Selected aptamer specially combing 5-8F cells based on automatic screening instrument
Zhukng Guo, Bijing Jin, Yil Fng, Yn Dng, Zhu Chn, Hui Chn, Song Li,Frnklin Wng-Ngn Chow, Polly H.M.Lung, Hnming Wng, Li Ci,Nongyu H,,∗
a State Key Laboratory of Bioelectronics, School of Biological Science and Medical Engineering, Southeast University, Nanjing 210096, China
b Hunan Key Laboratory of Biomedical Nanomaterials and Devices, Hunan University of Technology, Zhuzhou 412007, China
c Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, China
dGuangzhou Wondfo Biotech Co., Ltd., Guangzhou 510641, China
e Guangzhou Wondfo iCubate Biotech Co., Ltd., Guangzhou 510641, China
ABSTRACT Since the concept of aptamer emerged, many scientists have launched a rich field of research around it.However, few nucleic acids aptamer which use cell as target can be put into practical applications.We believe that a great deal of this lies in the complexity and irreproducibility of aptamer screening experiments themselves.The complexity is due to the cumbersome processes and the technical requirements for laboratory personnel, whereas irreproducibility arises from the fact that the starting point of such screens is nucleic acid libraries with random fragments, and that different libraries directly determine the differences or even the success or failure of screening results.The complexity and irreproducibility mentioned above, in turn, lead to the inability of this experiment to unfold on a large scale, which naturally cannot lead to excellent results for practical applications.In response to this problem, our group has developed an instrument for automated screening of tumor cell nucleic acid aptamers and characterized the properties of nucleic acid aptamers obtained using this instrument in a comprehensive manner.
Keywords:Aptamer Cell-SELEX Automatic instrument Tumor Efficient screening
Nasopharyngeal carcinoma (NPC) refers to the malignant tumors that occur in the top and lateral walls of the nasopharyngeal cavity, and the incidence is one of the highest malignant tumors of the ear, nose, and throat.In our country, NPC tends to occur in Guangxi and Guangdong most and has obvious regional distribution characteristics.
At present, the most widely used screening mean for NPC in clinic is serological detection of EBV (Epstein-Barr virus),i.e., detecting VCA (virus capsid antigen) IgA and EA (early intracellular antigen) IgA antibody titers of EBV with immunoenzymatic method.However, although VCA IgA antidetection has high sensitivity, it shows low accuracy, potentially leading to false-positive cases; For EA IgA, despite possessing higher detection accuracy, it has insufficient detection sensitivity and sometimes leads to undetectable cases.In addition, because of the special anatomical location of the nasopharynx (the surrounding important organs and its complex structures), surgery is not a very appropriate approach to treat NPC, and radiotherapy is the first-choice treatment for NPC.The 5-year survival rate of NPC patients without distant metastasis after comprehensive radiotherapy-based treatment reaches more than 80%, therefore, accurate early diagnosis is of great significance for the treatment of NPC [1–4].
Nanotechnology has found many biomedical applications, for example, drug delivery [5], nucleic acid extraction [6,7], biosensors [8] and tumor treatment [9–11].Nucleic acid aptamers are also nanomaterials, and they are functional oligonucleic acids that are able to identify specific target with high affinity, and are able to bind to the targets with a three-dimensional folded structure and, thereby, play the effect of recognition.Because of their similar recognition mechanism with antibodies, nucleic acid aptamers are often referred to as chemical antibodies [12–14] and has applications in many directions [15–18].Nucleic acid aptamers have unique advantages over antibodies, such as ease of chemical synthesis, little batch-to-batch difference, low immunogenicity, high environmental adaptability and better tissue penetration [19–21].
Since the advent of this technology, nucleic acid aptamers have been widely used for screening against different targets such as proteins [22–26], viruses [27–30] and cells [31–35].Among them,systematic evolution of ligands by exponential enrichment against cell (Cell-SELEX) appears as a new technology [36,37], it has attracted a lot of attentions and leads to many related studies[38–42].However, the practical application of this technology always has difficulties.The Cell-SELEX experiment screening cycle is long while the screening results are uncontrollable, a screening experiment over many months may not necessarily yield a nucleic acid aptamer with sufficient affinity and, in addition, this experiment is highly demanding for operator expertise, treatment of any step in the experiment will have significant impact on the final results [43–45].In order to find solutions to these problems, our group put forward the idea of an automated nucleic acid aptamer screening instrument and completed the scaffolding and verification of the instrument [46–50], this paper will start from NPC cells to demonstrate the feasibility of using this instrument for automated screening of nucleic acid aptamers targeting tumor cells.
Human NPC cells (5-8F cells) were used as target cells during this aptamer screen, and human normal nasopharyngeal epithelial cells (NP69 cells) were used as negative cells.
Human hepatocellular carcinomas cells (HepG2 cells), human lung cancer cells (A549 cells), human gastric cancer cells (HGC-27 cells), human cervical cancer cells (Hela cells), Michigan cancer foundation cells (MCF-7 cells) were used in subsequent validation experiments.
Cell culture conditions, reagents used, PCR amplification system settings and parameter settings are shown in Supporting information.
In this experiment, the automatic aptamer screening instrument built by our research group was performed for aptamer screening,reagents-station controls are shown in Fig.1.The machine screening steps are set as follows:
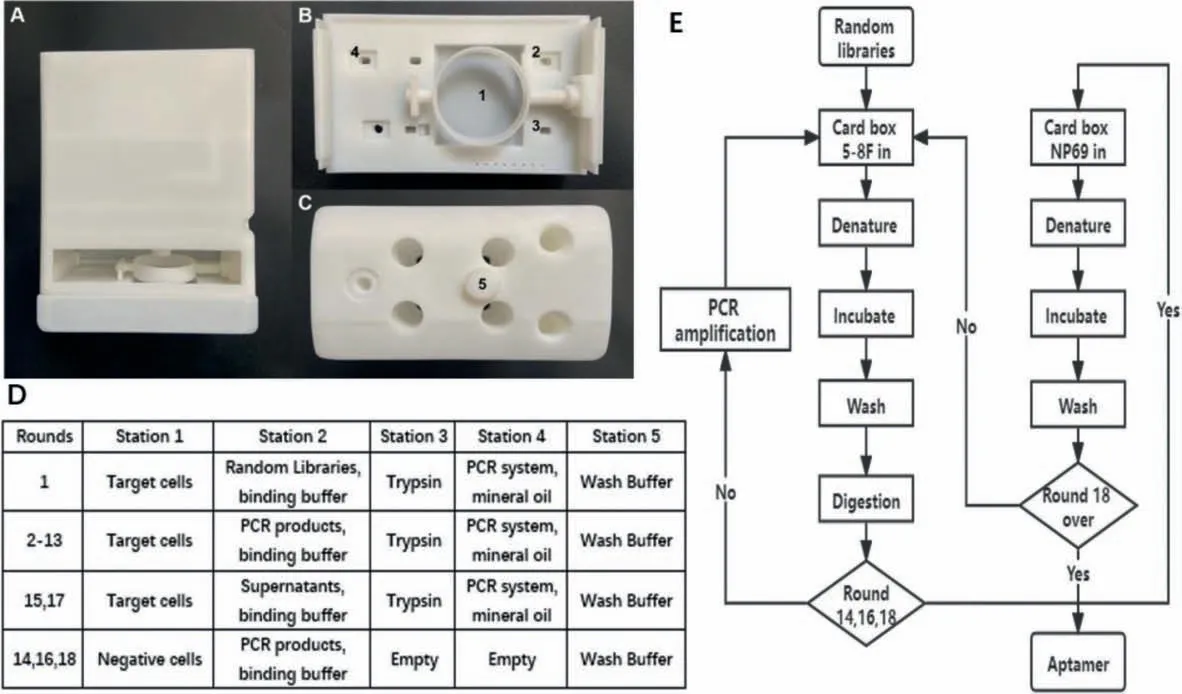
Fig.1.(A) Cartridge front view; (B) cartridge base top view and (C) cartridge lid top view.(D) Reagents-station controls.(E) Screening flow settings.
(1) Mix the random library with ultrapure water to reach the final concentration of 100 μmol/L.Add it into the prepared PCR tube, denature at 95 °C for 10 min, place it on ice for 10 min,mix it with the screening binding solution, and then add it to station 2.
(2) Take 5-8F cells that have reached logarithmic growth stage in a 35 mm petri dish and move it to station 1.Suck and discard the culture solution, use the screening wash buffer for cleaning,1 mL of cleaning solution was added each time, shake for 1 min,then discard the wash buffer.
(3) Absorb the mixture of random library and binding buffer in station 2, transfer it to station 1, shake the petri dish with a motor,and incubate the mixture with cells for 60 min.
(4) After incubation, add wash buffer for cleaning, use 1 mL cleaning solution each time, keep the petri dish shaking for cleaning for 1 min, then discard the cleaning solution.
(5) After cleaning, absorb the trypsin digestive solution from station 3, add it to station 1, absorb the digested liquid with a pipette gun, and transfer it to station 4 for PCR amplification.
(6) Hold station 4 at 4°C after finishing the PCR amplification,save the product and use it as the library for the next round of screening.
We dissolved 0.225 g glucose and 250 μL MgCl2(1 mol/L) in 50 mL of 1×PBS buffer to prepare washing buffer, added 50 mg of bovine serum albumin and 5 mg of tRNA into washing buffer to prepare binding buffer.In addition, to obtain the highest affinity fragment possible within a limited screening round, a pressure screening condition control was introduced as the number of rounds increased, other details of screening flow settings are provided in Fig.1.Among the screening process, rounds 14, 16 and 18 were negative screenings, and we directly used the supernatant of the previous round as screening libraries.Therefore, in these rounds, there is no need for PCR amplification.
After 18 rounds of automatic screening, the aptamer obtained from instrumental screening were sequenced, as judged by the results, the random fragment of its middle part was: CCACCGGTCCGAATCCGTAGTACCGCCCGAAGACAGCGGT.To analyze the spatial structure of the screening-obtained nucleic acid aptamer, we combined the fixed sequences at both ends with the measured random sequences for secondary structure prediction using the mFlod website and the result is shown in Fig.2.
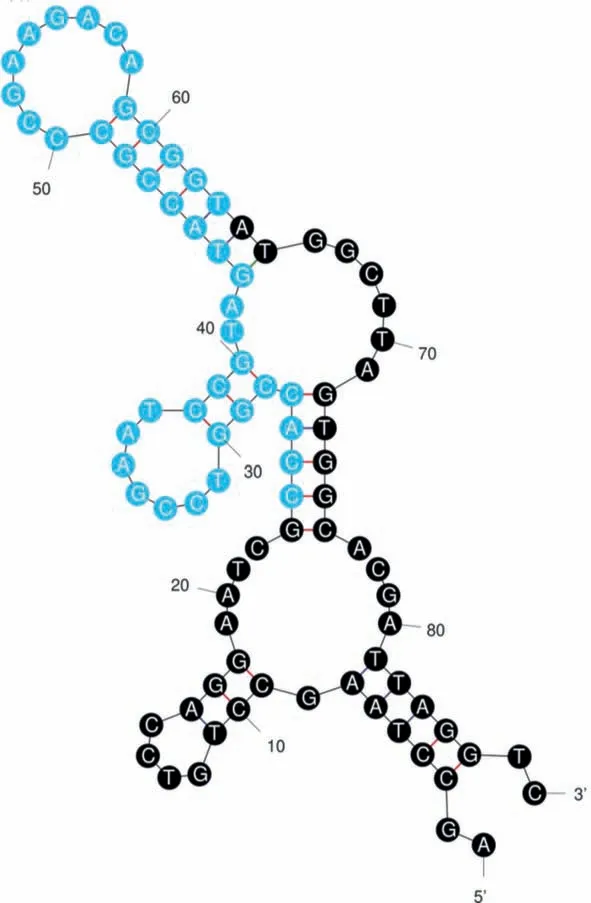
Fig.2.Secondary folding structure of our obtained nucleic acid aptamer.
With the free energyΔGas −26.89 kcal/mol, it can be seen that this strip of nucleic acid aptamer has some obvious stem loop structures, which facilitates its specific binding with cell surface proteins.
To verify the specificity of the resulting nucleic acid aptamer,we performed flow cytometry experiments and the results were in Fig.3, incubating the aptamer modified by 5′6-FAM fluorescence group with 5-8F cells and NP69 cells, respectively.The mean fluorescence intensity of each group of cells after incubation for 30 min was observed using flow cytometry to characterize the differences in the binding capacity of the nucleic acid aptamer to different cells.As can be seen in the lower panel, the average fluorescence intensity of 5-8F cells is significantly higher than that of NP69 cells.It was demonstrated that the aptamer obtained in this experiment was endowed with the ability to discriminate 5-8F cells from NP69 cells.
Further on, to demonstrate that this aptamer specifically recognizes 5-8F cells without binding other tumor cells, we conjugated fluorescent aptamer to 5-8F cells, HepG2 cells, A549 cells, hgc-27 cells, HeLa cells as well as MCF-7 cells.Flow cytometry was used to observe the mean fluorescence intensity of various cells after incubation for 30 min, and in turn, to characterize the difference of binding ability of nucleic acid aptamer to different cells.As can be seen from Fig.3, the fluorescence intensity exhibited by the 5-8F cell group after incubation with nucleic acid aptamer with modified fluorescence is significantly higher than those of all the other tumor cells, thus it can be stated that the aptamer obtained in this experimental screening are capable of specific recognition of 5-8F cells.
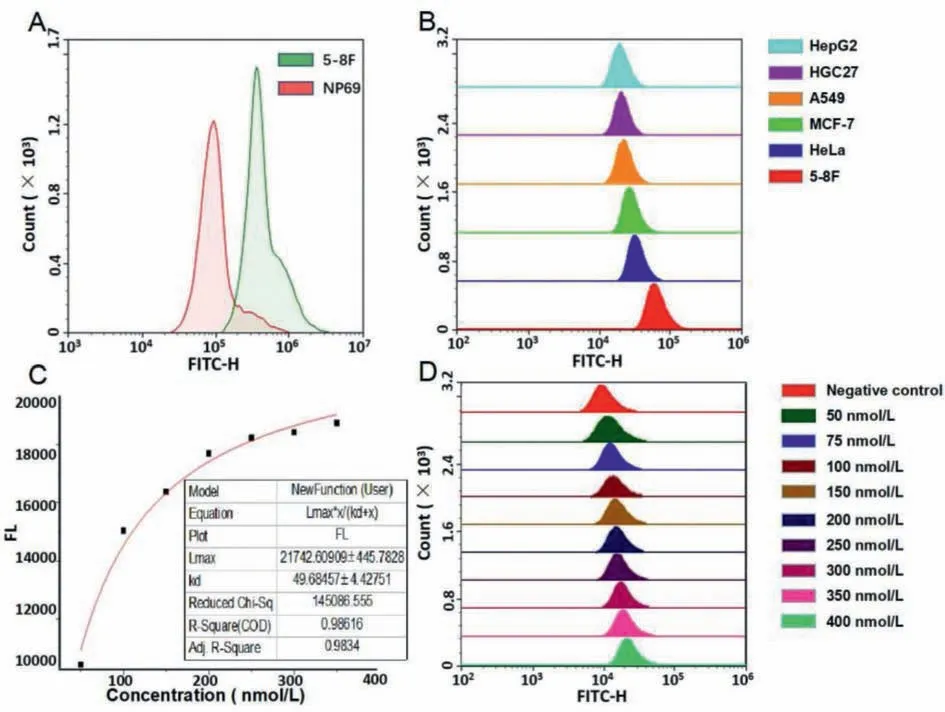
Fig.3.(A) Incubating the fluorescent aptamer with two types of cells, respectively, and measure the mean fluorescence intensity.(B) Incubating the fluorescent aptamer with six types of tumor cells, respectively, and measure the mean fluorescence intensity.(C) Curve fitting results.(D) Incubating the 5-8F cells with 10 different groups in various concentration of fluorescent aptamer, measure the mean fluorescence intensity.Color controls shows in right side legend.
To further make sure the verification of the binding ability of the obtained aptamer, we measured the dissociation constant Kdvalue of this strip of nucleic acid aptamer upon incubation with 5-8F cell.We set the concentration gradients of the fluorescencemodified nucleic acid aptamer at 50, 75, 100, 150, 200, 250, 300,350, 400 nmol/L in turn, and then incubated the aptamers of these different concentrations separately with the cell suspension with a same concentration of (because measuring the dissociation constant requires an excess of either of the two, here to make the cells excessive, control the number of cells to 3×105each group).Mean fluorescence intensity of cells in different aptamer concentration groups was measured using a flow cytometer, and the data are presented in Fig.3.
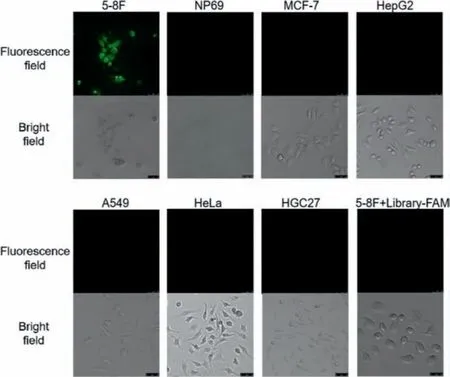
Fig.4.Incubating the various cells with fluorescent aptamer for 30 min, then clean them with wash buffer, photographed using a confocal microscope.
After obtaining the data, to calculate theKdvalue, curve fitting was performed in OriginPro 2017 software using equation:FL=Lmax×X/(Kd+X), FL stands for mean fluorescence intensity;Lmaxstands for saturated fluorescence intensity; X stands for the concentration of aptamer.As can be seen from the curve fitting results in the lower panel, this aptamer dissociation constantKdequals 49.68 ± 4.4, indicating its strong binding to the 5-8F cells.In addition, R-square and Adj.R-square can be seen as 0.99 and 0.98, meaning that the fitting result is credible in this experiment.
Finally, to get intuitive evidence for the specific recognition of our obtained nucleic acid aptamer, we used confocal microscopy to image 5-8F cells, NP69 cells, HepG2 cells, A549 cells, HGC-27 cells,HeLa cells and MCF-7 cells incubated with fluorescence-modified aptamers for observation.To confirm that 5-8F cells can only bind specifically to our obtained nucleic acid aptamer rather than to the initial libraries, we also designed an experimental group of 5-8F cells incubated with fluorescence-modified libraries.As can be seen from Fig.4, only 5-8F cells incubated with aptamer among all the experimental groups showed obvious fluorescence, demonstrating the specificity and sensitivity of our aptamer in recognition of 5-8F cells.
In this experiment, we successfully obtained a nucleic acid aptamer that can specifically recognize 5-8F cells.The more major implication, however, is to pass through a completely new way to complete Cell-SELEX.Although our instrument is currently at the experimental stage, it has been able to shorten the screening process stably to less than one month.In foreseeable future, we will further optimize this instrument.We will add cell storage module and automatic delivery module to this automatic screening instrument.In this way, we can store 20 cartridges loaded with cells and corresponding reagents in the instrument, and use conveyor belts to automatically move the cartridges to their corresponding stations before each round of screening.Through these two improvements, the screening process would again be massively shortened again.By 24-h continuous operation of the instrument on a daily basis, it will be possible to complete the entire screening process in two days (20 rounds, 2 h per round).Furthermore, with a small change on the fixing device in Fig.1B, once other types of targets are fixed in the cartridge, this instrument can also be used for other kinds of aptamer screening.This is of great significance to the whole aptamer screening system.A large number of aptamers will be obtained by this instrument, and quantitative changes will induce qualitative changes.Finally, we will be able to obtain more high affinity aptamers.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was financially supported by the National Key Research and Development Program of China (No.2018YFC1602905),National Natural Science Foundation of China (Nos.61871180,61527806).
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.cclet.2022.01.081.
 Chinese Chemical Letters2022年9期
Chinese Chemical Letters2022年9期
- Chinese Chemical Letters的其它文章
- A review on recent advances in hydrogen peroxide electrochemical sensors for applications in cell detection
- Rational design of nanocarriers for mitochondria-targeted drug delivery
- Emerging landscapes of nanosystems based on pre-metastatic microenvironment for cancer theranostics
- Radiotherapy assisted with biomaterials to trigger antitumor immunity
- Development of environment-insensitive and highly emissive BODIPYs via installation of N,N’-dialkylsubstituted amide at meso position
- Programmed polymersomes with spatio-temporal delivery of antigen and dual-adjuvants for efficient dendritic cells-based cancer immunotherapy
