A microfluidic surface-enhanced Raman scattering(SERS)sensor for microRNA in extracellular vesicles with nucleic acid-tyramine cascade amplification
Yue Zhao,Xiaoxing Fang,Min Bai,Jin Zhang,Huahang Yu,Feng Chen,Yongxi Zhao
Institute of Analytical Chemistry and Instrument for Life Science,The Key Laboratory of Biomedical Information Engineering of Ministry of Education,School of Life Science and Technology,Xi’an Jiaotong University,Xi’an 710049,China
1 These authors contributed equally to this work.
ABSTRACT Exosomal microRNA(miRNA)is an ideal candidate of noninvasive biomarker for the early diagnosis of cancer.Sensitive and accurate sensing of abnormal exosomal miRNA plays essential role for clinical promotion due to its close correlation with tumor proliferation and progression.Herein,a microfluidic surface-enhanced Raman scattering(SERS)sensor was proposed for an on-line detection of exosomal miRNA based on rolling circle amplification(RCA)and tyramine signal amplification(TSA)strategy.The microfluidic chip consists of a magnetic enrichment chamber,a serpentine fluidic mixer and a plasmonic SERS substrate functionalized with capture probes.The released miRNA activates the capture probe,triggers RCA reaction,and generates a large number of single-stranded DNA products to drive the catalysis of nanotags deposition via TSA,producing numerous“hot spots”to enhance the SERS signals.In merit of the microfluidics chip and nucleic acid-tyramine cascade amplification,the developed SERS sensor significantly improves the sensitivity for the exosomal miRNA assay,resulting in a limit of detection(LOD)as low as 1 pmol/L and can be successfully applied in the analysis of exosomes secreted from breast tumor cells,which demonstrates the potential utility in practical applications.
Keywords:Microfluidic chips Surface-enhanced Raman scattering(SERS)Exosome Tyramide signal amplification Nucleic acid amplification
Extracellular vesicles(EVs),a class of membrane-enclosed vesicles secreted from cells,have emerged as pivotal communication tools in cell to cell communications[1,2].Tumor-derived EVs,especially exosomes,carry abundant genetic information originated from their parental cells and offer the potential for early diagnosis and monitoring of tumors[3-5].Among these,abnormal exosomal microRNAs(miRNAs)are reported to be closely related to tumor proliferation,and become promising liquid biopsy biomarkers candidates for disease diagnosis[6,7].In the past decades,numerous well-established methods were developed to analyze exosomal miRNAs,such as Northern blotting and microarrays[8,9].However,due to the low abundance of miRNA in exosomes,the traditional methods are limited by their intrinsic modest sensitivity and large sample volumes.Reverse transcription polymerase chain reaction(RT-PCR)is the gold standard for quantification of miRNAs,which although provides an excellent sensitivity,greatly suffers from laborious manipulation and time-consuming procedures,such as exosomes lysis,RNA extraction,reverse transcription and thermal cycling amplification[10,11].
To date,considerable efforts have been devoted to pursuing a good sensitivity and rapid readout for miRNA sensing[12-15].Surface-Enhanced Raman Scattering(SERS)has garnered tremendous attention for the analysis of trace biomolecules due to its fast response and superior sensitivity[16,17].The most common mechanism to improve the sensitivity of SERS sensors is to create“hot spots”structures and result in a local electromagnetic field enhancement[18,19].SERS nanotags have always been combined with metal substrates to produce high-density“hot spots”and plasmonic enhancement[20].The sandwich structure,which is assembled by capture probe,target miRNA and SERS nanotags,exhibits good performance on the SERS detection of tumorrelated miRNA[21-23].However,this kind of sandwich complex is formed in hybridization manner and vulnerable to the degradation of miRNA,resulting a false negative result.Recently,nucleic acids amplification has been involved in the SERS detection of miRNA.In these sensors,miRNA serves as a trigger to the amplification reaction,and subsequently be released from the system with no degradation influence on the enhanced SERS signals[24,25].Owing to nucleic acids amplification,the recognition event of a single target molecule can be converted into a number of sequence vacancy for binding probes,obtaining an amplified signal output.For instance,rolling circle amplification(RCA)produces tandem periodic sequence units,which could capture lots of SERS nanotags and make contribution to the electromagnetic enhancement on the substrate[26,27].However,the numerous SERS nanotags are separated between repetitive sequences and have no proper gap between neighboring nanoparticles to produce“hot spots”.Tyramine signal amplification(TSA)is a powerful enzyme-mediated amplification method,which utilizes horseradish peroxidase(HRP)to catalyze the deposition of tyramine-reporter conjugates[28,29].In this way,the SERS nanotags can be enriched and aggregated,generating localized enhancement of signal with geometric scale[30,31].However,due to the small size and buoyant density of exosomes in complex biological sample,the convenient and accurate quantification of exosomal miRNA remains a challenge.Currently viable methods such as ultracentrifugation and ultrafiltration require tedious workflows with high expenditure,which may result in poor recovery,large sample consumption and potential contamination[32,33].Microfluidics approaches have been extensively exploited to address these limitations,and the on-chip enrichment of exosomes streamlined with molecular profiling has been successfully developed[34-37].Nevertheless,the rigorous lysis condition introduced from sample pre-treatment steps are incompatible with the downstream nucleic acids amplification[38,39].So it is imperative to develop a microfluidic platform for the analysis of exosomal miRNA.
Herein,we propose a microfluidic approach for SERS detection of exosomal miRNAviaRCA cascaded with TSA.In this work,we integrate the enrichment of exosomes,subsequent on-line lysis and miRNA detection into a microfluidic platform.Once the primer modified on the substrate could be activated by the released target miRNA,the biotin labeled probe hybridizes with RCA products and recruits streptavidin-HRP(SA-HRP)with precisely controlled density,which could catalyze the deposition of tyramine-labeled SERS nanotags in the presence of H2O2and achieve a cascade signal amplification.
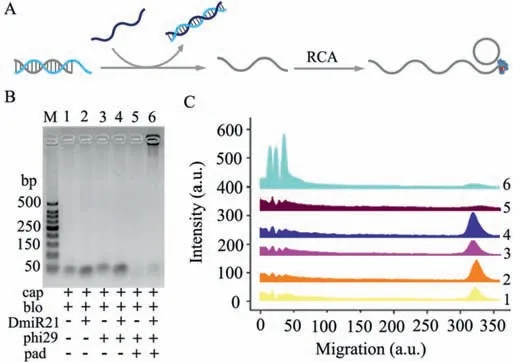
Fig.1.(A)Schematic illustration of the target-activated RCA reaction.(B)Gel electrophoresis results show the specificity of DmiR-21 powered signal amplification.(C)Analysis of gel electrophoresis characterization of(B).
As illustrated in Scheme 1,the microfluidic chip consists of three functional compartments:a magnetic enrichment zone,a serpentine fluidic mixer for exosomes lysis and RNA release,and an RNA capture zone for on-chip SERS analysis.Briefly,the magnetic beads pretreated with CD63 aptamer are introduced through inlet 2 accompanied with the exosomes containing sample,and the exosomes can be retained within the magnetic enrichment zone under the magnetic force.Then the lysis buffer is added through inlet 1 to incubate the captured exosomes,and miRNA could be fully released during lysis buffer filling the chamber and flows into the serpentine channel.The probes functionalized on the capture zone are blocked with complementary sequence in advance.The released exosomal miRNA possesses more complementary sequences with the block probe,which favors toehold-mediated strand displacement reaction to activate the masked RCA primer and produces a concatenated long single-stranded DNA.Multiple biotinlabeled probes hybridize to the tandem repeated sequences of RCA products and recruit numerous SA-HRP enzyme conjugates on the substrate of chip.In the presence of H2O2,HRP converts tyramine to a short-lived and reactive oxidized intermediate,which could bind covalently to adjacent HRP.Subsequently,TSA process is conducted to catalyze the deposition of tyramine-modified SERS nanotags and the cascade amplification are acquired from the sensor.
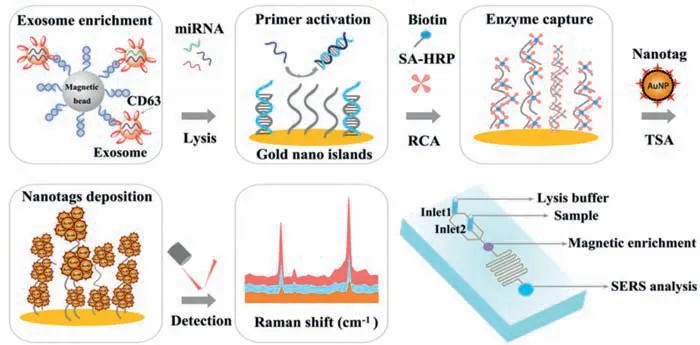
Scheme 1.Schematic diagram of the microfluidic SERS sensor for the detection of exosomal miRNA with nucleic acid-tyramine cascade amplification.
Given that the RCA is initiated by a strand displacement process,the design of capture(cap),block(blo)and padlock(pad)probes were firstly investigated(Fig.S1 in Supporting information).As shown in the Fig.1A,the cap probes are pre-sealed with the blo probes and cannot be used as primers for initiating RCA.Upon addition of DmiR-21(DNA sequence mimicking miR-21),its more complementary sequences with blo probe favors DNA branch migration.The released free cap probe serves as a primer for RCA,the products of which are analyzed by gel electrophoresis.As shown in Figs.1B and C,the RCA products triggered by DmiR-21 are specifically observed in lane 6 with an extremely low mobility.In contrast,no band appears in the absence of DmiR-21,pad probes or polymerase in lane 1 to lane 5,indicating no RCA reaction occurs in the control experiment.
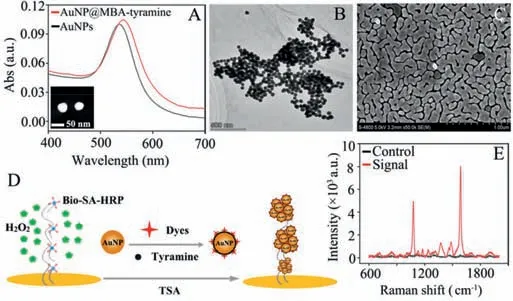
Fig.2.(A)UV-vis spectra of AuNPs(black)and AuNPs functionalized with 4-MBA and tyramine(red).Inset:TEM image of the AuNPs.(B)TEM image of SERS nanotags recruited on RCA products.(C)SEM image reveals the morphology of the nanostructured gold islands with abundant nanogaps.(D)Schematic illustration of the SERS sensing with nucleic acid-tyramine cascade amplification.(E)SERS responses with(red)and without(black)the DmiR-21.
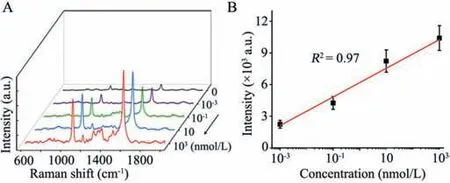
Fig.3.(A)SERS measurements of DmiR-21 with various concentrations.(B)The linear relationship between signal intensity of the band at 1583 cm-1 and DmiR-21 concentrations.The error bars are calculated from three trials.
The SERS nanotags were prepared according to the previous method with minor modifications.The gold nanoparticle(AuNP)was functionalized with 4-mercaptobenzoic acid(4-MBA)as reporter molecules and further conjugated with tyramine to be AuNP@MBA-tyramine.As shown in Fig.2A,the linking between AuNPs with 4-MBA and tyramine results in a redshift of absorbance maximum wavelength about 5 nm compared with naked AuNP.These dispersed SERS nanotags perform a quick response to the catalysis of HRP in the presence of H2O2,and form aggregation with high-density(Fig.2B).The glass slide was covered with gold nanoclusters as the plasmonic substrate according to seedmediated growth,which is characteristic of numerous nanogaps“hotspots”to provide electromagnetic field enhancement for SERS analysis(Fig.2C).The capture probe immobilized onto the abovesynthesized substrate responses to the target,initiates RCA and TSA process,yielding hundreds of tandem catalytic sites for the aggregation of AuNP@MBA-tyramine(Fig.2D).Thus,the cascadeamplified SERS signal can be acquired from the sensor(Fig.2E)with good reproducibility(Fig.S2 in Supporting information).
In the merit of the above properties,we evaluated the quantitative performance of the developed SERS sensor with different concentrations of DmiR-21.It can be observed in Fig.3A,the intensity of the SERS signal strengthens gradually with increasing concentration of the target,posing an obvious positive correlation.Based on the intensity of the characteristic peak at 1583 cm-1,a calibration curve was achieved within the logarithm of the target DmiR-21 concentration ranging from 1 pmol/L to 1 μmol/L(Fig.3B).The near regression equation was Y = 6187.7+1359.6lgX(R2= 0.97).As expected,DmiR-21 at the concentration of 1 pmol/L can be easily and well distinguished against the blank noise,promising the extraordinary potential for the analysis of exosomal miRNA.
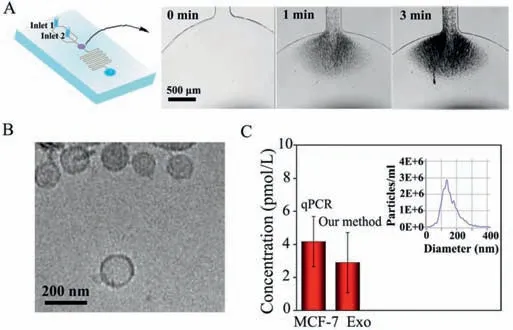
Fig.4.(A)Schematic diagram of the microfluidic device.The sequential snapshots showing the process of bead aggregates on the magnetic field during continuous flow in chip.(B)Cryo TEM images of MCF-7 exosomes.(C)The quantitative results of the proposed SERS sensor and qPCR for exosomal miR-21 from MCF-7(inset image shows the concentration and size distribution of the MCF-7 exosomes analyzed by NTA).Error bars represent standard deviations for measurements from three independent experiments.
The SERS sensor was further used for the detection of exosomal miRNAs in human breast carcinoma cell samples(MCF-7)for practical applications.First,the sample pre-mixed with CD63 aptamer-labeled magnetic beads was introduced through inlet 2 and retained within the magnetic enrichment chamber(Fig.4A).Cryo-transmission electron microscope(Cryo-TEM),atomic force microscopy(AFM)and dynamic light scattering(DLS)(Fig.S3 in Supporting information)were performed to study the morphologies and size of the exosomes.As shown in Fig.4B,the exosomes in the field of view are saucerlike with a mean diameter around 150 nm.Subsequently,we measured the concentration and average diameter of exosomes with nanoparticle tracking analysis(NTA)(Fig.4C,inset),which is determined as 5 × 107particles/mL,and the average diameter was 138 nm,which is consistent with the previous literature.Then the lysis buffer injected through inlet 1 and filled the magnetic enrichment chamber,resulting gradual disruption to the membrane of exosomes.The lysate and hybridization buffer were mixed and simultaneously flowed into the serpentine channel,so as to capture the released miRNA for SERS analysis.Followed by the nucleic acid-tyramine cascade amplification,the concentration of exosomal miR-21 secreted from MCF-7 cells can be obtained.Afterward,quantitative polymerase chain reaction(qPCR)was used in parallel to assess the accuracy of our proposed SERS sensor.As shown in Fig.4C,the result of SERS sensor was very consistent with qPCR(Fig.S4 in Supporting information),demonstrating that our proposed SERS sensor possesses high accuracy and practical potential.
To conclude,a microfluidic SERS sensor based on nucleic acidtyramine cascade amplification is constructed for the analysis of exosomal miRNA.With the merits of microfluidic chips,nucleic acids amplification,TSA and SERS analysis,the enrichment,lysis of exosomes and release,detection of miRNA can be streamlined on an integrated system,avoiding sample consumption and greatly improving the quantitative accuracy.With the introduction of RCA and TSA,target miRNA produces tandem periodic long single-stranded DNA,recruits numerous catalytic units and causes a large number of nanotags’deposition on the SERS substrate.Due to the above superiorities,SERS sensor displays ultrahigh sensitivity with the limit of detection(LOD)down to 1 pmol/L and can be applied on the on-chip detection of miR-21 in the exosomes derived from breast cancer cells.Moreover,the proposed microfluidic SERS sensor has the potential to detect multiple targets to provide a research foundation for exosomes analysis.Consequently,we envision this proposed approach to be a potential tool in a number of biomedical applications where the sensitive and accurate detection of low-abundance targets is required.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research was supported by the National Natural Science Foundation of China(Nos.31671013,22004096,21874105 and 21705124),the China Postdoctoral Science Foundation(Nos.2019M663658 and 2020T130096ZX),the Natural Science Basic Research Program of Shaanxi(Nos.2020JQ-020,2020JQ-021 and 2018JC-001),the Fundamental Research Funds for the Central Universities(No.xzy012020034)and“Young Talent Support Plan”of Xi’an Jiaotong University.
Supplementary materials
Supplementary material associated with this article can be found,in the online version,at doi:10.1016/j.cclet.2021.08.047.
 Chinese Chemical Letters2022年4期
Chinese Chemical Letters2022年4期
- Chinese Chemical Letters的其它文章
- Key progresses of MOE key laboratory of macromolecular synthesis and functionalization in 2020
- Small nanoparticles bring big prospect:The synthesis,modification,photoluminescence and sensing applications of carbon dots
- Cell membrane-coated nanoparticles for immunotherapy
- Diketopyrrolopyrrole-derived organic small molecular dyes for tumor phototheranostics
- Exosome based miRNA delivery strategy for disease treatment
- Recent advances in targeted stimuli-responsive nano-based drug delivery systems combating atherosclerosis
