The mitochondria-localized protein OsNDB2 negatively regulates grain size and weight in rice
Mingxin Guo,Jiajia Liu,Linlin Hou,Suna Zhao,Nana Zhang,Lili Lu,Xusheng Zhao
College of Life Sciences,Luoyang Normal University,Luoyang 471934,Henan,China
Keywords:Rice OsNDB2 Type II NAD(P)H dehydrogenase Allelic mutants Grain size
ABSTRACT Grain size is a determinant of rice grain yield.In plants,mitochondria supply energy for cellular metabolism via the mitochondrial electron transport chain.Here we report that OsNDB2,which encodes a putative rotenone-insensitive type II NAD(P)H dehydrogenase(ND),negatively regulates grain size and weight in rice.Six ND members representing three major types of rice were identified,and the predicted OsNDB2 protein was localized to mitochondria.Contents of OsNDB2 transcripts were higher in young panicles and leaf blades.OsNDB2 overexpression reduced grain length,grain width,and 1000-grain weight and moderately influenced plant height,while knockout of OsNDB2 increased grain size and 1000-grain weight.Allelic mutations of OsNDB2 were associated with diverse grain appearances.Cellular observations revealed that variations in grain size of transgenic lines were caused by change in cell expansion but not cell proliferation in spikelet hulls.Our study sheds light on OsNDB2 function and provides a new potential breeding approach for increasing rice grain size and weight.
1.Introduction
The plant mitochondrial electron transport chain(mtETC)is located in the inner mitochondrial membrane and is composed of four multi-subunit complexes:complex I(NADH dehydrogenase),complex II(succinate dehydrogenase),complex III(cytochrome c reductase),and complex IV(cytochrome c oxidase).Plant mitochondria also possess an alternative respiratory pathway(AP)that can bypass the classical electron transport(ETC)pathway from complexes I or II to complex IV.This bypass requires rotenoneinsensitive type II NAD(P)H dehydrogenases(NDs),ubiquinone,and alternative oxidase(AOX)[1].NDs are encoded by a small gene family divided into three subgroups in plants:NDA,NDB,and NDC.NDA and NDC are located on the internal surfaces of the mitochondrial inner membrane,while NDB is external[2].
In Arabidopsis,NDs include two NDA members(AtNDA1 and AtNDA2),four NDB members(AtNDB1-AtNDB4),and an NDC member(AtNDC1)[3].All ND members have a FAD/NAD-binding domain.NDB members also harbor an EF hand-containing domain that directly mediates Ca2+activation of Arabidopsis NDB1[4-6].Suppression of both AtNDA1 and AtNDA2 affects metabolism and plant growth[7].Knockdown of AtNDB1 affects central metabolism and vegetative growth but increases tolerance to ammonium toxicity[8,9].AtNDB2 functions in external NADH oxidation in isolated mitochondria and drought stress[3].Given the low expression levels of AtNDB3 in most tissues,little is known about its function in Arabidopsis[4,10].Knockdown of AtNDB4 confers increased tolerance to salinity stress,and knockout of AtNDB4 by T-DNA insertion leads to changes in growth rate,root:shoot ratios and leaf area[11].AtNDC1 regulates the biosynthesis of vitamin K1in Synechocystis and Arabidopsis by performing the penultimate step[12].
This study identified the ND members in rice,and phylogenetic analysis indicated that the OsNDs could be divided into three subgroups.To date,no studies have been published reporting the biological functions of ND members in rice.In this study,we focused on OsNDB2 and investigated its functions using transgenic experiments.OsNDB2 overexpression reduced grain size,while knockout of OsNDB2 using the CRISPR/Cas9 system increased grain size.Allelic mutants generated by CRISPR/Cas9 showed diverse grain phenotypes.
2.Materials and methods
2.1.Vector construction and transgenic experiments
For an overexpression experiment,the full-length coding sequence of OsNDB2 was cloned from a geng(japonica)variety,Kongyu 131(KY131)cDNA library and ligated into the binary vector pZH2Bi.For the CRISPR/Cas9 system,a guide RNA was designed to target the fifth exon and the plant expression vector was constructed as previously described[13].All plant transformation vectors were introduced into KY131 by Agrobacterium tumefaciensmediated transformation.Primers used for vector construction are listed in Table S1.
2.2.RNA extraction and quantitative RT-PCR analysis
Total RNA was extracted using a TaKaRa MiniBEST Plant RNA Extraction Kit(TaKaRa,Dalian,China).For reverse transcription,1μg RNA and oligo(dT)primers were used to synthesize firststrand cDNA in a 20-μL reaction using a TaKaRa PrimeScript II 1st Strand cDNA Synthesis Kit(TaKaRa).Quantitative RT-PCR(qPCR)was performed using TaKaRa SYBR Premix Ex Taq II(TaKaRa)with a Bio-Rad CFX96 machine(Bio-Rad,Hercules,CA,USA).Three repeats were performed for each gene.The OsActin gene was used as an internal control for normalization.Primers used for qPCR are listed in Table S1.
2.3.Protein subcellular localization
The OsNDB2 coding sequence was cloned in frame with GFP under the CaMV 35S promoter,and OsNDB2-GFP was transiently expressed in rice protoplasts.For confocal microscopy,the localization pattern of OsNDB2-GFP was examined under a confocal laser scanning microscope Leica TCS SP5(Leica,Weztlar,Germany).GFP was excited at a wavelength of 488 nm,and the emission filters were 500-530 nm.Primers used to construct the OsNDB2-GFP vector are listed in Table S1.
2.4.Scanning electron microscopy(SEM)
Outer spikelet hull surfaces were observed and imaged by SEM Phenom Pro(Phenom,Eindhoven,the Netherlands).The cell length and width of outer epidermal cell of lemmas were measured using the accompanying software.
2.5.Statistical analysis
Excel 2010 software was used for all statistical tests.Differences between the KY131 and transgenic lines were tested with Student’s two-tailed t-test.
2.6.Accession numbers
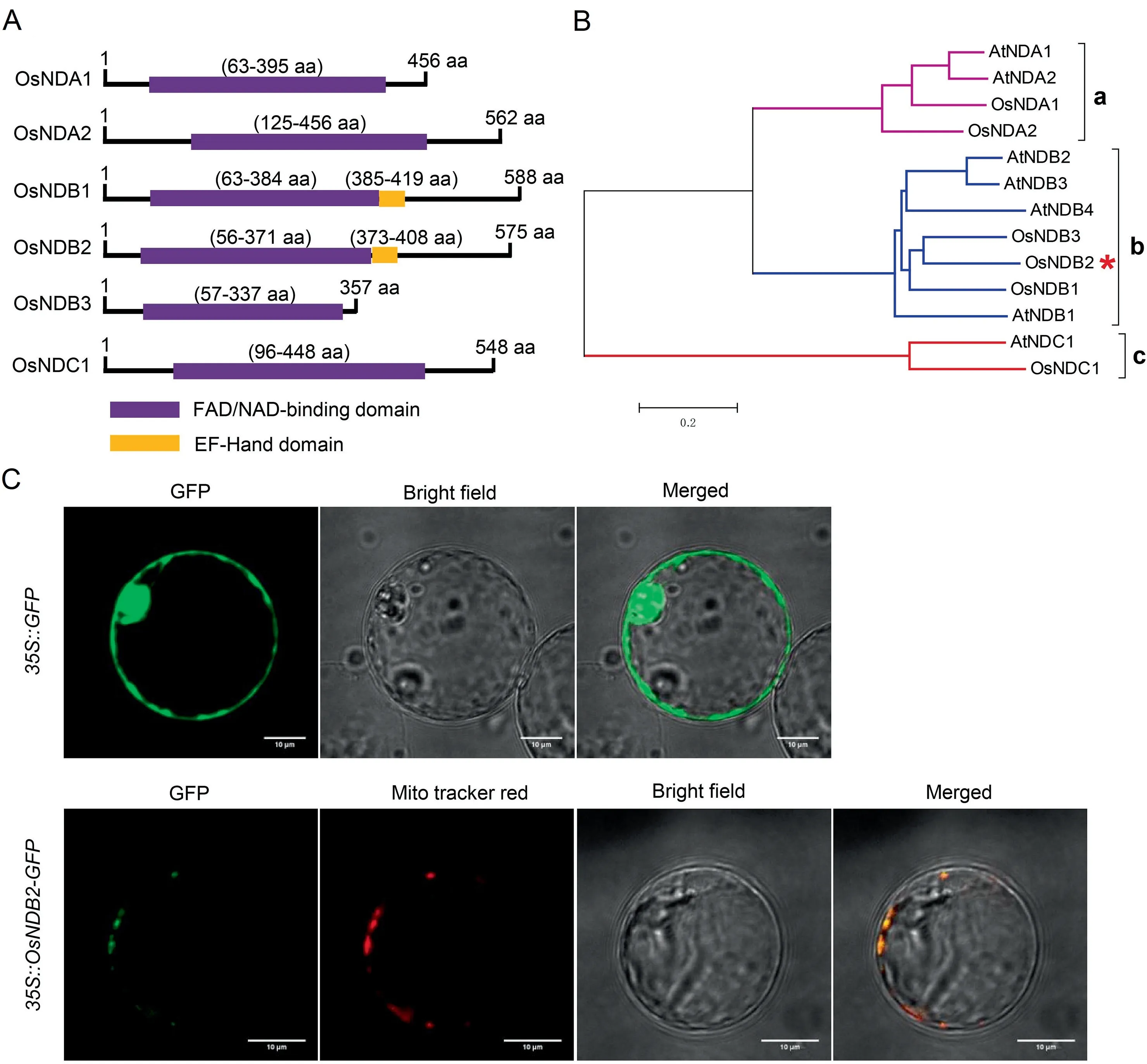
Fig.1.Phylogenetic analysis of type II NAD(P)H dehydrogenases(NDs)from rice and Arabidopsis and subcellular localization of OsNDB2.(A)Domain annotations of six NDs members in rice.(B)Phylogenetic tree of AtNDs and OsNDs.(C)Empty vector 35S::GFP and the 35S::OsNDB2-GFP fusion were transiently expressed in rice protoplasts.A mitochondrial tracker indicated that the OsNDB2-GFP fusion protein was expressed specifically in mitochondria.
The genes used in this study are available in RAP-DB(https://rapdb.dna.affrc.go.jp/)and TAIR(https://www.arabidopsis.org/)databases under accession numbers:OsNDA1(Os01g0830100),OsNDA2(Os07g0564500),OsNDB1(Os06g0684000),OsNDB2(Os05g0331200),OsNDB3(Os08g0141400),OsNDC1(Os06g0214 900),AtNDA1(At1g07180),AtNDA2(At2g29990),AtNDB1(At4g282 20),AtNDB2(At4g05020),AtNDB3(At4g21490),AtNDB4(At2g208 00),AtNDC1(At5g08740).
3.Results
3.1.Identification,domain analysis,and phylogenetic tree of OsNDs
BLASTP searches were conducted against the RAP-DB database,and six ND members were identified.To verify the initial results,domain searches of OsNDs were conducted to confirm the presence of the conserved domain using the InterPro database(https://www.ebi.ac.uk/interpro/).All six candidate members harbored the FAD/NAD-binding domain(Fig.1A).In addition to this conserved domain,OsNDB1and OsNDB2 were found to carry the EFHand domain(Fig.1A).To clarify the classification of the OsND genes and better understand the potential biological functions of OsNDs from well-studied NDs in Arabidopsis,13 ND proteins comprising seven proteins from Arabidopsis and six from rice were used to construct a phylogenetic tree(Fig.1B).OsND members could be divided into three subgroups:a(OsNDA1 and OsNDA2),b(OsNDB1,OsNDB2,and OsNDB3),and c(OsNDC1)(Fig.1B).
3.2.Subcellular localization of OsNDB2
To identify the subcellular location of OsNDB2,a 35S::OsNDB2-GFP vector was introduced into rice protoplasts.Given that OsNDs are components of the alternative respiratory pathway in plant mitochondria,we expected that OsNDB2 would be located in mitochondria.The OsNDB2-GFP and a mitochondrial tracker were coexpressed in rice protoplasts by transient transformation.OsNDB2-GFP was localized in mitochondria,whereas GFP on its own was localized in the cytoplasm(Fig.1C).Thus,OsNDB2 is a mitochondria-localized protein.
3.3.Overexpression of OsNDB2 reduced grain size and weight in rice
To characterize the expression pattern of OsNDB2,quantitative RT-PCR was performed with tissues from root,stem,leaf blade,leaf sheath,and young panicle.OsNDB2 expression was the highest in leaf blades and young panicles(Fig.2A).To investigate the biological functions of OsNDB2,we generated OsNDB2 overexpression(OE)lines in the KY131 background.Three OE lines were chosen and characterized in detail(Fig.2B).The plant heights of OE lines were moderately less than that of KY131 at the three-leaf and grain-filling stages(Figs.2C and D,S2).OsNDB2 overexpression significantly reduced grain size,including length and width(Fig.2EG).Compared with KY131,the grain lengths of three OE lines decreased by 10.1%,10.8%,and 11.8%,respectively.The grain widths of the three OE lines decreased by 8.6%,10.9%,and 10.9%,respectively.Similarly,the 1000-grain weights of three OE lines also decreased(Fig.2G)by 10.7%,18.7%,and 29.0%,respectively.
3.4.Knockout of OsNDB2 increases grain size and weight in rice
To further investigate how OsNDB2 affects grain size,we generated knockout mutants with the CRISPR/Cas9 system(CRI).Three CRI mutant lines were chosen for subsequent analyses.One carried a 15-bp deletion and the other two had an A or T insertion in the fifth exon(Fig.3A).The mutant protein ndb2-1 still harbored two domains,whereas the FAD/NAD-binding domain lacked five amino acid residues from Ile331 to Met335 compared with the wild-type(WT)protein(Fig.3B).For the ndb2-2 and ndb2-3 proteins,a one-base insertion at 3100 bp in the genomic region resulted in a frameshift leading to a truncated protein(348 aa),and the mutant proteins lacked an EF-Hand domain and retained a truncated FAD/NAD-binding domain(Fig.3B).In contrast to the OE lines,the CRI mutants showed no significant changes in plant height at the seedling and grain-filling stages(Figs.S1,S2).However,OsNDB2 mutations affected grain size in different ways.The CRI-1 showed no change in grain length,but the grain width increased by 10.9%(Fig.3C-E).In CRI-2 and CRI-3 mutants,the grain lengths increased significantly by 10.6% and 11.0%,respectively,but the grain widths showed no change compared with KY131(Fig.3C-E).The 1000-grain weights of the three CRI lines increased by 9.6%,7.8%,and 8.8%(Fig.3E).
3.5.OsNDB2 regulates cell expansion in spikelet hulls
To dissect the cellular mechanism underlying the phenotypic variation in grain size between transgenic and KY131 plants,we investigated spikelet hull cells of KY131,OE-2 and CRI-3.In line OE-2,the outer epidermal cells in lemmas were shorter and thinner than those of KY131 lemmas(Fig.4A-C).In line CRI-3,the outer epidermal cells in lemmas were longer than those of KY131 lemmas but showed similar width to KY131 lemmas(Fig.4A-C).The cell numbers in lemmas of transgenic lines(OE-2 and CRI-3)were similar to that in KY131 lemmas in both longitudinal and transverse directions(Fig.4D,E).These results indicate that OsNDB2 regulates grain size by affecting cell expansion in spikelet hulls.
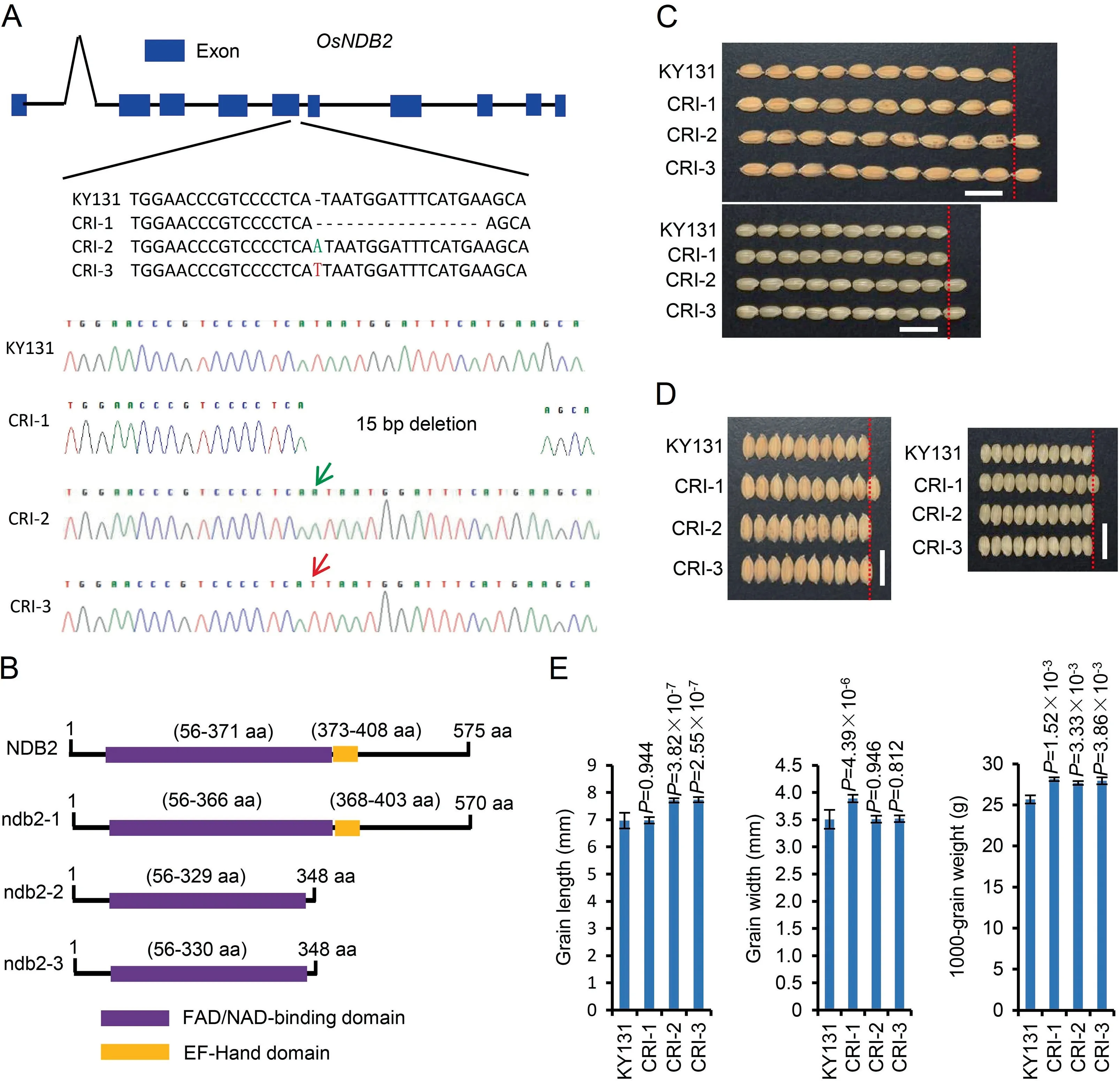
Fig.3.Knockout of OsNDB2 with CRISPR/Cas9(CRI)system increases grain size and weight in rice.(A)Gene structure of OsNDB2 and sequences at target sites in T1 plants produced by CRISPR/Cas9 system.(B)Domain annotations of NDB2 and the three mutant proteins ndb2-1,ndb2-2,and ndb2-3.(C,D)Grain morphology of KY131 and three CRI lines.Scale bars,1 cm.(E)Statistics of grain length,grain width,and 1000-grain weight in KY131 and three CRI lines.Values are means±SD,n=10 for grain length and width and n=3 for 1000-grain weight.Differences between KY131 and CRI lines were tested with Student’s two-tailed t-test.

Fig.4.OsNDB2 regulates grain size by affecting cell expansion in spikelet hulls.(A)Scanning electron microscopy images of the outer surfaces of KY131,OE-2 and CRI-3.(B,C)Mean cell length(B)and width(C)of outer epidermal cells in KY131,OE-2 and CRI-3,lemmas.(D,E)Outer epidermal cell number in longitudinal(D)and transverse(E)directions in KY131,OE-2 and CRI-3.Scale bars in(A),100μm.Values in(B)and(C)are means±SE,n=300 cells.Values in(D)and(E)are means±SD,n=10 spikelets.Differences between the KY131 and transgenic lines were tested with Student’s two-tailed t-test.
4.Discussion
This study performed genome-wide identification and characterization of ND members in rice.Six OsND genes representing three major types(OsNDA,OsNDB,OsNDC)were identified,and the subgroup b member OsNDB2 was functionally characterized in detail.Our results indicate that OsNDB2 negatively regulates grain size in rice.OsNDB2 encodes external rotenone-insensitive NADPH dehydrogenase,a core component of the alternative respiratory pathway in plant mitochondria.In plants,mitochondria are the main organelles that produce ATP for cellular metabolism via respiratory oxidation of organic acids and transfer of electrons to O2via the mtETC[14].Manipulating core components in the mitochondrial electron transport system could change the energy supply and cellular metabolism and alter plant growth and development.A recent review[15]summarized that several signaling pathways control seed size through maternal tissues and zygotic tissues,including or involving the ubiquitin-proteasome pathway,G-protein signaling,mitogen-activated protein kinase(MAPK)signaling,phytohormone perception and homeostasis,transcriptional regulators,and the HAIKU(IKU)pathway.Our study provides a new method of increasing crop seed size by CRISPR/Cas9-mediated genome editing that targets core components of the mtETC in plants.
Allelic mutants of OsNDB2 generated by gene editing showed distinct grain appearances(Fig.3C,D).The allelic mutant CRI-1 with deletion of five amino acid residues in the FAD/NADbinding domain had wider grains but similar grain length compared to KY131.The other two allelic mutants(CRI-2 and CRI-3),each with a one-base-pair insertion in the fifth exon,retained only a truncated FAD/NAD-binding domain and showed longer grains but similar grain width compared to KY131(Fig.3C,D).Similarly,four OsGS3 alleles conferred different grain sizes,including long,medium,and short[16].Loss of function of OsMAPK6 resulting from a single-nucleotide deletion in the sixth exon reduced grain size[17],but a gain-of-function mutant of OsMAPK6 resulting from a G-to-A mutation in the fourth exon formed long grains[18].A new OsGS2 allele generated by gene editing targeted the miR396 recognition site and produced larger grains than the wild-type OsGS2 allele[19].Thus,alleles may confer different phenotypes for one trait.Rapid advances in genomeediting systems enable researchers to generate allelic mutants,which can be used in breeding programs.The simultaneous gene editing of OsNDB2 and other negative regulators mediating grain size will likely generate larger grain.Alternatively,allelic mutants of OsNDB2 crossed with other mutants with wide or long grain in KY131 could likely generate wider or longer grain than a single mutant.Thus,this study clarifies the functions of OsNDB2 and provides a potential breeding approach for increasing rice grain size.
CRediT authorship contribution statement
Mingxin Guo:Conceptualization,Methodology,Investigation,Writing-Original Draft,Writing-review & editing.Jiajia Liu:Investigation,Data curation.Linlin Hou:Investigation,Data curation.Suna Zhao:Investigation,Data curation.Nana Zhang:Investigation,Data curation.Lili Lu:Investigation,Data curation.Xusheng Zhao:Funding acquisition,Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by Luoyang Key Science and Technology Innovation Program(2101016A).
Appendix A.Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2022.07.016.
- The Crop Journal的其它文章
- Profiling seed soluble sugar compositions in 1164 Chinese soybean accessions from major growing ecoregions
- Integrated linkage mapping and genome-wide association study to dissect the genetic basis of zinc deficiency tolerance in maize at seedling stage
- Delaying application time of slow-release fertilizer increases soil rhizosphere nitrogen content,root activity,and grain yield of spring maize
- Effects of increasing panicle-stage N on yield and N use efficiency of indica rice and its relationship with soil fertility
- Yield sustainability of winter wheat under three limited-irrigation schemes based on a 28-year field experiment
- Identification of a stable major QTL for fresh-seed germination on chromosome Arahy.04 in cultivated peanut(Arachis hypogaea L.)

