contribute to increasing disease resistance.However,excessive expression of CERK1-V also leads to defects in plant growth by regulating photosynthesis,auxin response,GA pathways and inducing the ER-stress pathway.
5.Conclusions
Heterologous expression of CERK1-V in wheat increased fungal disease resistance.The excessively high expression level of CERK1-V showed dual effects on plant defense and plant growth.Optimized overexpression led to high resistance without impairment of plant growth.This study revealed the mechanism of CERK1-V mediated resistance.We propose CERK1-V as a gene resource for increasing multiple fungal disease resistance.
CRediT authorship contribution statement
Anqi Fan:Performed experiment,data interpretation and prepare original draft.Luyang Wei:Performed experiment,data collection and analysis,and prepare original draft.Xu Zhang:Data analysis and methodology.Jia Liu:Data analysis and methodology.Li Sun:Data curation and edit manuscript.Jin Xiao:Data curation and edit manuscript.Yajia Wang:Data analysis and methodology.Haiyan Wang:Data curation and edit manuscript.Jian Hua:Review and editing.Ravi P.Singh:Review and editing.Zongkuan Wang:Funding acquisition,Project administration,Data curation,Writing-review & editing.Xiue Wang:Funding acquisition,Project administration,Writing-review & editing,Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This study was supported by the National Natural Science Foundation of China(31661143005,31801350 and 32011530167),National Key Research and Development Program of China(2016YFD0102001-004),and China Postdoctoral Science Foundation(2018M642266).We acknowledge Tao Wang(Nanjing Agricultural University)for SA and H2O2concentration measurement.
Appendix A.Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2022.02.005.
参考文献[1]J.D.G.Jones,J.L.Dangl,The plant immune system,Nature 444(2006)323-329.[2]S.Robatzek,D.Chinchilla,T.Boller,Ligand-induced endocytosis of the pattern
recognition receptor FLS2 in Arabidopsis,Gene Dev.20(2006)537-542.
[3]C.Zipfel,G.Kunze,D.Chinchilla,A.Caniard,J.D.G.Jones,T.Boller,G.Felix,Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation,Cell 125(2006)749-760.
[4]E.Gubaeva,A.Gubaev,R.L.Melcher,S.Cord-Landwehr,R.Singh,N.E.El Gueddari,B.M.Moerschbacher,‘Slipped Sandwich’model for chitin and chitosan perception in Arabidopsis,Mol.Plant-Microbe Interact.31(2018)1145-1153.
[5]A.Miya,P.Albert,T.Shinya,Y.Desaki,K.Ichimura,K.Shirasu,Y.Narusaka,N.Kawakami,H.Kaku,N.Shibuya,CERK1,a LysM receptor kinase,is essential for chitin elicitor signaling in Arabidopsis,Proc.Natl.Acad.Sci.U.S.A.104(2007)19613-19618.
[6]T.Liu,Z.Liu,C.Song,Y.Hu,Z.Han,J.She,F.Fan,J.Wang,C.Jin,J.Chang,J.M.Zhou,J.Chai,Chitin-induced dimerization activates a plant immune receptor,Science 336(2012)1160-1164.
[7]Y.Cao,Y.Liang,K.Tanaka,C.T.Nguyen,R.P.Jedrzejczak,A.Joachimiak,G.Stacey,The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1,eLife 3(2014)e03766.
[8]T.Shimizu,T.Nakano,D.Takamizawa,Y.Desaki,N.Ishii-Minami,Y.Nishizawa,E.Minami,K.Okada,H.Yamane,H.Kaku,N.Shibuya,Two LysM receptor molecules,CEBiP and OsCERK1,cooperatively regulate chitin elicitor signaling in rice,Plant J.64(2010)204-214.
[9]H.Kaku,Y.Nishizawa,N.Ishii-Minami,C.Akimoto-Tomiyama,N.Dohmae,K.Takio,E.Minami,N.Shibuya,Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor,Proc.Natl.Acad.Sci.U.S.A.103(2006)11086-11091.
[10]B.Gong,J.Xue,N.Zhang,L.Xu,X.Yao,Q.Yang,Y.Yu,H.Wang,D.Zhang,J.Li,Rice chitin receptor OsCEBiP is not a transmembrane protein but targets the plasma membrane via a GPI anchor,Mol.Plant 10(2017)767-770.
[11]J.Wan,K.Tanaka,X.Zhang,G.H.Son,L.Brechenmacher,T.H.Nguyen,G.Stacey,LYK4,a Lysin motif receptor-like kinase,is important for chitin signaling and plant innate immunity in Arabidopsis,Plant Physiol.160(2012)396-406.
[12]B.Liu,J.F.Li,Y.Ao,J.Qu,Z.Li,J.Su,Y.Zhang,J.Liu,D.Feng,K.Qi,Y.He,J.Wang,H.B.Wang,Lysin motif-containing proteins LYP4 and LYP6 play dual roles in peptidoglycan and chitin perception in rice innate immunity,Plant Cell 24(2012)3406-3419.
[13]R.Willmann,H.M.Lajunen,G.Erbs,M.A.Newman,D.Kolb,K.Tsuda,F.Katagiri,J.Fliegmann,J.J.Bono,J.V.Cullimore,A.K.Jehle,F.Götz,A.Kulik,A.Molinaro,V.Lipka,A.A.Gust,T.Nürnberger,Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection,Proc.Natl.Acad.Sci.U.S.A.108(2011)19824-19829.
[14]K.Yamada,K.Yamaguchi,T.Shirakawa,H.Nakagami,A.Mine,K.Ishikawa,M.Fujiwara,M.Narusaka,Y.Narusaka,K.Ichimura,Y.Kobayashi,H.Matsui,Y.Nomura,M.Nomoto,Y.Tada,Y.Fukao,T.Fukamizo,K.Tsuda,K.Shirasu,N.Shibuya,T.Kawasaki,The Arabidopsis CERK1-associated kinase PBL27 connects chitin perception to MAPK activation,EMBO J.35(2016)2468-2483.
[15]K.Yamada,K.Yamaguchi,S.Yoshimura,A.Terauchi,T.Kawasaki,Conservation of chitin-induced MAPK signaling pathways in rice and Arabidopsis,Plant Cell Physiol.58(2017)993-1002.
[16]G.Galotto,I.Abreu,C.Sherman,B.Liu,M.Gonzalez-Guerrero,L.Vidali,Chitin triggers calcium-mediated immune response in the plant model Physcomitrella patens,Mol.Plant-Microbe Interact.33(2020)911-920.
[17]M.Boudsocq,M.R.Willmann,M.McCormack,H.Lee,L.Shan,P.He,J.Bush,S.H.Cheng,J.Sheen,Differential innate immune signalling via Ca2+sensor protein kinases,Nature 464(2010)418-422.
[18]M.Coca,B.San Segundo,AtCPK1 calcium-dependent protein kinase mediates pathogen resistance in Arabidopsis,Plant J.63(2010)526-540.
[19]C.Huang,Y.Yan,H.Zhao,Y.Ye,Y.Cao,Arabidopsis CPK5 phosphorylates the chitin receptor LYK5 to regulate plant innate immunity,Front.Plant Sci.11(2020)702.
[20]M.Figueroa,K.E.Hammond-Kosack,P.S.Solomon,A review of wheat diseases a field perspective,Mol.Plant Pathol.19(2018)1523-1536.
[21]J.Kourelis,R.A.L.van der Hoorn,Defended to the Nines:25 years of resistance gene cloning identifies nine mechanisms for R protein function,Plant Cell 30(2018)285-299.
[22]W.Li,Y.Deng,Y.Ning,Z.He,G.Wang,Exploiting broad-spectrum disease Resistance in crops:from molecular dissection to breeding,Annu.Rev.Plant Biol.71(2020)575-603.
[23]S.G.Krattinger,E.S.Lagudah,W.Spielmeyer,R.P.Singh,J.Huerta-Espino,H.McFadden,E.Bossolini,L.L.Selter,B.Keller,A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat,Science 323(2009)1360-1363.
[24]J.W.Moore,S.Herrera-Foessel,C.Lan,W.Schnippenkoetter,M.Ayliffe,J.Huerta-Espino,M.Lillemo,L.Viccars,R.Milne,S.Periyannan,X.Kong,W.Spielmeyer,M.Talbot,H.Bariana,J.W.Patrick,P.Dodds,R.Singh,E.Lagudah,A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat,Nat.Genet.47(2015)1494-1498.
[25]R.Buschges,K.Hollricher,R.Panstruga,G.Simons,M.Wolter,A.Frijters,R.van Daelen,T.van der Lee,P.Diergaarde,J.Groenendijk,S.Topsch,P.Vos,F.Salamini,P.Schulze-Lefert,The barley Mlo gene:a novel control element of plant pathogen resistance,Cell 88(1997)695-705.
[26]J.Fu,H.Yu,X.Li,J.Xiao,S.Wang,Rice GH3 gene family:regulators of growth and development,Plant Signal.Behav.6(2011)570-574.
[27]G.Li,J.Zhou,H.Jia,Z.Gao,M.Fan,Y.Luo,P.Zhao,S.Xue,N.Li,Y.Yuan,S.Ma,Z.Kong,L.Jia,X.An,G.Jiang,W.Liu,W.Cao,R.Zhang,J.Fan,X.Xu,Y.Liu,Q.Kong,S.Zheng,Y.Wang,B.Qin,S.Cao,Y.Ding,J.Shi,H.Yan,X.Wang,C.Ran,Z.Ma,Mutation of a histidine-rich calcium-binding-protein gene in wheat confers resistance to Fusarium head blight,Nat.Genet.51(2019)1106-1112.
[28]Z.Su,A.Bernardo,B.Tian,H.Chen,S.Wang,H.Ma,S.Cai,D.Liu,D.Zhang,T.Li,H.Trick,P.St.Amand,J.Yu,Z.Zhang,G.Bai,A deletion mutation in TaHRC confers Fhb1 resistance to Fusarium head blight in wheat,Nat.Genet.51(2019)1099-1105.
[29]H.Wang,S.Sun,W.Ge,L.Zhao,B.Hou,K.Wang,Z.Lyu,L.Chen,S.Xu,J.Guo,M.Li,P.Su,X.Li,G.Wang,C.Bo,X.Fang,W.Zhuang,X.Cheng,J.Wu,L.Dong,W.Chen,W.Li,G.Xiao,J.Zhao,Y.Hao,Y.Xu,Y.Gao,W.Liu,Y.Liu,H.Yin,J.Li,X.Li,Y.Zhao,X.Wang,F.Ni,X.Ma,A.Li,S.S.Xu,G.Bai,E.Nevo,C.Gao,H.Ohm,L.Kong,Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat,Science 368(2020)eaba5435.
[30]H.Li,Z.Dong,C.Ma,X.Tian,Z.Qi,N.Wu,B.Friebe,Z.Xiang,Q.Xia,W.Liu,T.Li,Physical mapping of stem rust resistance gene Sr52 from Dasypyrum villosum based on ph1b-induced homoeologous recombination,Int.J.Mol.Sci.20(2019)4887.
[31]A.Yildirim,S.S.Jones,T.D.Murray,R.F.Line,Evaluation of Dasypyrum villosum populations for resistance to cereal eyespot and stripe rust pathogens,Plant Dis.84(2000)40-44.
[32]H.He,S.Zhu,R.Zhao,Z.Jiang,Y.Ji,J.Ji,D.Qiu,H.Li,T.Bie,Pm21,encoding a typical CC-NBS-LRR protein,confers broad-spectrum resistance to wheat powdery mildew disease,Mol.Plant 11(2018)879-882.
[33]L.Xing,P.Hu,J.Liu,K.Witek,S.Zhou,J.Xu,W.Zhou,L.Gao,Z.Huang,R.Zhang,X.Wang,P.Chen,H.Wang,J.D.G.Jones,M.Karafiátová,J.Vrána,J.Bartoš,J.Dolezˇel,Y.Tian,Y.Wu,A.Cao,Pm21 from Haynaldia villosa encodes a CC-NBSLRR protein conferring powdery mildew resistance in wheat,Mol.Plant 11(2018)874-878.
[34]Z.Liu,Q.Sun,Z.Ni,T.Yang,R.A.Mcintosh,Development of SCAR markers linked to the Pm21 gene conferring resistance to powdery mildew in common wheat,Plant Breed.5(1999)215-219.
[35]I.Jakobson,H.Peusha,L.Timofejeva,K.Järve,Adult plant and seedling resistance to powdery mildew in a Triticum aestivum×Triticum militinae hybrid line,Theor.Appl.Genet.112(2006)760-769.
[36]D.Fu,C.Uauy,A.Distelfeld,A.Blechl,L.Epstein,X.Chen,H.Sela,T.Fahima,J.Dubcovsky,A novel kinase-START gene confers temperature-dependent resistance to wheat stripe rust,Science 323(2009)1357-1360.
[37]Y.Ishida,M.Tsunashima,Y.Hiei,T.Komari,Wheat(Triticum aestivum L.)Transformation Using Immature Embryos,Springer,New York,NY,USA,2014,pp.189-198.
[38]K.Shirasu,K.Nielsen,P.Piffanelli,R.Oliver,P.Schulze-Lefert,Cell autonomous complementation of mlo resistance using a biolistic transient expression system,Plant J.17(1999)293-299.
[39]S.Audic,J.M.Claverie,The significance of digital gene expression profiles,Genome Res.7(1997)986-995.
[40]Q.Xie,A.P.Sanz-Burgos,H.Guo,J.A.García,C.Gutiérrez,GRAB proteins,novel members of the NAC domain family,isolated by their interaction with a geminivirus protein,Plant Mol.Biol.39(1999)647-656.
[41]M.Jiang,J.Zhang,Effect of abscisic acid on active oxygen species,antioxidative defence system and oxidative damage in leaves of maize(Zea mays)seedlings,Plant Cell Physiol.42(2001)1265-1273.
[42]D.H.Lee,H.W.Choi,B.K.Hwang,The pepper E3 ubiquitin ligase RING1 gene,CaRING1,is required for cell death and the salicylic acid-dependent defense response,Plant Physiol.156(2011)2011-2025.
[43]J.Gao,F.Wang,J.Sun,Z.Tian,H.Hu,S.Jiang,Q.Luo,Y.Xu,D.Jiang,W.Cao,T.Dai,Enhanced Rubisco activation associated with maintenance of electron transport alleviates inhibition of photosynthesis under low nitrogen conditions in winter wheat seedlings,J.Exp.Bot.22(2018)5477-5488.
[44]B.P.Ngou,H.Ahn,P.Ding,J.D.Jones,Mutual potentiation of plant immunity by cell-surface and intracellular receptors,Nature 592(2021)110-115.
[45]M.Yuan,Z.Jiang,G.Bi,K.Nomura,M.Liu,Y.Wang,B.Cai,J.Zhou,S.Y.He,X.Xin,Pattern-recognition receptors are required for NLR-mediated plant immunity,Nature 592(2021)105-109.
[46]Y.Hong,Q.Liu,Y.Cao,Y.Zhang,D.Chen,X.Lou,S.Cheng,L.Cao,The OsMPK15 negatively regulates Magnaporthe oryza and Xoo disease resistance via SA and JA signaling pathway in rice,Front.Plant Sci.10(2019)752.
[47]A.Kasprzewska,Plant chitinases-regulation and function,Cell Mol.Biol.Lett.8(2003)809-824.
[48]G.Y.Yu,G.J.Muehlbauer,Benzothiadiazole-induced gene expression in wheat spikes does not provide resistance to Fusarium head blight,Physiol.Mol.Plant Pathol.59(2001)129-136.
[49]R.Makandar,J.S.Essig,M.A.Schapaugh,H.N.Trick,J.Shah,Genetically engineered resistance to Fusarium head blight in wheat by expression of Arabidopsis NPR1,Mol.Plant-Microbe Interact.19(2006)123-129.
[50]B.P.Thomma,K.Eggermont,I.A.Penninckx,B.Mauch-Mani,R.Vogelsang,B.P.Cammue,W.F.Broekaert,Separate jasmonate-dependent and salicylatedependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens,Proc.Natl.Acad.Sci.U.S.A.95(1998)15107-15111.
[51]B.P.Thomma,K.Eggermont,K.F.Tierens,W.F.Broekaert,Requirement of functional ethylene-insensitive 2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea,Plant Physiol.121(1999)1093-1101.
[52]M.Berrocal-Lobo,A.Molina,R.Solano,Constitutive expression of ethyleneresponse-factor1 in Arabidopsis confers resistance to several necrotrophic fungi,Plant J.29(2002)23-32.
[53]J.Glazebrook,Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens,Annu.Rev.Phytopathol.43(2005)205-227.
[54]M.Gou,Z.Shi,Y.Zhu,Z.Bao,G.Wang,J.Hua,The F-box protein CPR1/CPR30 negatively regulates R protein SNC1 accumulation,Plant J.69(2012)411-420.
[55]D.F.Klessig,H.W.Choi,D.A.Dempsey,Systemic acquired resistance and salicylic acid:past,present,and future,Mol.Plant-Microbe Interact.31(2018)871-888.
[56]D.Wang,K.Pajerowska-Mukhtar,A.H.Culler,X.Dong,Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway,Curr.Biol.17(2007)1784-1790.
[57]L.Gallego-Giraldo,L.Escamilla-Trevino,L.A.Jackson,R.A.Dixon,Salicylic acid mediates the reduced growth of lignin down-regulated plants,Proc.Natl.Acad.Sci.U.S.A.108(2011)20814-20819.
[58]C.Ruberti,Y.Lai,F.Brandizzi,Recovery from temporary endoplasmic reticulum stress in plants relies on the tissue-specific and largely independent roles of bZIP28 and bZIP60,as well as an antagonizing function of BAX-Inhibitor 1 upon the pro-adaptive signaling mediated by bZIP28,Plant J.93(2018)155-165.
[59]L.Shan,P.He,J.Li,A.Heese,S.C.Peck,T.Nürnberger,G.B.Martin,J.Sheen,Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity,Cell Host Microbe 4(2008)17-27.
[60]T.Xiang,N.Zong,Y.Zou,Y.Wu,J.Zhang,W.Xing,Y.Li,X.Tang,L.Zhu,J.Chai,J.M.Zhou,Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases,Curr.Biol.18(2008)74-80.
[61]H.Kaku,N.Shibuya,Molecular mechanisms of chitin recognition and immune signaling by LysM-receptors,Physiol.Mol.Plant 95(2016)60-65.
[62]M.Hayafune,R.Berisio,R.Marchetti,A.Silipo,M.Kayama,Y.Desaki,S.Arima,F.Squeglia,A.Ruggiero,K.Tokuyasu,A.Molinaro,H.Kaku,N.Shibuya,Chitininduced activation of immune signaling by the rice receptor CEBiP relies on a unique sandwich-type dimerization,Proc.Natl.Acad.Sci.U.S.A.111(2014)E404-E413.
[63]S.Tanaka,A.Ichikawa,K.Yamada,G.Tsuji,T.Nishiuchi,M.Mori,H.Koga,Y.Nishizawa,R.O’Connell,Y.Kubo,HvCEBiP,a gene homologous to rice chitin receptor CEBiP,contributes to basal resistance of barley to Magnaporthe oryzae,BMC Plant Biol.10(2010)288.
[64]W.Lee,J.J.Rudd,K.E.Hammond-Kosack,K.Kanyuka,Mycosphaerella graminicola LysM effector-mediated stealth pathogenesis subverts recognition through both CERK1 and CEBiP homologues in wheat,Mol.Plant-Microbe Interact.27(2014)236-243.
[65]F.Lu,H.Wang,S.Wang,W.Jiang,C.Shan,B.Li,J.Yang,S.Zhang,W.Sun,Enhancement of innate immune system in monocot rice by transferring the dicotyledonous elongation factor Tu receptor EFR,J.Integr.Plant Biol.57(2015)641-652.
[66]H.J.Schoonbeek,H.H.Wang,F.L.Stefanato,M.Craze,S.Bowden,E.Wallington,C.Zipfel,C.J.Ridout,Arabidopsis EF-Tu receptor enhances bacterial disease resistance in transgenic wheat,New Phytol.206(2015)606-613.
[67]S.Lacombe,A.Rougon-Cardoso,E.Sherwood,N.Peeters,D.Dahlbeck,H.P.van Esse,M.Smoker,G.Rallapalli,B.P.H.J.Thomma,B.Staskawicz,J.D.G.Jones,C.Zipfel,Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance,Nat.Biotechnol.28(2010)365-369.
[68]J.N.Tripathi,J.Lorenzen,O.Bahar,P.Ronald,L.Tripathi,Transgenic expression of the rice Xa21 pattern-recognition receptor in banana(Musa sp.)confers resistance to Xanthomonas campestris pv.Musacearum,Plant Biotechnol.J.12(2014)663-673.
[69]B.Schwessinger,O.Bahar,N.Thomas,N.Holton,V.Nekrasov,D.Ruan,P.E.Canlas,A.Daudi,C.J.Petzold,V.R.Singan,R.Kuo,M.Chovatia,C.Daum,J.L.Heazlewood,C.Zipfel,P.C.Ronald,W.Ma,Transgenic expression of the dicotyledonous pattern recognition receptor EFR in rice leads to liganddependent activation of defense responses,PLoS Pathog.11(2015)e1004809.
[70]M.G.Saghir,Taxonomy and phylogeny in Triticeae:a historical review and current status,Adv.Plants Agric.Res.3(2016)139-143.
- The Crop Journal的其它文章
- Profiling seed soluble sugar compositions in 1164 Chinese soybean accessions from major growing ecoregions
- The mitochondria-localized protein OsNDB2 negatively regulates grain size and weight in rice
- Integrated linkage mapping and genome-wide association study to dissect the genetic basis of zinc deficiency tolerance in maize at seedling stage
- Delaying application time of slow-release fertilizer increases soil rhizosphere nitrogen content,root activity,and grain yield of spring maize
- Effects of increasing panicle-stage N on yield and N use efficiency of indica rice and its relationship with soil fertility
- Yield sustainability of winter wheat under three limited-irrigation schemes based on a 28-year field experiment

