DNA hypomethylation promotes learning and memory recovery in a rat model of cerebral ischemia/reperfusion injury
Guang Shi , Juan Feng Ling-Yan Jian, Xin-Yu Fan,
Abstract Cerebral ischemia/reperfusion injury impairs learning and memory in patients.Studies have shown that synaptic function is involved in the formation and development of memory, and that DNA methylation plays a key role in the regulation of learning and memory.To investigate the role of DNA hypomethylation in cerebral ischemia/reperfusion injury, in this study, we established a rat model of cerebral ischemia/reperfusion injury by occlusion of the middle cerebral artery and then treated the rats with intraperitoneal 5-aza-2′-deoxycytidine, an inhibitor of DNA methylation.Our results showed that 5-aza-2′-deoxycytidine markedly improved the neurological function, and cognitive, social and spatial memory abilities, and dose-dependently increased the synaptic density and the expression of SYP and SHANK2 proteins in the hippocampus in a dose-dependent manner in rats with cerebral ischemia/reperfusion injury.The effects of 5-aza-2′-deoxycytidine were closely related to its reduction of genomic DNA methylation and DNA methylation at specific sites of the Syp and Shank2 genes in rats with cerebral ischemia/reperfusion injury.These findings suggest that inhibition of DNA methylation by 5-aza-2′-deoxycytidine promotes the recovery of learning and memory impairment in a rat model of cerebral ischemia/reperfusion injury.These results provide theoretical evidence for stroke treatment using epigenetic methods.
Key Words: cognitive memory; DNA methylation; DNMT1; hippocampus; ischemia/reperfusion; social memory; spatial memory; TET1; transient middle cerebral artery occlusion; 5-aza-2′-deoxycytidine
Introduction
Stroke is the second leading cause of death and disability in the world(Campbell and Khatri, 2020).Most strokes (87%) are ischemic strokes(Benjamin et al., 2017), which often lead to cerebral ischemia/reperfusion(I/R) injury.Motor, speech, behavior and cognitive impairment are common complications of cerebral ischemia.The main manifestations of cognitive impairment are inattention and memory decline.These prevalent symptoms have led most recent studies to focus on cerebral I/R injury-induced changes in cognitive memory (Sun et al., 2020; Lee and Choi, 2021).
Synaptic plasticity is the neurobiological basis of learning and memory, with synaptophysin (SYP) and SH3 and multiple ankyrin repeats protein 2 (SHANK2)being two closely related proteins.SYP is a calcium-binding glycoprotein present in nerve presynaptic vesicle membranes that reflects the distribution and density of synapses (Ma et al., 2017; Liu et al., 2021; Lu et al., 2022).SHANK2 is a postsynaptic density protein that plays an important role in neuronal synaptic development, postsynaptic membrane receptor anchoring and intercellular signal transmission (Zaslavsky et al., 2019).In addition, other synapse-related proteins, such as neurexins, homer protein homolog 1 and matrix metalloproteinase-9, also play important roles in synaptic plasticity.Studies have shown that the expression of synaptic proteins is reduced after cerebral I/R injury (Alawieh et al., 2020; Zhang et al., 2021); however, the molecular mechanism of this process is still unclear.
Epigenetics refers to the process by which stable changes in biological phenotype or gene expression occur through DNA methylation, histone modification, chromosome remodeling or non-coding RNA, but without changing the DNA sequence (Cavalli and Heard, 2019; Guo et al., 2020).DNA methylation sites in eukaryotic genomes are mainly located on CG dinucleotide(CpG) islands.DNA methyltransferase (DNMT) catalyzes the conversion of cytosine (C) to 5-methylcytosine (5mC) and inhibits gene expression.In contrast, DNA demethylase (TET) converts 5mC to 5-hydromethylcytosine(5hmC) and upregulates gene expression.Furthermore, a recent study has shown that epigenetic modification of synaptic genes is involved in the process of learning and memory; the study reported that DNA methylation ofBdnf
promoter IV was increased after cognitive impairment (Yu et al.,2021).The expression of immediate early genes such asc-Fos
,Egr1
andArc
are very susceptible to DNA methylation (Poon et al., 2020).These results suggest that the reversible and dynamic regulation of DNA methylation is an important factor that mediates the changes in neuronal plasticity.In addition,DNA methylation may also be involved in the occurrence and development of neuropsychiatric diseases such as stroke, epilepsy and schizophrenia (Boison and Rho, 2020; Magwai et al., 2021; Wang et al., 2021).Studies have shown that DNA methylation of S-adenosylhomocysteine hydrolase (Zhao et al.,2020) and arachidonate 5-lipoxygenase-activating protein (Bie et al., 2021)may increase the risk of stroke.These studies indicated that the degradation of synaptic proteins induced by cerebral I/R injury may be mediated by DNA methylation.On the basis of the above research, we hypothesized that cerebral I/R injury alters the expression of DNMTs and TETs, increases DNA methylation ofSyp
andShank2
at the CpG island, and finally, causes learning and memory dysfunction.The purpose of this study was to investigate epigenetics underlying the mechanism of cerebral I/R injury-induced learning and memory deficits.Methods
Animals
Male Sprague-Dawley rats (specific pathogen-free, 180-220 g, 6 weeks old) were used in this study.The rats were housed with food and water ad libitum in a room at 23 ± 1°C and humidity at 50 ± 10% under a 12-hour light/dark cycle.All procedures were approved by the Laboratory Animal Care Committee of China Medical University (approval No.P2021PS221K) on March 1, 2021.All experiments were designed and reported according to the Animal Research: Reporting ofIn Vivo
Experiments (ARRIVE) guidelines (Percie du Sert et al., 2020).Anesthesia in the study was induced with 5.0% isoflurane(Hebei Yipin, Shijiazhuang, China) and maintained with 2.5% isoflurane.All procedures followed the NIH Guidelines for the Care and Use of Laboratory Animals.The experimental design was shown in Figure 1.Sprague-Dawley rats were randomly divided into four groups: Sham + Sal, tMCAO + Sal, tMCAO +L_5-aza and tMCAO + H_5-aza.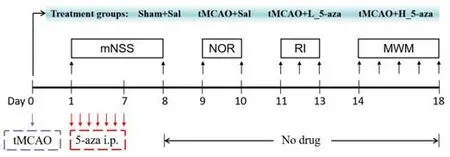
Figure 1|Experimental design of the present study.
Transient middle cerebral artery occlusion
A transient middle cerebral artery occlusion (tMCAO) rat model was used to simulate cerebral I/R injury.After 12 hours of fasting, the right middle cerebral artery was exposed and the internal carotid artery and external carotid artery were separated.The proximal ends of the external carotid artery and common carotid artery were ligated with a suture.The internal carotid artery and common carotid artery were temporarily blocked using artery clamps.A 4-0 monofilament nylon suture with a rounded tip was gently inserted into the internal carotid artery.The threading was stopped when slight resistance was felt (about 18 mm from the bifurcation of the external carotid artery).After 120 minutes of occlusion, reperfusion was performed by rapidly retracting the suture (Chen et al., 2001).Forty-four rats were used in the experiment.Eight rats in the sham group underwent the same procedure without occlusion.Five out of 12 rats died in the tMCAO + Sal and tMCAO + L_5-aza groups.Four out of 12 rats died in the tMCAO + H_5-aza group.
Drug treatment
All rats (n
= 6 per group) underwent the tMCAO surgery on day 0.Rats in the Sham + Sal and tMCAO + Sal groups were treated with saline.Rats in the tMCAO + L_5-aza and tMCAO + H_5-aza groups were treated with 5-aza-2′-deoxycytidine (5-aza; an inhibitor of DNA methylation) at either a low (L,0.5 μg/g) or high (H, 1.0 μg/g) concentration.5-aza (Cat# A119533; Aladdin,Shanghai, China) was dissolved and diluted in saline.5-aza or normal saline (1.0 mL) was intraperitoneally injected once per day for 7 consecutive days.Rats were injected with 5-aza or saline at 11:00 a.m.from day 1 to day 7.All rats underwent behavioral tests from day 1 to day 18, and the hippocampus was collected at 1 hour after the final Morris water maze on day 18.The ipsilateral hippocampus (right hemisphere) was collected for transmission electron microscopy.The contralateral hippocampus (left hemisphere) was isolated for real-time polymerase chain reaction (PCR), western blot and DNA methylation analysis.Neurological deficit assessment
The modified neurological severity score (mNSS) was used to evaluate neurological function on days 1 and 8.The mNSS evaluation is a composite score of motor tests, sensory tests, beam balance tests, reflex absence and abnormal movements (Chen et al., 2001).The scores range from 0 to 18, with higher scores suggesting greater neurological injury.
Novel object recognition
The novel object recognition (NOR) test was used to assess cognitive memory(Sawangjit et al., 2018) on days 9 and 10.The experiment comprised two phases in which each rat was allowed to explore two different objects for 10 minutes each.In the familiarization phase (day 9), each rat was placed in a plastic box (70 cm in length, width and height).Two objects (A1 and A2) of different shape and color were placed in a diagonal position about 10 cm apart against the wall.In the test phase (day 10), one familiar object (A2)was removed from the apparatus and another novel object (B) was placed in the same position.To eliminate the effect of odors, 75% ethanol was used to clean the objects between each session.Exploration time was recorded when the nose was directed towards the object at a distance of less than 2 cm, but not standing on or close to the object.
Resident-intruder test
Social memory was determined using the resident-intruder test (RI)(Potasiewicz et al., 2020) on days 11-13.This experiment was performed in a home cage where each rat was housed 30 minutes prior to the test.On day 11, one naive intruder rat (the same age and sex as the resident rat)was introduced into the resident’s cage.Both rats were placed back to their respective cages after each session.The experiment was repeated once using the same intruder rat on day 12.The session started by introducing an unfamiliar intruder rat into the cage on day 13.The interaction time was recorded for 5 minutes.Interactions were defined as sniffing, touching and following the intruder rat.
Morris water maze test
Spatial memory was assessed using the Morris water maze test (Rochais et al., 2020) on days 14-18.The maze consisted of a circular water tank (180 cm in diameter), which was conceptually divided into four quadrants.A hidden platform (8 cm in diameter) was located in the middle of the second quadrant and submerged 2 cm below the water surface.Each rat was allowed to swim freely for 90 seconds to find the platform.After each trial, the rats were placed on the platform for 15 seconds.Each rat was tested four times per day(days 14-17), and the average escape latency was calculated.The platform was removed on day 18 and each rat was placed in the water at the fourth quadrant to look for the platform for 90 seconds.The number of platform crossings and the time spent in each quadrant were recorded.
Transmission electron microscopy
Rats were fully anesthetized and then transcardial perfusion was performed using normal saline solution.Right hippocampal tissue was cut into slices(1 mm) and fixed in 1% osmium tetroxide for 1 hour.The hippocampus was dehydrated in a graded ethanol series, embedded in Polybed 812 resin (Polysciences, Warrington, PA, USA), and stained with lead citrate.The synaptic structures were observed and photographed by transmission electron microscopy (JEM-1200EX; Jeol Ltd., Akishima, Japan).The synaptic thickness was determined quantitatively according to a previous study(DeFelipe et al., 1999).
Real-time PCR
To explore the synapse-associated genes, PubMed data were analyzed by R software, and a heatmap was constructed with the results.These synapseassociated genes were confirmed by real-time PCR.Briefly, total RNA was isolated from the left hippocampus with a TRIzolTM Plus RNA Purification Kit(Cat# 12183555; ThermoFisher, Waltham, MA, USA).The cDNA was prepared by reverse transcription using the Maxima First Strand cDNA Synthesis Kit(ThermoFisher).Expression of mRNA was measured using real-time PCR(Stratagene Mx3000P; Agilent Technologies, Bӧblingen, Germany) and a Taq DNA Polymerase Kit (Cat# K1036; Apexbio, Houston, TX, USA).After an initial denaturation step of 95°C for 30 seconds, 40 cycles of PCR were performed.Each cycle consisted of a melting step at 95°C for 15 seconds and an annealing step at 60°C for 1 minute.The comparative Ct (2) method (Livak and Schmittgen, 2001) was used to analyze the relative mRNA expression, which was normalized to Gapdh.The primer sequences for each gene were listed in Table 1.
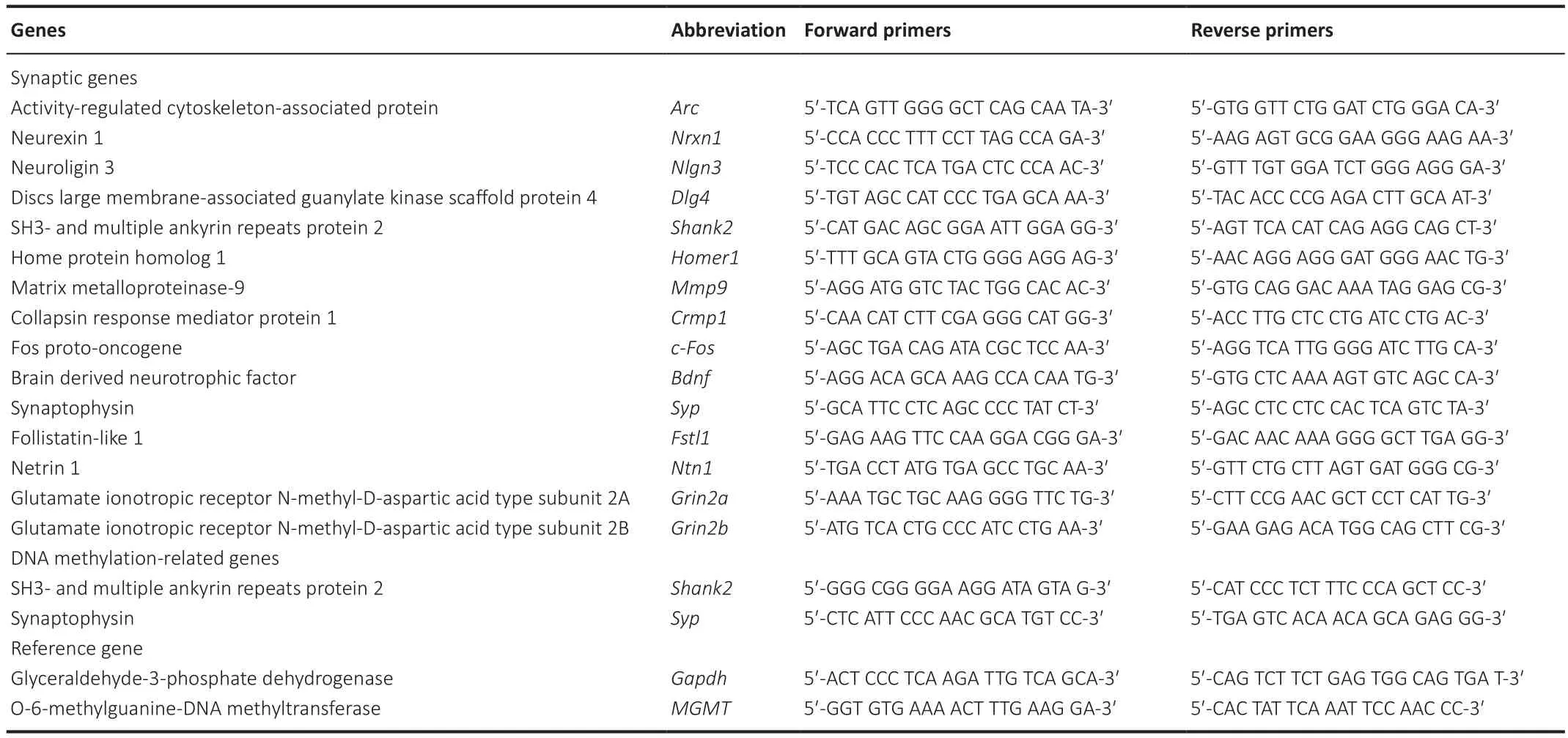
Table 1 |List of the different primers used in the study
Western blot analysis
The hippocampal lysate was prepared on ice.Protein samples were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto a nitrocellulose membrane.Each membrane was blocked with 5% non-fat milk and incubated using the corresponding primary antibody(anti-SHANK2 mouse monoclonal, Abcam, Cat# ab94575, RRID:AB_10675273;anti-SYP rabbit monoclonal, Abcam, Cat# ab32127, RRID: AB_2286949; anti-DNMT1 rabbit monoclonal, Abcam, Cat# ab188453, RRID: AB_2877711; anti-TET1 rabbit monoclonal, Abcam, Cat# ab191698, RRID: AB_2858250; anti-GAPDH rabbit polyclonal, Affinity, Pottstown, PA, USA; Cat# AF7021, RRID:AB_2839421) at a concentration of 1:1000.The membranes were thawed at 22°C for 45 minutes on the following day and the primary antibody was washed out.The membranes were incubated with the secondary antibody(goat anti-rabbit IgG (H+L) horseradish peroxidase, Affinity, Cat# S0001, RRID:AB_2839429) at 22°C for 2 hours.Protein bands were visualized by enhanced chemiluminescence (Cat#MA0186-1; Meilunbio, Dalian, China) and quantified with ImageJ software (v1.8.0; National Institutes of Health, Bethesda, MD,USA) (Schneider et al., 2012).
Measurement of 5mC and 5hmC
Genomic DNA was extracted from the hippocampus using a Quick-DNA Tissue kit (Cat# MA0106; Meilunbio).The level of 5mC and 5hmC was measured with a 5mC and 5hmC DNA enzyme-linked immunosorbent assay kit (Cat#d5325 and Cat# d5425; Zymo Research Corp., Orange County, CA, USA)according to the manufacturer’s instructions.Briefly, 100 ng of each DNA was added to a PCR tube.Then, 5mC or 5hmC coating buffer was added until the final volume reached 100 μL, and then the DNA was denatured at 98°C for 5 minutes.Each well was washed with 200 μL of 5mC or 5hmC enzyme-linked immunosorbent assay buffer and incubated at 37°C for blocking.Then, 100 μL of anti-5mC or anti-5hmC antibody was added and incubated at 37°C for 1 hour.HRP Developer (Cat# d5325 and Cat# d5425; Zymo Research Corp.) was added to each well and the absorbance was measured at 405 nm.The results were expressed in percent 5mC or 5hmC in a DNA sample calculated by a standard curve.
Measurement of gene-specific DNA methylation
CpG islands ofShank2
andSyp
were identified using Methprimer (www.urogene.org/methprimer/) (Li and Dahiya, 2002).The OneStep qMethyl-PCR kit (Cat# d5310; Zymo Research Corp.) was used to detect region-specific DNA methylation via amplification of 5mC in the CpG dinucleotide context.In brief, 20 ng of each DNA sample was incubated with either test or reference reaction mixtures at 37°C for 2 hours.DNA in the test reaction was digested with enzymes, whereas DNA in the reference was not.Real-time PCR was performed using the following parameters: initial denaturation at 95°C for 10 minutes; 40 cycles of PCR with a melting step at 95°C for 30 seconds,an annealing step at 55°C for 1 minute and an extension step at 72°C for 1 minute; and final extension at 72°C for 7 minutes.The methylation level was calculated using the equation 100 × 2, where ∆Ct is equal to the average Ct value from the test reaction minus the average Ct value from the reference reaction.The primer sequences are shown in Table 1.Statistical analysis
Data and statistical analyses complied with the recommendations for experimental design and analysis in pharmacology (Curtis et al., 2018).Data were analyzed by researchers who were blind to the origin of the data.Data were expressed as mean ± standard error of the mean (SEM).The behavioral tests, mRNA and protein expression, and DNA methylation level were analyzed using one-way analysis of variance with a Bonferroni correction for multiple comparisons.The Bonferroni correction was only applied if theF
-value achievedP
< 0.05.The mNSS were analyzed with a Kruskal-Wallis one-way analysis of variance on ranks followed by Dunn’s test for multiple comparisons.Correlations between DNMT1, 5mC level and DNA methylation at Shank2 and Syp were analyzed using Pearson correlation analysis in SPSS 13.0 software (SPSS Inc., Chicago, IL, USA).A heatmap analysis of gene expression was performed by R (R3.3.1 for Windows, Oakland, New Zealand)(R Core Team, 2021).Statistical analysis was performed using SPSS 13.0 software.The level of significance was set at aP
< 0.05.Results
5-aza ameliorates memory deficits in cerebral I/R injury rats
To evaluate the effects of 5-aza on learning and memory in tMCAO rats,behavioral tests (NOR, RI and Morris water maze) were performed on days 9-18.The mNSS were higher in the tMCAO + Sal, tMCAO + L_5-aza and tMCAO + H_5-aza groups than those in the Sham + Sal group (P
< 0.001 for each comparison) on day 1, indicating that cerebral I/R injury induced neurological deficits at 24 hours after reperfusion.5-aza treatment significantly (P
< 0.05) reduced the cerebral I/R-induced increase in mNSS in a dose-dependent manner on day 8 (Figure 2A).The tMCAO + Sal group had markedly reduced exploration time of B on day 10 (P
< 0.01; Figure 2B)and interaction time with R2 on day 13 (P
< 0.05; Figure 2C) compared with the Sham + Sal group.The tMCAO + Sal group had a longer escape latency than that of the Sham + Sal group on day 17 (P
< 0.05; Figure 2D), and fewer platform crossings (P
< 0.01; Figure 2E) and reduced time spent in Q2 (P
<0.05; Figure 2F) on day 18.Treatment with high dose of 5-aza significantly increased exploration time of B (P
< 0.01), interaction with R2 (P
< 0.05),platform crossings (P
< 0.01) and time spent in Q2 (P
< 0.05), and decreased escape latency (P
< 0.05) in tMCAO rats (Figure 2B-F).5-aza inhibits the hippocampal synaptic damage induced by cerebral I/R injury
To assess whether synaptic function was involved in learning and memory deficits, the expression of synaptic proteins, mRNA, and synaptic density were observed at 1 hour after the behavioral tests.The expression of SHANK2 and SYP was significantly downregulated (SHANK2:P
< 0.05, SYP:P
< 0.001;Figure 3A and B), and synaptic density was reduced remarkably (P
< 0.001;Figure 3C) in the Sham + Sal group compared with the tMCAO + Sal group.A heatmap analysis also showed that cerebral I/R injury markedly reduced the expression of 15 synapse-associated genes (Figure 3D).In general,5-aza dose-dependently inhibited cerebral I/R injury-induced decreases in synaptic density in the ipsilateral hippocampus (P
< 0.01), and the expression of synaptic proteins (SHANK2:P
< 0.05, SYP:P
< 0.001) and mRNA in the contralateral hippocampus (Figure 3).5-aza blocks the cerebral I/R injury-induced increase in genomic DNA methylation
To explore whether epigenetics was associated with the regulation of synaptic protein expression, genomic DNA methylation was measured.As shown in Figure 4, the tMCAO had significantly upregulated DNMT1 expression (P
<0.001) and 5mC level (P
< 0.05) compared with those in the Sham + Sal group.Furthermore, TET1 expression (P
< 0.01) and 5hmC level (P
< 0.05) in the tMCAO + Sal group were lower than those in the Sham + Sal group.Treatment with 5-aza dose-dependently decreased DNMT1 expression (P
< 0.001) and 5mC level (P
< 0.05 orP
< 0.01), but not TET1 and 5hmC, in the contralateral hippocampus.5-aza prevents the hypermethylation of synaptic genes in the hippocampus of cerebral I/R injury rats
To investigate whether DNA methylation was related to the protein expression changes, gene-specific DNA methylation in the contralateral hippocampus was assessed.As shown in Figure 5A and B,Shank2
andSyp
each had one CpG island (GC% content >50).The tMCAO + Sal group had markedly increased DNA methylation atShank2
(P
< 0.05) andSyp
(P
< 0.01) compared with that of the Sham + Sal group.Treatment with high dose of 5-aza reduced the tMCAO-induced increase in DNA methylation atShank2
andSyp
(P
< 0.05;Figure 5C).There was a strong correlation between DNMT1, 5mC level and DNA methylation atShank2
andSyp
(Figure 5D).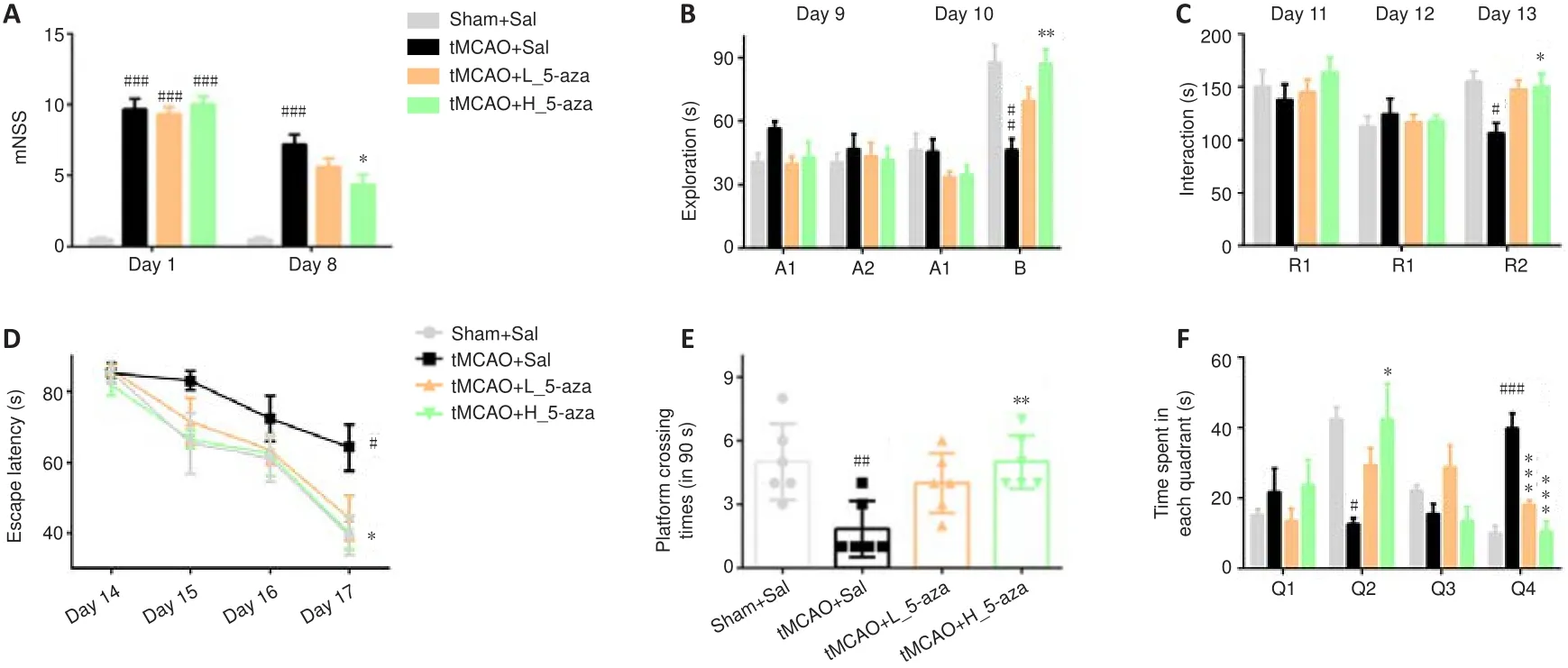
Figure 2|5-aza ameliorates the memory deficits in cerebral ischemia/reperfusion injury rats.
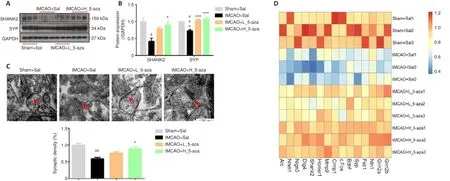
Figure 3| 5-aza inhibits the hippocampal synaptic damage induced by cerebral ischemia/reperfusion injury.
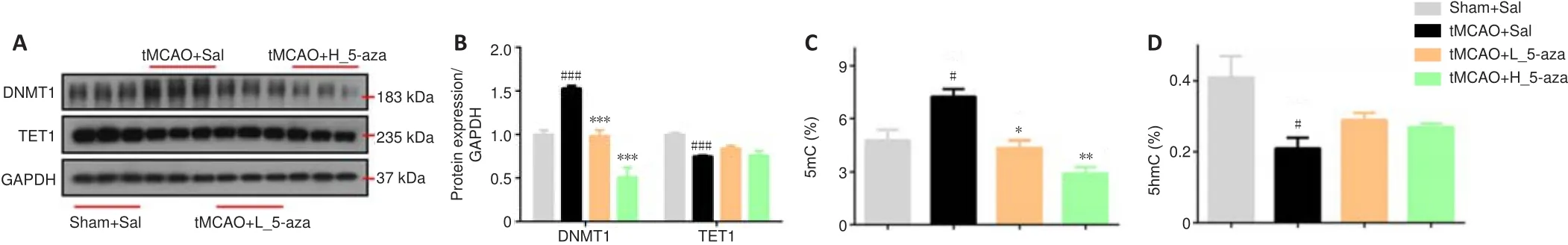
Figure 4| 5-aza blocks the cerebral ischemia/reperfusion injury-induced increases in genomic DNA methylation.

Figure 5|5-aza prevents the hypermethylation at the synaptic genes in cerebral ischemia/reperfusion injury rats.
Discussion
The clinical manifestations after thrombolysis mainly involve memory and cognitive impairment, or intelligence impairment syndrome with language,vision, emotion and personality disorders.Therefore, we aimed to use a tMCAO rat model to comprehensively evaluate the alterations of neurological function and memory induced by cerebral I/R injury.The mNSS method systematically assesses multiple physiological indexes in rats such as reflex,tactile function and balance (Han et al., 2018).Our study found that cerebral I/R injury reduced the mNSS of rats on days 1 and 8, suggesting that ischemia for 2 hours induced cerebral I/R injury.Subsequently, cognitive, social and spatial memory in cerebral I/R-injured rats were determined by the NOR,RI and Morris water maze tests, respectively.The results showed that the exploration time of a novel object B and unfamiliar rat R2 were markedly decreased in tMCAO rats.In addition, platform crossings and time spent in Q2 were markedly reduced in tMCAO rats.These results indicate that cerebral I/R injury induced neurological, cognitive, social and spatial memory impairments.Similar to the findings in our study, transient global ischemia for 5 or 10 minutes reduced learning and spatial memory assessed by the stepdown and Y maze tests (Liu et al., 2020; Guan et al., 2021).Furthermore, Sun et al.(2021) showed that a 90-minute tMCAO induced cognitive impairment as assessed by an avoidance task.Our work demonstrated for the first time that cerebral I/R injury also damages social memory in rats.
The hippocampus is the main component of the limbic system in mammalian brains.Synaptic proteins in the hippocampus play an important role in the formation of learning and memory.For example, neurexins (NRXN1)and neuroligins (NLGN3) are transmembrane synaptic proteins that form the neurexin/neuroligin trans-synaptic complex.In particular, NLGNs bind to SHANK through postsynaptic density protein-95, and home protein homolog 1, Arc, c-Fos and brain-derived neurotrophic factor are wellestablished regulators of neuronal excitability.Matrix metalloproteinase-9 also participates in synaptic plasticity by controlling the shape of dendritic spines.These are critical regulatory roles that follistatin-like 1, collapsin response mediator proteins and netrin-1 play during neuritogenesis and neurotransmitter secretion.In contrast, GluN2A and GluN2B play a central role in excitotoxic neuronal death caused by ischemic stroke.Our results showed that the expression of synaptic genes was decreased significantly in cerebral I/R-injured rats.Similarly, significant decreases in synaptic density and SHANK2 and SYP expression were also found in the hippocampus.Previous studies have found a significant loss of hippocampal neurons and NeuN-immunopositive cells and a reduction of postsynaptic density protein-95 and vesicular glutamate transporter 1 that were induced by cerebral I/R injury(Zaric et al., 2018; Guan et al., 2019; Lee et al., 2021).These results confirmed that synaptic dysfunction is an important factor that can lead to learning and memory impairments.Although the reduction of synaptic proteins in cerebral I/R-injured rats has been reported in previous studies, the molecular mechanism remains unclear.
DNA (de)methylation plays an important role in the development of the central nervous system (Poon et al., 2020).DNA methylation and demethylation are mainly catalyzed by DNMT1 and TET1, respectively.Our results showed that the expression of DNMT1 was significantly upregulated,whereas the expression of TET1 was significantly downregulated after cerebral I/R injury.In addition, we found that the trends of hippocampal 5mC and 5hmC levels were remarkably similar to those of DNMT1 and Tet1.These results suggest that I/R injury can lead to genomic DNA hypermethylation.Previous studies reported that focal cerebral ischemia upregulated global DNA methylation and reduced DNMT1 expression in neurons conferring neuroprotection (Tang and Zhuang, 2019).Ischemic stroke injury has been shown to induce a 3-fold increase in -CH3 group incorporation and a 4-fold decrease in TET1 level (Tang and Zhuang, 2019; Zhou et al., 2021).Nevertheless, genomic DNA hypermethylation does not indicate whether the expression of a particular synaptic gene will be downregulated.Therefore, we selectedSyp
andShank2
, which are biomarkers of presynaptic and postsynaptic membranes, respectively, to detect alterations in DNA methylation in gene sequences.Using CpG prediction software, we predicted 0 to +2000 bp after the transcription start site and found a CpG island (GC%content > 50%) atSyp
andShank2
.The results indicated that cerebral I/R injury resulted in DNA hypermethylation of both Syp and Shank2 and demonstrated that the downregulation of SYP and SHANK2 expression induced by cerebral I/R injury was indeed due to the hypermethylation ofSyp
andShank2
.Furthermore, we found that the levels of DNA methylation atSyp
andShank2
were strongly correlated with DNMT1 and 5mC, indicating that the DNA hypermethylation of Syp and Shank2 was mediated by DNMT1.It is believed that synaptic damage is irreversible, but recent studies have confirmed that synapses in the adult mammalian brain can be re-established(Augusto-Oliveira et al., 2019; Wang et al., 2020).These results provide new evidence for the treatment of ischemic stroke.5-aza, a nonspecific inhibitor of DNMTs, has been shown to induce DNA demethylation in rodents (Colwell et al., 2021; Jiang et al., 2021).In the present study, we found that 5-aza enhanced synaptic density and synaptic proteins expression in cerebral I/R-injured rats.Significant decreases in the levels of DNMT1 and 5mC were observed after 5-aza treatment, and resulted in DNA hypomethylation atSyp
andShank2
and upregulation of SYP and SHANK2.Finally, in terms of behavioral effects, we found that 5-aza reduced mNSS and escape latency,and elevated the exploration time of novel object B and unfamiliar rat R2.A previous study showed that 5-aza reduced cerebral infarct volume and ameliorated ischemic brain injury in mice (Endres et al., 2000).Another study showed that 5-aza treatment increased metallothionein expression, which contributed to neuroprotection after ischemic stroke (Park et al., 2013).The effect of 5-aza observed in cerebral I/R injury helps to verify the concept that synaptic damage can be reversed in the hippocampusin vivo
.Currently,most treatments for stroke mainly focus on neuroprotective therapy, such as inhibition of excitatory amino acid toxicity, calcium overload, inflammatory response or neuronal necrosis, apoptosis and autophagy.The corresponding drugs can only treat a single target.Increases in DNA methylation after stroke occur at the global level and at individual gene promoters, suggesting that inhibition of DNA methylation by 5-aza could be a multi-target treatment for stroke.There were three limitations in the current study: 1) only male rats were used to investigate the effect of 5-aza, 2) DNMT (DNMT3a, DNMT3b) and TET (TET2,TET3) were not measured in the experiment, and 3) there was no group that was administered 5-aza alone.
The present study investigated the epigenetic mechanism underlying cerebral I/R injury-induced impairments of spatial, cognitive and social memory.Here, we demonstrated that 5-aza treatment reduced overall genomic DNA methylation and methylation at specific synaptic genes in tMCAO rats.Furthermore, repeated 5-aza treatment blocked cerebral I/R injury-induced decreases in synaptic density and synaptic genes expression in the hippocampus.Finally, the damage to learning and memory induced by cerebral I/R injury was ameliorated by 5-aza treatment.Together, these findings suggest that 5-aza protects hippocampal synaptic function in cerebral I/R-injured rats by inhibiting global and gene-specific DNA methylation.
Acknowledgments:
We are grateful to Gui-Feng Zhao (Shengjing Hospital of China Medical University) for feeding the rats.
Author contributions:
Study conception and design, and manuscript drafting: GS; data collection: GS, JF; data analysis and interpretation: GS, LYJ;manuscript revision: XYF.All authors read and approved the final manuscript.
Conflicts of interest:
The authors declare no conflict of interest.
Availability of data and materials:
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Open access statement:
This is an open access journal, and articles are distributed under the terms of the Creative Commons AttributionNonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
Open peer reviewers:
Claudia Espinosa-Garcia, Emory University, USA;Jiangshan Deng, Shanghai Jiao Tong University Affiliated Sixth People’s Hospital,China.
Additional file:
Open peer review report 1.
- 中国神经再生研究(英文版)的其它文章
- Potential physiological and pathological roles for axonal ryanodine receptors
- Roles of constitutively secreted extracellular chaperones in neuronal cell repair and regeneration
- Melatonin, tunneling nanotubes, mesenchymal cells,and tissue regeneration
- MicroRNAs as potential biomarkers in temporal lobe epilepsy and mesial temporal lobe epilepsy
- Notice of Retraction
- Emerging roles of GPR109A in regulation of neuroinflammation in neurological diseases and pain

