An excimer ‘ON–OFF’switch based on telomeric G-quadruplex and rGO for trace thrombin detection
Long Zho, Frid Ahmed, Hi Xiong,∗
a Institute for Advanced Study, Shenzhen University, Shenzhen 518060, China
b College of Physics and Optoelectronic Engineering, Shenzhen University, Shenzhen 518060, China
ABSTRACT In the recent decade, GO has emerged as an amazing 2D nanomaterial for developing DNA-based biosensors due to its fluorescence quenching properties, whereas similar research based on rGO was reported rarely.Herein, a novel multi-pyrene functionalized G-rich DNA probe based on the screened rGO showed much higher fluorescence quenching efficiency and excimer emission than that of universal GO.Different from the universal thrombin detection of the G4-forming aptamer-TBA(GGTTGGTGTGGTTGG), the original telomeric sequence is used in this study.The excimer emission “ON-OFF” switch amplified the response of thrombin detection is as low as 50 units.Furthermore, for four pyrene moieties that are sited in a crowded steric circumstance, the melting temperature (Tm) values and molecular dynamics simulations showed a positive effect on duplex G-quadruplex or GDNA·cDNA stability, without disturbing its helix structure.
Keywords:G-quadruplex rGO Multi-pyrene excimer Biosensor Thrombin detection
Since the first demonstration of a fluorescent probe based on graphene oxide (GO) complexed with single-stranded DNA (ssDNA)to bind complementary oligonucleotide sequences in 2009 [1], ss-DNA/GO complexes, owing to their binding and quenching properties, have been widely applied to detect small molecules, metal ions, and biomolecules as well as for drug-delivery [2–5].Generally, negatively charged ssDNA is adsorbed on the GO surface by‘π-π’stacking between the DNA nucleobase moieties and GO carbon rings.Reduced graphene oxide (rGO) can be synthesized by the chemical reduction of GO, rGO possesses a high degree of aromaticity, which can facilitate a ‘π-π’stacking interface [6–9].As a result, the different oxygen contents of rGO have various quenching efficiencies for the fluorescence intensity of labeled ssDNA and aptamers, even if the examples are deficient [10,11].
Furthermore, modified pyrene fluorophores exhibiting an emission excimer at red-shifted absorption are extensively being developed as desirable tools in diagnostics and nanomedicine [12].Compared to various fluorophores, the excimer emission of pyrene derivatives is especially generated from aπ-stack dimer between a pyrene unit in an electronically excited state and another in the ground state [13–15].In 1996, the Kool group first reported the direct attachment of pyrene to deoxyribose, as pyrene is a chromophore replacing nucleobases [16].Owing to dimer stacking with a tremendous overlap in theirπ-orbitals, the earliest pyrenecontaining multichromophores exhibit efficient excitation energy transfer and electron transfer [17,18].To detect complementary DNA/RNA and the multi-labeling of RNA or LNA, more pyrene chromophores with non-nucleoside linkers have been incorporated into DNA in the last two decades [19–23].
G-quadruplexes are secondary structures of nucleic acids, which are composed of two or more stacked G-quartets with four guanine residues paired together through Hoogteen-like hydrogen bonds.Endogenous G-quadruplex models have been confirmed in the promoter regions of numerous oncogenes and the human telomeric sequence, and they have been regarded as potential targets for anticancer drugs at the telomeric level with the inhibition of telomerase activity overexpressed in tumor cells [24–26].Furthermore,the unique properties of G-quadruplexes have been investigated in some important reconstructions/reconfigurations and applications, including DNA origami, biosensing nanostructures, nanodevices, nanocarriers for disease therapeutics, and the detection of pathogens, including the causative agent of COVID-19 [27–32].
In 2012, Xionget al.employed tripropargylated nucleosides to construct three-armed (Y-shaped) dendronized DNAviathe‘stepwise and double click’approach [33].In 2016, Kim and coworkers attached pyrene-modified monomers as dangling residues of the G-quadruplex.Red-shifted fluorescence emission was observed upon the addition of K+[34].Herein, oligonucleotides containing single or multiple residues of tripropargylated 2′-deoxyuridine were prepared by the solid-phase synthesis of its phosphoramidites.Thereafter, the artificial ssDNA incorporating multi-pyrene moieties at the center or terminal sites could be obtainedviathe ‘click’chemistry.Furthermore, the feasibility of using pyrene-functionalized G-rich human telomeric ssDNA as a fluorescence probe based on synthetic rGO was investigated.To optimize the sensitivity of the G-rich probe, the oxygen content of rGO was varied by analyzing the fluorescence quenching efficiency and excimer emission for multi-pyrene modification.Based on these results, various thrombin concentrations were detected.
According to previous studies, the 5-[di(prop–2-ynl)amino]prop–1-ynyl derivative of 2′-deoxyuridine (2) was prepared from 5-iodo-2′-deoxyuridine (1) and 6-fold of tripropargylamine using the catalysts [Pd0(PPh3)4] and CuI.The Sonogashira cross-coupling reaction was performed to obtain nucleoside (2)in 71% yield.Further, the ‘double click’ reaction was carried out with nucleoside (2) containing two terminal triple bonds and 1-azidomethylpyrene (3) in the presence of CuSO4and sodium ascorbate (Scheme S1 in Supporting information).The corresponding phosphoramidite (5) was the final product, and all intermediates were characterized using1H and13C NMR spectroscopy (Scheme S2 in Supporting information).
Scheme 1.‘Click’reactions were performed on oligonucleotides and 1-azidomethylpyrene (3).
Modified oligonucleotides incorporating single or multiple residues of (2) were synthesized and constructed using standard solid-phase synthesis.The crude oligonucleotides were detritylated and purified by agarose gel electrophoresis or reversed-phase HPLC.To attach pyrene to the nucleobases on ssDNA, the ‘double click’reaction was also performed on the nucleoside moiety (2)and 1-azidomethylpyrene (3) (Scheme 1).Oligonucleotides incorporating artificial residues (4) were confirmed by LC-ESI-TOF mass spectrometry (Table S1 and Fig.S1 in Supporting information).
The melting temperature (Tm) values of monomolecular Gquadruplexes or bimolecular duplexes incorporating single or multiple residues of (4) were measured by ultraviolet (UV) thermal denaturation (Figs.S2-S4 in Supporting information).Tmmeasurements showed that modifications with one or two moieties of(4), bearing oligonucleotides with two or four pyrene units, exhibited a positive effect on duplex or G-quadruplex stability.Indeed, an increase inTmvalues for the unmodified oligonucleotides(GDNA-1 for G4 andGDNA-1·CDNA for duplex) is shown in Table 1.As a comparison, the attachment to oligonucleotides of single or multiple residues of (2) demonstrated a negative effect on duplex stability, with a decrease in theTmvalues for the unmodified oligonucleotides (Table S2 in Supporting information).To determine whetherGDNA formed G-quadruplexes in the PBS buffer(20 mmol/L, pH 7.0),GDNA-10 was chosen for CD analysis.The CD spectrum with a maximum band at approximately 264 nm and a minimum at approximately 241 nm indicated a quadruplex with all the strands oriented parallel to each other (Fig.S5 in Supporting information) [35–38].

Table 1 Tm values of G-quadruplexes and pyrene oligonucleotide duplexes with mono- or multi-pyrene residues.
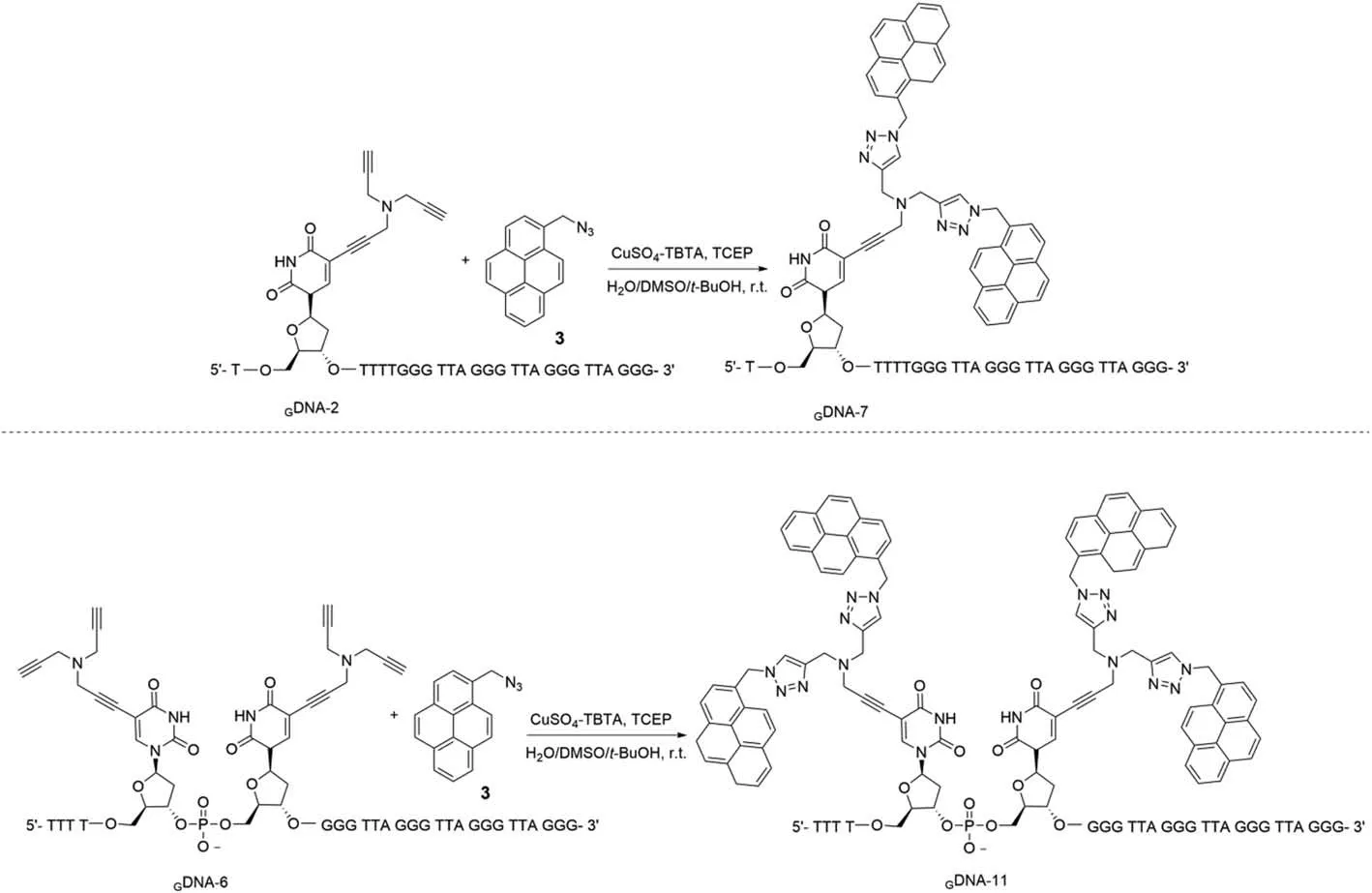
Molecular dynamics simulations at the MM+ force field (HyperChem 8.0 Professional; Hypercube Inc.) were conducted on the 27-mer duplexesGDNA-6·CDNA,GDNA-8·CDNA, andGDNA-11·CDNA containing one or two modification sites in the duplex (Fig.1).Fig.1A shows a duplex containing two dU residues displaced by the nucleoside (2) residue at the proximal position.Molecular modeling indicates that the proximal nucleoside (2) moieties bearing terminal alkynes seem to interfere with the DNA helix and arenot well adapted to the major groove.For the crowded steric situation, triazole rings of the modified derivative (4) at one or two proximal sites are not drawn while representing the stacking interactions of the nucleobase pairs (Figs.1B and C).Molecular modeling indicated that despite the presence of four pyrene moieties in the crowded steric circumstance, all residues were well accommodated in the major groove without disturbing the DNA helix.
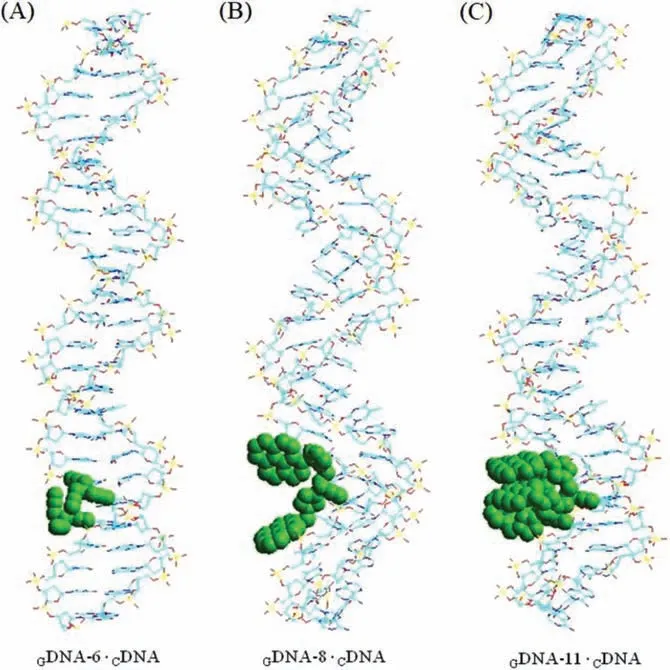
Fig.1.Molecular models of (A) duplex 5′-d(TTT T22 GGG TTA GGG TTA GGG TTA GGG) (GDNA-6) · 3′-d(AAA CCC AAT CCC AAT CCC AAT CCC) (CDNA), (B) duplex 5′-d(TTT TT4 GGG TTA GGG TTA GGG TTA GGG) (GDNA-8) · (CDNA) and (C) duplex 5′-d(TTT T44 GGG TTA GGG TTA GGG TTA GGG) (GDNA-11) · (CDNA).The molecular dynamics models were simulated by using energy minimized AMBER calculations.The green balls are diaplayed as the modification sites.
The oxygen contents of GO and rGO were measured by X-ray photoelectron spectroscopy (XPS).The GO samples showed characteristic peaks for carboxyl groups and C–O single bonds.After reduction by NaBH4(different reduction times), the oxygen content of rGO gradually decreased, as shown in Figs.S6 and S7 (Supporting information).rGO with different oxygen contents has different water solubilities.Indeed, rGO-5 and rGO-6 demonstrated poor dispersion in water.The size of GO and rGO on the mica flakes was confirmed by AFM.The thickness of GO and rGO is approximately 1.0±0.2 nm.Considering the overestimation and the oxygen-containing group, the obtained thickness of approximately 1.0 nm for GO or rGO reasonably indicates a single-layer (Fig.S7B).
Owing to the different aromatic structures and oxygen content, GO and rGO have different adsorption capacities for ssDNA.In 20 mmol/L PBS buffer (pH 7.0), ssDNA was adsorbed on the GO or rGO surface in the form of a G-quadruplex.To investigate the fluorescence quenching efficiency, various concentrations of GO and rGO (0–50 μg/mL) were treated withGDNA-10 (Fig.S8 in Supporting information).Upon interaction with GO or rGO, the fluorescence intensity ofGDNA-10 was substantially quenched.When 2 μg/mL GO or rGO was added to theGDNA-10 in the PBS buffer,fluorescence quenching efficiencies of GO, rGO-1, rGO-3, and rGO-4 were below 20%.With a gradual increase in the GO or rGO concentration, the fluorescence intensity was further reduced.The rGO samples with a less oxygen content, rGO-5 and rGO-6, could not disperse well in water and aggregated rapidly, and therefore, they could not be used to investigate the interaction withGDNA-10.
Upon the addition of 30 μg/mL rGO-2 in 1 μmol/L solution ofGDNA-10, the fluorescence intensity was quenched by 94% within 5 min, whereas the quenching of the fluorescence intensity ofGDNA-10 (1 μmol/L) was approximately 84% by the addition of GO or other rGOs (Fig.2A).Upon increasing the concentration of rGO-2 over 30 μg/mL, the fluorescence intensity ofGDNA-10 (1 μmol/L)was quenched by more than 95% (Fig.2B).When the concentration of rGO-2 was increased to 50 μg/mL, the fluorescence quenching efficiency ofGDNA-10 reached 100%.The other rGO or GO samples did not show complete quenching even at higher concentrations(50 μg/mL).For comparison, the quenching efficiency ofGDNA with different amounts of GO or rGO was also tested in 75 mmol/L Tris–HCl buffer at pH 7.5 (Figs.S9 and S10 in Supporting information).The optimized rGO-2 concentration of 30 μg/mL was selected for fluorescence quenching withGDNA in subsequent assays.
A series of fluorescent quenching assays was performed onGDNA with different pyrene-labeled nucleoside positions after adding rGO-2 (30 μg/mL) (Fig.2C).The fluorescence intensity ofGDNAs (GDNA-7,GDNA-8, andGDNA-11) incorporating nucleoside(4) on the exterior of a G-quadruplex core was stronger than that ofGDNAs containing the modified nucleosides inside the quadruplex region (GDNA-9 andGDNA-10).Usually, quenching effects are caused by guanine residues acting as the strongest quencher.When ssDNA forms a G-quadruplex structure in the PBS buffer,the pyrene moieties are rapidly involved in ‘π-πstacking’with the G-tetrads, and the fluorescence intensity ofGDNA with pyrenemodified nucleosides inside the G-quadruplex core decreases significantly.These findings are also in agreement with earlier observations of pyrene modifications in DNA duplexes [39–41].GDNA-11, incorporating two proximal derivatives, nucleoside 4 with four pyrene moieties in a crowded steric situation, triggered a redshifted fluorescence excimer emission (λem=478 nm).Upon the addition of 30 μg/mL rGO-2, the fluorescence quenching efficiency of allGDNAs was more than 90% (Fig.2D).The fluorescence quenching efficiency of differentGDNA adsorbed on rGO-2 was also tested in 75 mmol/L Tris–HCl buffer (pH 7.5, Fig.S9).In the Tris–HCl buffer, the fluorescence intensity ofGDNA-11 was 2-fold higher than that of GDNAs (fromGDNA-7 toGDNA-10).AsGDNAs assume a random-coil structure in the Tris–HCl buffer, the pyrene moieties cannot rapidly form the ‘π-πstack’with the G-tetrads.These results showed that rGO-2 had a good quenching efficiency forGDNA in the Tris–HCl buffer.
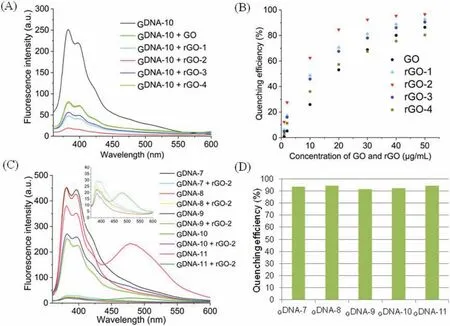
Fig.2.(A) Fluorescence spectra of GDNA-10 before and after adding GO or rGO (30 μg/mL) in PBS buffer (20 mmol/L, pH 7.0).(B) Fluorescence quenching efficiency (F0-F)/F0 of GDNA-10 in terms of different concentrations of GO and rGO. F0 and F are the fluorescence intensity before and after the addition of GO or rGO (λex=340 nm, GDNA-10 concentration=1 μmol/L).(C) Fluorescence spectra of GDNA before and after adding rGO-2 (30 μg/mL) in PBS buffer (20 mmol/L, pH 7.0).(D) The fluorescence quenching efficiency (F0-F)/F0 at 340 nm of λex and concentration with 1 μmol/L of GDNA.
Detecting the content of thrombin in tumor cells is of considerable significance for studying cancer cell proliferation and cancer diagnosis [42].In 2010, Liet al.reported a graphene FRET aptasensor for thrombin detection.FAM aptamers are suitable for commercial use [43].Using DNA intercalating dyes as FRET reporters,a quantum dot-aptamer beacon was successfully used for labelfree thrombin detection [43].Using a novel signal amplification strategy, Tanget al.developed a thrombin detection assay using a chiral supramolecular assembly with a physiological K+background in 2017 [44].In contrast to the universal thrombin detection of the G4-forming aptamer-TBA(GGTTGGTGTGGTTGG), an original telomeric sequence was used in this study.Herein, we developed a new pyrene-labeled G-quadruplex and rGO-based biosensing platform for thrombin detection.The fluorescence recovery of theGDNA-11 and rGO-2 complexes is depicted upon the addition of different amounts of thrombin (Figs.3A-C).Except for the fluorescence emissions at 381 nm and 395 nm, the excimer fluorescence at 478 nm also increased with the increasing concentration of thrombin.Compared with typical commercial dyes, these results can avoid the interference of many background signals for practical applications.The detection limit was 50 units of thrombin in a total volume of 1 mL.The addition of thrombin leads to fluorescence recovery owing to the formation of quadruplex-thrombin complexes, which have a weak affinity to rGO and push the dyes away from the rGO surface.
Fig.3.(A) Fluorescence spectra of the GDNA-11 and rGO-2 via different concentrations of thrombin in 20 mmol/L of PBS buffer (pH 7.0, λex=340 nm, GDNA-11 concentration=1 μmol/L, rGO-2 concentration=30 μg/L).(B) The amplified fluorescence spectra of the GDNA-11 and rGO-2 after adding thrombin (from 0 to 500 units/mL).(C) Fluorescence intensity spectra of the GDNA-11 and rGO-2 after adding thrombin (from 0 to 500 units/mL,λex=340 nm, GDNA-11 concentration=1 μmol/L,rGO-2 concentration=30 μg/L).(D) AFM images including height profiles of GDNA-11 and rGO-2 senor.(E) The amplified images of GDNA-11 and rGO-2 senor.Scale size: 5 μm×5 μm.
Atomic force microscopy (AFM) was performed to observe the structure of the rGO-GDNA sensor.The typical images (Fig.3D)displayed a few white areas on the rGO surface because of the presence ofGDNA, with a thickness of less than 10 nm, for the rGO-GDNA biosensor.TheGDNA sequences were uniformly distributed on the rGO surface without apparent selectivity, which is in good agreement with a previous report [45].Besides, the details ofGDNA-11 are magnified and shown in Fig.3E.
In this study, a series of mono- and multi-pyrene-labeled Gquadruplex sequences in different buffers were constructed.The ssDNA absorbed on GO or rGO can be effectively protected against enzymatic degradation and biological interferencein vivo.Furthermore, the adsorption of ssDNA on GO or rGO surfaces results in the exposition of ssDNA nucleic digestion and desorption of dsDNA,desorbed from the (r)GO by a complementary strand, to nuclease digestionin vitro.In the G-quadruplex form, the fluorescence intensity of multi-pyrene functional DNA probes decreases owing to the fluorescence quenching of stacked pyrene moieties, whileTmvalues show increased stability.Importantly, the G-quadruplex form with consecutive pyrene modifications (GDNA-11) exhibited strong excimer emission.Further, the fluorescence quenching of DNA based on GO and rGO with different oxygen contents was investigated in PBS or Tris–HCl buffers.Furthermore, the morphology of the rGO-based aptasensor assembled with the pyrene-labeledGDNA was determined by AFM.
In this study, negatively charged rGO demonstrated a better fluorescence quenching efficiency for DNA aptamer compared to GO.As an optimized result, the G-quadruplex form with consecutive pyrene modifications (GDNA-11) based on rGO-2 was selected as a biosensor to detect thrombin.Moreover, the application of such kind of optical or electrical G-quadruplex rGO-biosensor in cancer cell recognition will open the possibility of diagnostics and other diverse nano-medical applications.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgments
This work is supported by the Science and Technology Innovation Commission of Shenzhen, China (Nos.KQJSCX20180328095517269 and JCYJ20210324095607021), and Top Young Talent of the Pearl River Talent Recruitment Program,China.
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.cclet.2022.02.048.
 Chinese Chemical Letters2022年9期
Chinese Chemical Letters2022年9期
- Chinese Chemical Letters的其它文章
- A review on recent advances in hydrogen peroxide electrochemical sensors for applications in cell detection
- Rational design of nanocarriers for mitochondria-targeted drug delivery
- Emerging landscapes of nanosystems based on pre-metastatic microenvironment for cancer theranostics
- Radiotherapy assisted with biomaterials to trigger antitumor immunity
- Development of environment-insensitive and highly emissive BODIPYs via installation of N,N’-dialkylsubstituted amide at meso position
- Programmed polymersomes with spatio-temporal delivery of antigen and dual-adjuvants for efficient dendritic cells-based cancer immunotherapy
