Next-generation sequencing-based analysis of the effect of N6-methyldeoxyadenosine modification on DNA replication in human cells
Juan Wang,Yuwei Sheng,Ying Yang,Xiaoxia Dai,Changjun You
Molecular Science and Biomedicine Laboratory, State Key Laboratory of Chemo/Biosensing and Chemometrics,College of Chemistry and Chemical Engineering,Hunan Provincial Key Laboratory of Biomacromolecular Chemical Biology,Hunan University, Changsha 410082, China
ABSTRACT N6-methyldeoxyadenosine(6mdA)modification is considered as a new epigenetic mark that may play important roles in various biological processes.However,it remains unclear about the effect of 6mdA on DNA replication in human cells.Herein,we combined next-generation sequencing with shuttle vector technology to explore how 6mdA affects the efficiency and accuracy of DNA replication in human cells.Our results showed that 6mdA neither blocked DNA replication nor induced mutations in human cells.Moreover,we found that the depletion of translesion synthesis DNA polymerase(Pol) κ,Pol η,Pol ι or Pol ζ did not significantly change the biological consequences of 6mdA during replication in human cells.The negligible impact of 6mdA on DNA replication is consistent with its potential role in epigenetic gene expression.
Keywords:N6-methyldeoxyadenosine DNA replication Next-generation sequencing Shuttle vector technology Translesion synthesis DNA polymerase
Nucleic Acid methylation represents an important type of nucleic acid modifications that may play crucial roles in diverse biological processes in living organisms[1,2].Of them,5-methylcytosine and its oxidative derivates(e.g.,5-hydroxymethylcytosine)are well-established epigenetic marks dynamically occuring in DNA and RNA[3,4].In addition,N6-methyladenine modification in RNA has been acknowledged as a novel epigenetic modification involved in regulation of various biological activities such as tissue development,cellular DNA damage response,and stem cell fate determination[5,6].Methylation atN6position of deoxyadenosine(dA)has also been characterized as a dominant internal modification in bacteria since the discovery ofN6-methyldeoxyadenosine(6mdA)five decades ago(Fig.1)[7].With the recent development of highly sensitive detection techniques,6mdA has also been observed in several eukaryotes,such asChlamydomonas[8],C.elegans[9],Drosophila[10,11],and human[12–15].Increasing evidence has indicated the possible involvement of 6mdA methylation in development,stress response and diseases including neurodegenerative disorders and cancers[11,14–18].
Understanding the biological significance of 6mdA necessitates the investigation about how this modified nucleoside affects the flow of genetic information during DNA replication.It has been reported that 6mdA reduced the rate of DNA replication mediated by the Klenow fragment ofEscherichia coli(E.coli)DNA polymerase I and the large fragment ofBstDNA polymerasein vitro[19].6mdA was also found to partially inhibitin-vitroDNA replication by human DNA polymerase(Pol)ηand Polι,which play important roles in the translesion synthesis(TLS)of various types of DNA adducts[20,21].In addition,since the 6mdA methyltransferase METTL4 can modulate the mitochondrial DNA copy number,the presence of 6mdA has been hypothesized to affect mitochondrial DNA replication in mammals[14].To date,the biological effects of 6mdA during genomic DNA replication in mammalian cells remain elusive.
We have previously reported the use of shuttle vector technology,together with liquid chromatography-tandem mass spectrometry(LC-MS/MS)-based analysis,for successful evaluation of the biological consequences and repair of multiple types of DNA adducts during replication in mammalian cells[22,23].It has also been reported that the next-generation sequencing(NGS)methodology is capable of determining the replicative bypass and mutagenic properties of DNA adductsin vitroand inE.colicells,which are consistent with those obtained from LC-MS/MS and other traditional approaches but at a much higher throughput[24–26].Herein,we combined our traditional shuttle vector technology with NGS to investigate the biological consequences of 6mdA methylation during replication in human cells(Figs.2A and B).We also investigated the potential roles of the major TLS DNA polymerases,including Polκ,Polη,Polιand Polζ,in the replication bypass of 6mdA in human cells.
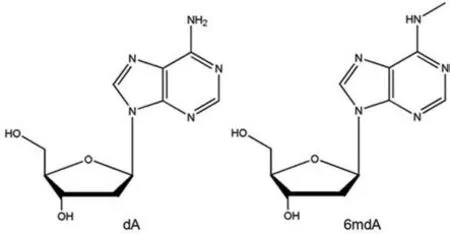
Fig.1.Chemical structures of deoxyadenosine(dA)and N6-methyldeoxyadenosine(6mdA).
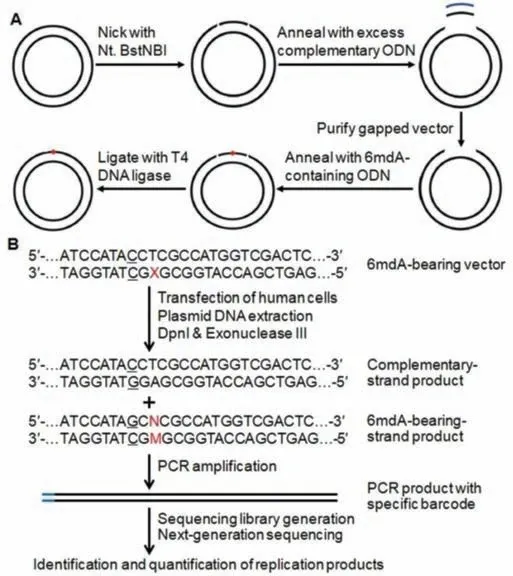
Fig.2.Experimental outline.(A)A schematic diagram illustrating the procedures for the construction of the plasmid harboring a site-specifically incorporated 6mdA.(B)NGS-based strategy for assessing the impact of the 6mdA on DNA replication.‘X’indicates 6mdA,and the C/C mismatch site is underlined.‘M’represents the nucleobase formed at the initial 6mdA site after replication,and ‘N’designates the paired nucleobase of ‘M’in the complementary strand.
Using the gapped vector-based method[25,26],we first constructed the double-stranded pTGFP-Hha10 shuttle vector housing a site-specifically incorporated 6mdA(Fig.2A).We also introduced C/C mismatched bases near the modified nucleoside site so as to differentiate the replication products of 6mdA-bearing strand from that of the unmodified complementary strand,which was used as an internal control as described elsewhere[23,27].In this vein,introduction of mismatched base pair has been accepted as an efficient strategy to distinguish the replication products of the modified nucleoside-bearing and unmodified control strands,which may be replicated at distinct efficiencies in mammalian cells[23,27].
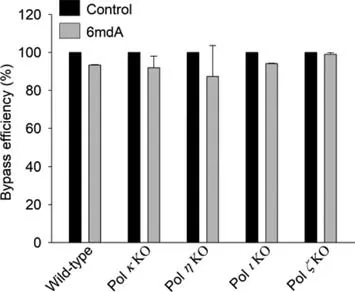
Fig.3.Bypass efficiencies of 6mdA in wild-type HEK293T cells and the relevant TLS polymerase-knockout(KO)cells.The data represent the means and standard deviations of results from three independent experiments.
The 6mdA-bearing pTGFP-Hha10 plasmid was transfected into HEK293T cells as well as the isogenic cells deficient in Polκ,Polη,Polιor Polζfor the replication study.After 24 h of incubation,the progenies of 6mdA-bearing plasmid were isolated from human cells,and residual unreplicated plasmid DNA was removed by a combined treatment of DpnI and exonuclease III[22,23].The DNA region of interest was PCR amplified and indexed with samplespecific barcodes,which designed the respective five host cell lines and the three biological replications.The 15 sets of tagged PCR products were combined and adenylated at the 3′-end,and then ligated to PE adapters.The ligation products were subsequently amplified with PE PCR primers,and the resulting PCR product was purified and subjected to NGS analysis(Fig.2B,Table S1 in Supporting information).
We obtained a total of ~2 million valid NGS reads for the replication products of 6mdA-bearing plasmid from triplicate replication experiments in five host cell lines(Tables S2 and S3 in Supporting information).The effect of 6mdA on replication efficiency can be determined by the ‘bypass efficiency’,which was calculated by dividing the total number of sequencing reads from 6mdAcarrying strand with that obtained from its unmodified complementary strand.The quantification data showed that the bypass efficiencies of 6mdA varied from ~87.3% to 99.0% in HEK293T cells as well as the isogenic cells deficient in Polκ,Polη,Polιor Polζ(Fig.3),which suggested that the presence of 6mdA did not substantially inhibit DNA replication in human cells.
Using the NGS-based assay,the effect of 6mdA on replication fidelity can be determined by the ‘base substitution frequency’,that is,the relative distribution of nucleobase(A,T,C or G)frequencies opposite the original modified site from 6mdA-carrying strand.It turned out that the mutation frequencies of A →C,A →G or A →T arising from the replication of the 6mdA-carrying strand in wild-type and TLS polymerase-deficient HEK293T cells were generally occurring at frequencies of 0.01%–0.03%(Fig.4A),which were similar to the background error rates(0.01%–0.07%)identified from the unmodified complementary strand(Fig.4B).These results indicated that 6mdA did not compromise the fidelity of DNA replication in human cells.
As discussed above,DNA 6mdA modification has recently attracted substantial research interests regarding their genomic distribution patterns and potential biological functions in eukaryotes[7,28].In this study,we have demonstrated that 6mdA did not inhibit DNA replication or induce mutations in human HEK293T cells using a next-generation sequencing-based cellular replication assay.In addition,replication bypass of 6mdA is also highly efficient and accurate in human cells that are deficient in any of the four TLS polymerases examined in our study.These results are inconsistent with the observation that 6mdA can impede considerablyin vitroDNA replication mediated by several distinct DNA polymerases including the Klenow fragment ofE.coliDNA polymerase I as well as human Polηand Polι[19–21].These differences could be,at least in part,attributed to the fact that thein vitroreplication studies were carried out with a single-type DNA polymerase,whereas the cellular DNA replication often involved the collaborative participation of different DNA polymerases and their auxiliary proteins[26,29].
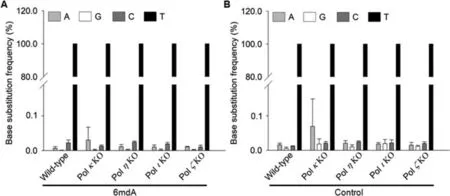
Fig.4.Mutation frequencies of 6mdA(A)and control(B)in wild-type HEK293T cells and the relevant TLS polymerase-knockout(KO)cells.The data represent the means and standard deviations of results from three independent experiments.
It has been suggested that DNA 6mdA modification may act as a new epigenetic mark that is associated with the regulation of gene expression[30].In this respect,the enrichment of 6mdA around the transcription start sites or transposon regions was found to be correlated with increased transcriptional activity[8,10,31].It was also reported that 6mdA can impact transcription by interfering with RNA polymerase elongation,changing chromatin structure,or modifying transcription factor binding affinities[32,33].Since epigenetic DNA modifications generally do not compromise the flow of genetic information during replication[34],our findings that 6mdA has a negligible effect on DNA replication in human cells is in keeping with its potential role in epigenetic regulation of gene expression.
Similar as 6mdA,it was reported that the replication of 5-methyldeoxycytidine(5mdC)and its oxidation products,including 5-hydroxymethyl-dC,5-formyl-dC and 5-carboxyl-dC,are highly efficient and accurate in human cells,consistent with the notion that these oxidized cytidine derivatives may serve as novel epigenetic marks.These epigenetic DNA modifications do not perturb Watson-Crick base pairing,which may account for their noncytotoxic effects in human cells[4,34].On the other hand,theN1 andN3 positions of dA are also among the major methylation sites in DNA,and the resulting DNA adducts 1mdA and 3mdA are highly cytotoxic because they can impair Watson-Crick interactions and strongly block DNA replication in human cells[35,36].
In conclusion,we have employed NGS,together with shuttle vector technology,to quantatively assess how 6mdA affects the fidelity and efficiency of DNA replication in human cells.Our results showed that 6mdA has a negligible impact on DNA replication in human cells,which is consistent with its potential role as an epigenetic mark.It is worth noting that we only evaluated the effect of 6mdA on DNA replication in a single sequence context,and it is possible that the biological consequences of 6mdA during replication may be infuenced by sequence contexts.In this vein,previous studies have shown that DNA sequence contexts could affect the repair and replicative bypass of several DNA lesions such as the ultraviolet light-induced 6-4 photoproduct[37,38].In addition,it has been recently reported that 6mdA can be modified by ALKBH1 protein to giveN6-hydroxymethyl-dA in mammalian genomic DNA[39],and it would be interesting to investigate how this new type of chemical modification modulates the process of DNA replication.in mammalian cells.A more complete understanding of the biological consequences of 6mdA also requires a further assessment of the modulation activities of cellular repair proteins such as 6mdA demethylases in mammalian cells.The shuttle vector-and NGS-based strategy described in this study should be applicable for quantitatively assessing the potential effects of sequence contexts and cellular repair activities on the replication of 6mdA as well as other DNA adducts such asN6-hydroxymethyl-dA and the oxidation products of 5mdC in the future.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Prof.Yinsheng Wang for kindly providing the initial pTGFPHha10 vector and TLS polymerase knockout cell lines used in the present study.This work was supported by the National Natural Science Foundation of China(Nos.21807030,21907028),the Science and Technology Innovation Program of Hunan Province(No.2019RS2020),Natural Science Foundation of Hunan Province(No.2020JJ5046),and the Fundamental Research Funds for the Central Universities(Nos.531118010061,531118010259).
Supplementary materials
Supplementary material associated with this article can be found,in the online version,at doi:10.1016/j.cclet.2021.08.066.
 Chinese Chemical Letters2022年4期
Chinese Chemical Letters2022年4期
- Chinese Chemical Letters的其它文章
- Key progresses of MOE key laboratory of macromolecular synthesis and functionalization in 2020
- Small nanoparticles bring big prospect:The synthesis,modification,photoluminescence and sensing applications of carbon dots
- Cell membrane-coated nanoparticles for immunotherapy
- Diketopyrrolopyrrole-derived organic small molecular dyes for tumor phototheranostics
- Exosome based miRNA delivery strategy for disease treatment
- Recent advances in targeted stimuli-responsive nano-based drug delivery systems combating atherosclerosis
