A photopatterned SERS substrate with a sandwich structure for multiplex detection
Yuan Xue,Duo Liu,Xuebin Wang,Yanxin Xiang,Shengjie Du,Kai Ye,Chunyan Bao,Linyong Zhu
Shanghai Key Laboratory of Functional Materials Chemistry,School of Chemistry &Molecular Engineering,East China University of Science &Technology,Shanghai 200237,China
Keywords:SERs detection Photopatterning Sandwich structure Multiplex detection Quantitative detection
ABSTRACT Substrate photopatterning has provided versatile applications in biomedical fields.Herein,an universal and efficient photoligation reaction has been used to prepare a patterned capture substrate for a sandwich SERS immunoassay.Photoirradiation induces mild and efficient immobilization of antibodies at the desired region of a gold surface,and the antibody-antigen interaction helps the substrate to capture the antigens in solution specifically.After exposing to SERS probes,i.e.,the gold nanoparticles labelled with both antibodies and intrinsically strong Raman reporters,multiple quantitative SERS determination of antigens can be achieved with high sensitivity and specificity.The limit of detection can be as low as 10−12 mol/L for four kinds of cancer biomarkers,which provides a promising method for the construction of highly sensitive and high-throughput SERS detection chip and the application of in vitro diagnosis.
Surface-enhanced Raman scattering (SERS) has become a promising analytical tool because of its strong fingerprint recognition ability,non-destructive performance and much higher sensitivity than fluorescence or UV–vis spectroscopy [1-4].Combining to immunoassays,SERS has been probed to have great potential in a high-throughput and trace detection of biomolecules,and gained a special interest in early detection and diagnostics of human diseases [5-7].At present,in addition to improving the sensitivity of SERS immunoassay,the research focus in this field also includes the following two aspects.One is whether the SERS labeled immunoprobe can form specific adsorption on the substrate.The other is how to realize simultaneous detection from single component to multiple components.
Numerous techniques have been introduced to design and fabricate various SERS-active sensors and substrates [8-14],among which the SERS-based sandwich immunoassay reported by Lipertet al.represents a simple and effective strategy [15,16].There are two key components in their platform: (1) the immune substrate,a flat gold substrate modified with capture antibodies,which can specifically extract and concentrate antigens from solution;and (2)the SERS immunoprobe,a kind of gold nanoparticles labeled with antibodies and Raman reporters,which can generate strong SERS signal when it is close to the gold substrate.The target antigens are therefore selectively captured on the substrate and subsequently recognized by the gold nanoparticlesviaantibody-antigen interaction to form a SERS-based sandwich immunoassay.By this strategy,they have realized the detection of immunoglobulin G (IgG)[17],free prostate specific antigen (f-PSA) [18],virus [19],bacteria[20],and potential pancreatic cancer markers [21]at low levels and even at single-binding events.With the great success in sensitivity and specificity,there is a growing demand for multiplex and highthroughput analysis of large quantities of analytes in a single sample,especially in the fields of clinical diagnosis,biochemical analysis and environmental monitoring [22].Although the characteristic SERS signal of analytes can be directly used to realize multiplex detection [23],it is not applicable in most cases due to the low scattering cross section and spectra overlap of most analytes.In an effort to address these limitations,SERS-based encoding protocols have been developed for multiplex detection [24-26],in which complex preparation and operation are often required.An alternative strategy to obtain multiplex detection is spatially controllable patterning of the substrate,in which antibodies are located in different areas [27-29].Compared to traditional microcontact printing,photopatterning represents a more facile and powerful tool for substrate modification because it allows remote control in a high spatiotemporal resolution [30,31].After sequentially photopatterning different antibodies in the defined areas of a SERS substrate,multiple SERS detection can be achieved.
Keeping this perspective in mind,we herein demonstrate a photopatterned SERS substrate with a sandwich structure for multiplex detection of biomolecules simultaneously (Scheme 1).A bifunctional molecule SNB was synthesized and modified on the gold substrate by Au-S bond (Au-SNB),which can pattern antibodies through a mild photoligation reaction [32,33].With the help of a mask,different antibodies can be patterned on the gold substrate by repeating the steps of irradiation (LED 365 nm),antibody soaking and washing up.Then,the patterned substrate can be used as the capture substrate for a sandwich SERS immunoassay,which involves two steps: capture of target biomarker from solution and indirect Raman detectionviathe recognition of gold nanoparticles labelled with both antibodies and intrinsically strong Raman reporters (the SERS probes).Thanks to the precise location of the antibodies on the substrate,each target biomarker in the analyte can be specifically captured at each region.Finally,the sensitive and concentration dependent SERS signal can be generated in the specific region after recognizing with the SERS probes,and only one Raman reporter is required to achieve multiple and noninterferences signal.
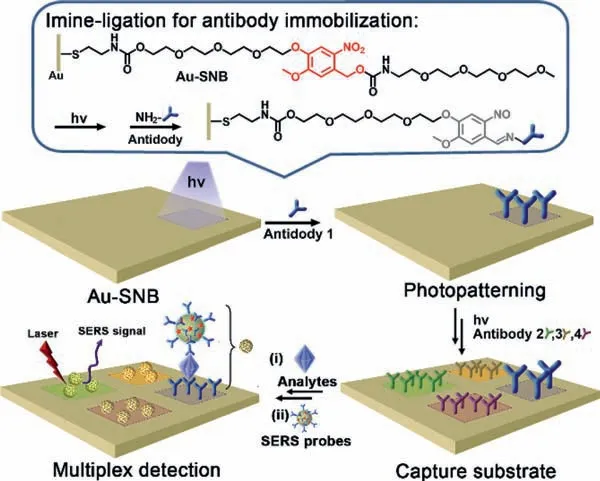
Scheme 1.Schematic representation of a sandwich SERS active substrate for multiplex detection prepared by sequential photopatterning of the capture substrate.
Considering the instability of mercapto groups,bifunctional molecule SNB is preserved in the form of disulfide precursor 2SNB(Figs.S1 and S2 in Supporting information).2SNB can be reduced to SNB in the presence of tris(2-carboxyethyl)phosphine(TCEP) (Fig.S3 in Supporting information).The introduction of two tetraethylene glycol groups to SNB is attributed to two aspects.One is to improve the water-solubility of SNB and the other is to have antifouling character to reduce the unspecific adsorption of biomolecules.Time resolved UV–vis absorption spectra confirm the efficient photolysis of SNB upon irradiation by a 365 nm lightemitting diode (LED) light source (10 mW/cm2) (Fig.S4 in Supporting information).Based on the photolysis mechanism of theonitrobenzyl group,the irradiation would induce the generation of 2-nitroso-5-benzaldehyde (Fig.S5 in Supporting information) and the active aldehyde can bind to amine-containing biomolecules by imine-ligation [32-34].As illustrated in Fig.1a,after the gold substrate (Au) was immersed in SNB solution,a self-assembled monolayer was formed on the substrate through Au-S bond (Au-SNB),which could be confirmed by the changed surface water contact angels (CAs) from 88.3° to 51.9°.Bovine serum albumin (BSA) was then selected as the model to evaluate the photoligation action between SNB and biomolecules.The reduced CA (38.1°) indicates the successful binding of BSA on the substrate (Au-BSA).X-ray photoelectron spectroscopy (XPS) was further carried out to characterize the substrates.As shown in Fig.1b (i),comparing to bare Au substrate,the signals of C 1s and O 1s for Au-SNB increase and a signal attributed to N 1s appears at 400 eV.For Au-BSA,these signals are greatly enhanced accompanied by the weakening of Au signals.Fig.1b (ii-v) shows are fine spectrograms of C 1s and S 2p of Au-SNB and Au-BSA by Gussian curve fitting,in which the C 1s spectrum can be deconvoluted into three subpeaks located at∼289.0,286.2,and 284.8 eV,which are,respectively,attributed to O-(C=O)-N,C-O and C-C bonds.After BSA binding,the contribution of C-C bond increases,and the main composition of S 2p changes from Au-S (162.0 eV) to S-S (163.5 and 162.3 eV) bond.All these results indicate that SNB and BSA are successfully deposited on the gold substrate as expected.
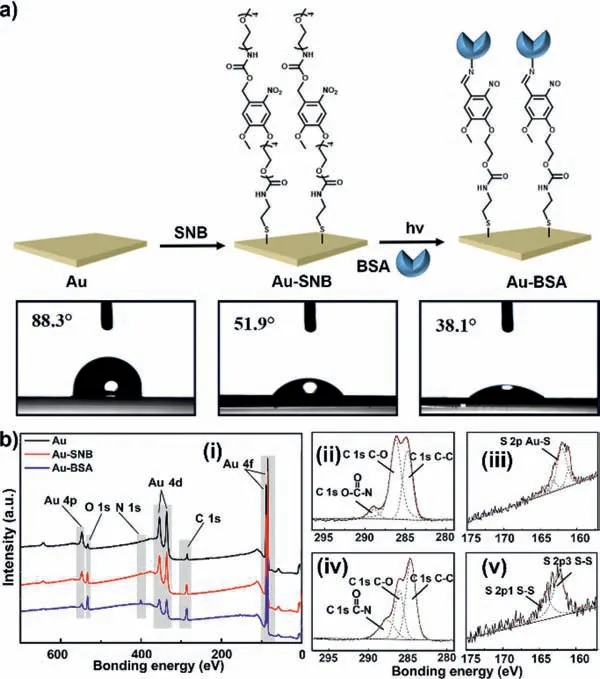
Fig.1.Preparation and characterization of capture substrates.(a) The preparation process of capture substrates and the corresponding CAs.Au: Au substrate,Au-SNB:SNB modified substrate,and Au-BSA: BSA modified substrate.(b) XPS survey spectra for Au,Au-SNB and Au-BSA (i): XPS high-resolution spectrum for C 1s (ii) and S 2p(iii) of Au-SNB,C 1s (iv) and S 2p (v) of Au-BSA,respectively.
Fig.2a shows the design and synthesis of SERS probes,which were prepared based on the optimized conditions as reported previously [35].Also shown are their transmission electron microscopy (TEM) structures (Fig.2b),UV-vis absorption and hydrodynamic size data by dynamic light scattering (DLS) (Fig.2c).The original gold nanoparticles (AuNPs) were prepared by sodium citrate aqueous solution system and have uniform size distribution with an average size of 38 nm (a maximum absorption at 532 nm).After sequentially labelled with Raman reporters (4-mercaptobenzoic acid,MBA) and linkers (NHS-PEG-SH,MW=2.0 kDa),AuNPs were stabilized with a layer of PEG molecules (mPEGSH,MW=3.0 kDa) with increased hydrodynamic diameter at 57 nm (AuNPs-PEG).Then,proteins or antibodies were chemically attached onto their surface by the action ofN-hydroxyl succinimide(NHS) to form SERS probes.Here,BSA was selected as the model,and the hydrodynamic diameter increases to 75 nm (Table S1 in Supporting information),accompanying with the broadening of the UV absorption peak of the SERS probes.In addition,FTIR−ATR spectra were used to characterize the nanoparticles (Fig.S6 in Supporting information).The appeared peaks at 1105 and 2885 cm−1,assigned to the C–O–C and CH2stretching vibration of PEG,respectively,confirm PEGylation of AuNPs,and the peak at 1659 cm−1assigned to the C=O stretching vibration of peptide bonds confirms BSA binding on the SERS probes.
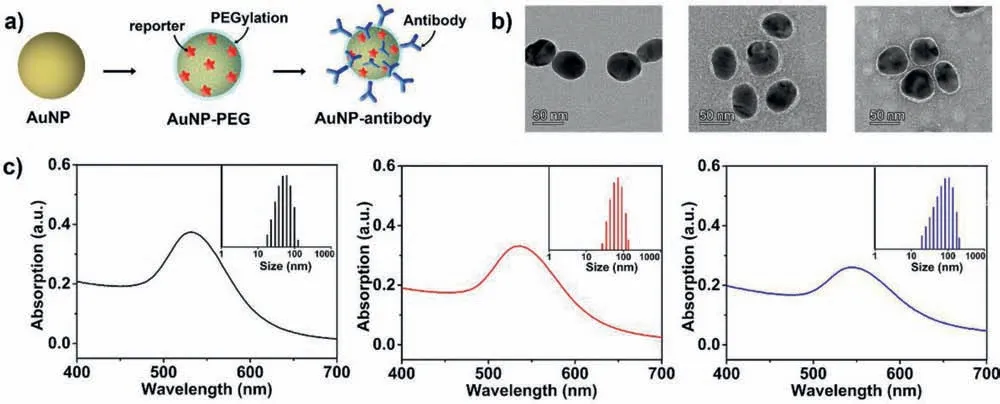
Fig 2.Preparation of SERS probes for the sandwich SERS immunoassay.(a) Schematic representation for the structures of an original gold colloids (AuNPs),a particle encoded with a Raman reporter and stabilized with a layer of mPEG-SH (AuNPs-PEG),and a particle modified with antibodies (SERS probe).(b) TEM images for the nanoparticles,which were negatively stained by phosphotungstic acid.(c) Optical absorption and DLS size data obtained from the particles as shown in (a).BSA was selected as the antibody model.

Fig 3.The sandwich SERS immunoassay.(a) Scanning electron microscope (SEM) of the capture substrate after PSA capture and SERS probes recognition,right is the enlarged image for the patterned area.(b) Comparison of SERS spectra for the different substrates.(c) Raman mapping image of anti-PSA patterned sandwich SERS substrate.The PSA concentration used in (a-c) is 0.5 μg/mL.(d) SERS spectra of sandwich SERS immunoassay of PSA at various concentrations.(e) Standard curve for PSA detection during 0.5-50.0 ng/mL with corresponding peak intensities at 1075 cm−1.
Firstly,to investigate the advantage of photopatterning,a representative antibody of prostate specific antigen (anti-PSA) was selected to be patterned on the capture substrate (Au-PSA) and labelled on the AuNPs for preparation of SERS probes.As expected,the sandwich based immunoresponse is confined to the desired areas (Fig.3a),that is,the SERS probes are evenly distributed in the irradiated area (a 1 mm × 1 mm square) of Au-PSA.It indicates that we can confine the SERS detection in patterned areas,which provides the possibility to realize simultaneous and multiplex SERS detection on the same substrate.Fig.3b shows the sandwich SERS spectrum of PSA (5 μL,0.5 μg/mL) in the patterned area and two distinct Raman bands,one at 1075 cm−1and the other at 1585 cm−1assigned to MBA ring-breathing vibration modes,are observed under 633 nm excitation.However,there is almost no detectable SERS signal in the same spectral region for the substrates without PSA or SERS probes.We also preformed Raman mapping to visualize the photopatterned sandwich test system (10 μm × 10 μm).As illustrated in Fig.3c,signals came from MBA at 1075 cm−1are localized in the patterned area with high fidelity,which further confirms the success and effectiveness of our photopatterning methodology.
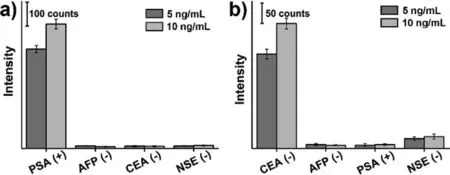
Fig 4.SERS response to specific (a) PSA and (b) CEA and non-specific antigens.
Then,the sensitivity of the singlet SERS sensing system was evaluated,in which the capture substrate and SERS probes were prepared with corresponding antibodies.Fig.3d shows the SERS spectra recorded for various concentrations of PSA and indicates that the Raman signal increases with the increase of concentrations.As illustrated in Fig.3e,a linear standard curve based on 1075 cm−1frequency of MBA was established by plotting SERS signal intensity against corresponding concentrations of PSA in the range of 0.5-50.0 ng/mL,and the limit of detection (LOD) of PSA was 0.19 ng/mL (6.7 pmol/L).It is significantly below the clinical cut-off value for the diagnosis of prostate cancer,since the normal value of PSA is usually less than 4 ng/mL in healthy people [36].In addition,sandwich SERS immunoassays of alpha fetoprotein (AFP),carcinoembryonic antigen (CEA) and neuron specific enolase (NSE) were also performed (Fig.S7 in Supporting information).All of them show obvious MBA Raman signals,and the signal intensity gradually increases with the increase of analyte concentrations.The LODs were 0.60 ng/mL (8.8 pmol/L),0.13 ng/mL (1.8 pmol/L),and 0.26 ng/mL (5.5 pmol/L) with a detection range of 1.0–100.0,0.5–50.0 and 1.0–100.0 ng/mL for AFP,CEA,and NSE,respectively,enabling SERS detection of biomarkers in human blood serum [37,38].To verify the specificity of our immunoassay,we have carried out cross reaction SERS detection.As shown in Fig.4,PSA and CEA can cause significant increase in SERS signal for anti-PSA (Fig.4a) and anti-CEA (Fig.4b) patterned substrates,respectively,while other non-specific antigens have only a weak response signal (≤8%).All these results confirm the sensitivity and specificity of our photopatterned sandwich SERS detection.
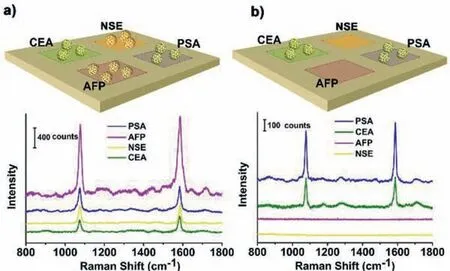
Fig 5.SERS multiplex immunoassay based on a capture substrate photopatterned with anti-PSA,anti-AFP,anti-CEA and anti-NSE in different areas (10 μm × 10 μm squares).(a) SERS spectra from different areas by adding of a mixture of four antigens at 5.0 ng/mL each.(b) SERS spectra detection of an analyte with two antigens(PSA and CEA) at 5.0 ng/mL.
In a final assessment,the multiplex SERS detection was explored by adding a combination of four kinds of antigens (PSA,AFP,CEA and NSA at 5.0 ng/mL each).A capture substrate photopatterned with their antibodies (anti-PSA,anti-AFP,anti-CEA and anti-NSE) in different areas (1 mm × 1 mm squares) and a mixture of SERS probes labelled with their antibodies were used for the sandwich multiplex SERS detection.As shown in Fig.5a,MBA Raman signals with different intensities are detected in the four patterned areas,and the intensities match the SERS signals corresponding to antigens at 5.0 ng/mL in the standard curves (Fig.3e,Figs.S7d-f in Supporting information).Meanwhile,an analyte containing PSA and CEA (5.0 ng/mL) was detected by using above SERS immunoassay.Fig.5b shows that SERS signals with comparable intensities appear in the areas patterned with anti-PSA and anti-CEA.However,no detectable signal is obtained in other two areas patterned with anti-AFP and anti-NSA.These results suggest that our SERS immunoassay has high specificity for different antigens and can achieve multiplex SERS detection without interference.Based on the fine spatial and temporal controllability of light,it can be imagined that our photoligation reaction can be used to construct SERS active microarray chips.
In conclusion,we have developed a novel sandwich SERS immunoassay substrate by using a photopatterned capture substrate.Combining with the SERS probes labelled with MBA Raman reporters and antibodies,the assay brings about the simultaneous,multiple,and quantitative determination of antigens with a limit of detection less than 1.0 ng/mL.The high sensitivity,specificity and spatiotemporal controllability provide a promising route for developing innovative early disease diagnosis and high-throughput biomarker screening assays.
Declaration of competing interest
There are no conflicts to declare.
Acknowledgments
This work is supported by Shanghai Science and Technology Commission (No.21ZR1415500) and the National Nature Science Foundation of China (NSFC,Nos.21907029,22171085).
Supplementary materials
Supplementary material associated with this article can be found,in the online version,at doi:10.1016/j.cclet.2021.09.016.
 Chinese Chemical Letters2022年3期
Chinese Chemical Letters2022年3期
- Chinese Chemical Letters的其它文章
- Direct catalytic nitrogen oxide removal using thermal,electrical or solar energy
- Construction and applications of DNA-based nanomaterials in cancer therapy
- Recent research progress of bimetallic phosphides-based nanomaterials as cocatalyst for photocatalytic hydrogen evolution
- Nanostructured materials with localized surface plasmon resonance for photocatalysis
- Recent progress of Pd/zeolite as passive NOx adsorber: Adsorption chemistry,structure-performance relationships,challenges and prospects
- Microfluidic methods for cell separation and subsequent analysis
