A modular single-cell pipette microfluidic chip coupling to ETAAS and ICP-MS for single cell analysis
Xing Wei,Meng Yang,Ze Jiang,Jinhui Liu,Xuan Zhang,Mingli Chen,Jianhua Wang
Research Center for Analytical Sciences,Department of Chemistry,College of Sciences,Northeastern University,Shenyang 110819,China
Keywords:Single-cell pipette Microfluidic chip Single-cell capture ETAAS ICP-MS CdTe QDs
ABSTRACT Accurate single-cell capture is a crucial step for single cell biological and chemical analysis.Conventional single-cell capturing often confront operational complexity,limited efficiency,cell damage,large scale but low accuracy,incompetence in the acquirement of nano-upgraded single-cell liquid.Flow cytometry has been widely used in large-scale single-cell detection,while precise single-cell isolation relies on both a precision operating platform and a microscope,which is not only extremely inefficient,but also not conducive to couple with modern analytical instruments.Herein,we develop a modular single-cell pipette(mSCP) microfluidic chip with high efficiency and strong applicability for accurate direct capture of single viable cell from cell suspensions into nanoliter droplets (30−1000 nL).The mSCP is used as a sampling platform for the detection of CdTe quantum dots in single cells with electrothermal atomic absorption spectrometry (ETAAS) for the first time.It also ensures precise single-cell sampling and detection by inductively coupled plasma mass spectrometry (ICP-MS).
Due to the existence of cell heterogeneity,the average information obtained from the cell populations may mask random diversity of individual cell responses,such as temporal variations in the content of vital substances at the single-cell level [1,2].Precise single-cell analysis can better understand the function and development of heterogeneous cell populations,thereby further clarifying a series of physiological mechanisms including cancer metastasis,stem cell differentiation,and immune response [3–5].Successful isolation of a single-cell from the cell population while maintaining cell viability and transferring it to a tiny detection volume is the first and most critical step for accurate single-cell analysis including PCR [6],cloning [7],and other single-cell biochemical assays [8–10].At present,cell suspension dilution [11],microscope operation (micromanipulation) [12],fluorescence sorting with cytometry [13],optical tweezers [14],inkjet printing [15],laser capture [16]and others [17]can achieve single-cell separation and acquisition.However,these strategies are not conducive to the demand for efficient single-cell analysis due to complex multi-device combination or expensive precision equipment.They are plagued by time-consuming pretreatment,complex operation,limited efficiency and deterioration of cell viability.In addition,the array-type single-cell capture and the single-cell focusing/encapsulating microfluidic chip based on fluid manipulation can realize fast and high-throughput single-cell processing.However,when it is necessary to isolate a specific required single cell,a complicated secondary operation is still required,which is unfavorable for non-continuous sampling detection methods,e.g.,electrothermal atomic absorption spectrometry (ETAAS),matrix assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF MS),and laser ablation inductively coupled plasma mass spectrometry (LA-ICPMS).
Microfluidic systems may well match the microstructure and cell size.It is able to accurately manipulate single cells with high detection sensitivity and high throughput at minimized reagent consumption.It has great potential in single cell precise manipulation and single cell analysis.Therefore,the development of a rapid,highly efficient,operationally simple and inexpensive strategy for cell manipulation is necessary for single-cell biochemical analysis.A hand-held single-cell pipet were reported to conveniently transfer single-cell into standard 96-/384-well plates,Petri dishes or vials [2].It consists of two pressurized channels,one Y-shaped microchannel,and one microscale hook.After efficient capture by the hooks,single cells could be released into nanoliter droplets.However,the microhook is designed as a structure with an opening downward.When the fluid in the channel is still,the cell captured will sink downward and leave the microhook structure due to no restrictive pressure.In the washing step,the cell captured is easily washed away.Herein,we report a novel single-cell capture chip–modular single-cell pipette (mSCP) that composes of a Z-shaped channel and a horizontal linked-microchannel,which successfully solves the stability of the cell-captured.The chip is able to separate single living cells quickly,conveniently and efficiently from cell suspension (Fig.1,Movie S1 in Supporting information).At the same time,the mSCP has been developed as a sampling platform for ETAAS and ICP-MS to achieve high-precision single-cell analysis/detection.This is the first time to use ETAAS for single cell analysis.
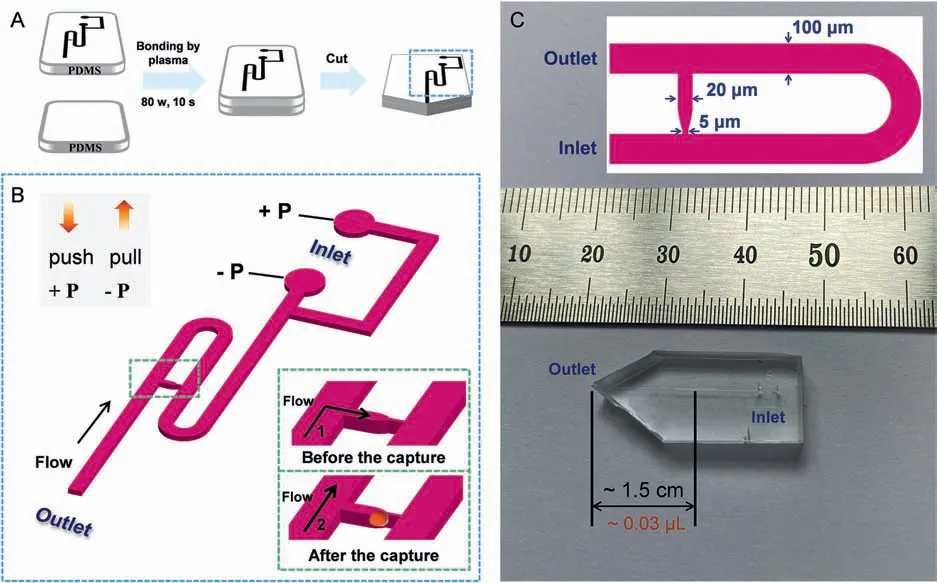
Fig.1.Preparation process and specific fluid channel details of the modular single-cell pipette (mSCP).(A) Schematic diagram of the mSCP chip preparation process,including lithography,casting,sealing and cutting.(B) The diagram of the mSCP chip channel structure,illustrating a linked-channel (capture unit) for the capture of single-cell.(C)The real picture and main structure dimensions of the mSCP chip.
The chip channel structure of mSCP is first designed through AutoCAD,and then fabricated by UV photolithography and polydimethylsiloxane (PDMS) molding/packaging techniques (The fabrication details are given in Fig.1A and Supporting information).The mSCP is a dual-channel pipet with a control unit consisting of two syringes.A negative-pressure channel (−P) operated by pulling one syringe and a positive-pressure channel (+P) operated by pushing another syringe (Fig.1B),and one cell is captured by the linkedmicrochannel during each operation.Two ports (−P and+P) are connected by one Y-shaped microchannel to the Z-shaped main channel.Near the outlet,a linked-microchannel perpendicular to the main channel is designed for cell capture.Among them,the main channel has a width of 100 μm,a design depth of 20 μm,and a total length of ∼2500 μm;the linked-microchannel has a wide gap (20 μm) and a narrow gap (5 μm);the exit of the linkedmicrochannel (wide part) is set atca.1.5 cm away from the tip of the mSCP (Fig.1C).
The cell transfer mechanism through mSCP employs a procedure similar to that of hand-held micro-pipette or micro-syringe liquid transfer in order to make the operation as simple as possible and maintain the system stable.Single-cell transfer and detection is achieved in five steps including preparation,aspiration/capture,cleanout,injection and transfer to detector (Fig.2 and Movie S1).(1) In preparation,the syringes connected to the two pressure channels are pre-filled with 300—500 μL of PBS solution,and they are then assembled on the −P and+P parts of the mSCP.A gentle push on the+P and −P syringes send the PBS into the Y-shaped microchannel to replace the air in the mSCP channel.A stable droplet is formed at the tip of the mSCP outlet.Due to the hydrophobicity of PDMS,a tiny spherical droplet is always obtained and can indicate the exit position of the chip.(2) In capture,the−P syringe is set to aspirate ∼100 μL of cell suspension (106−107cells/mL) into the main microchannel.Due to the channel size effect,the cells preferentially move into the direction of low flow resistance (higher flow rate).The size and shape of the linkedchannel are optimized to effectively capture single-cell in the size range of 10−20 μm.During the mSCP operation,the capture channel is in a horizontal state.This design is beneficial to avoid the failure of cell capture caused by the cells leaving the capture area between different steps of the mSCP movement (no flow rate and no pressure in the channel).(3) In cleanout,the tip of the mSCP is quickly immersed into the cell-free medium solution (PBS),the−P syringe is set to aspirate ∼300 μL of PBS into the main channel to flush the uncaptured cells into the −P syringe,and the channel is cleaned at the meantime.When the capture channel is occupied by one cell,the flow resistance of the capture channel becomes larger.After applying negative pressure in the channel,the fluid flows along the main channel without directly contacting the captured cell,which can greatly reduce the damage to the cells by the fluid shearing force.Therefore,the horizontal capture channel can further improve the cell activity during capture.Finally,the end of the chip is immersed in the cell-free liquid while moving it up and down several times to flush the attached cells on end of the chip.(4) In injection,the hydrophobicity of PDMS reduces cell adsorption to the extent as possible.By applying a gentle push to the+P syringe,the captured single-cell is easily released and injected into a nanoliter droplet formed at the tip of the mSCP.(5) Subsequently,the single-cell droplet can be conveniently transferred to the designated containers or detectors.During the mSCP operation,only one source of pressure is performed at each step.The whole process can be completed in ∼30 s.
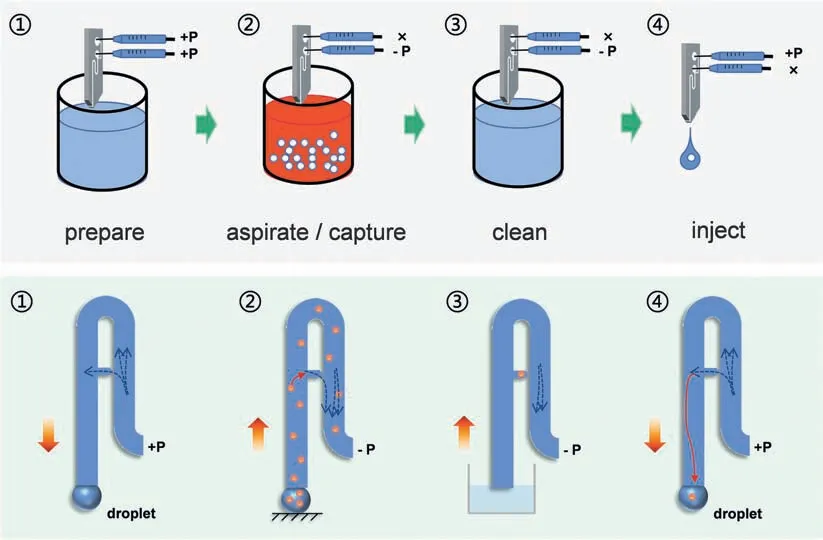
Fig.2.Work flow for single-cell capture/release by using mSCP.Single cell suspension droplets can be directly analyzed by ETAAS and ICP-MS.+P: provide positive pressure;−P: provide negative pressure;×: No operation.
In order to achieve rapid,simple operation and efficient live single-cell capture and release,we designed and developed a single-cell capture structure based on the principle of hydrodynamic flow [18–21].In a U-shaped channel,the linked-channel is designed based on the size effect and is perpendicular to the main channel,making it a trap in the main channel.Media flow and cells enter the trap from the top,where the majority of the flow will pass through the trap due to high resistance of the long main channel,and drag the cell into the trap.Once trapped,the cell will be maintained stably in the trap due to the pressure difference between the inlet and the outlet and the flow is re-directed to the main channel.In this process,the cells will preferentially pass through the capture gap [20].Since the width of the capture slit will be smaller than the size of the unit,a single unit will be stuck in front of the slit,and the negative pressure through the capture slit will be greatly reduced (flow resistance increased).Other cells cannot enter the capture chamber again,and can only change the direction of fluid movement and pass through the U-shaped main channel.To verify the single-cell capture efficacy,we simulate the velocity distribution in the channel of mSCP at the initial negative pressure of 2000 Pa with COMSOL Multiphysics (version 5.4).The numerical simulation demonstrates that before the cells are captured,the linked-channel has a large flow rate (fluid resistance is small),the cells are easily captured (Fig.S2A in Supporting information).When a cell occupies the linked-channel,the flow resistance is increased,the flow direction returns to the main channel,and the flow velocity around the captured cells is low (Fig.S2B in Supporting information).Therefore,the impact of fluid shear is limited,especially in the subsequent cleaning steps,which maintains the cell viability.
In this study,the capture and transfer of single cell is the most important issue for achieving desired results.In order to investigate the possibility of cell capture and transfer by this chip,the cells were stained with HepG2 (the diameter is 10–15 μm) as a model,single cell capture and release operations were performed on the chip.The capture and release of cells were observed under a fluorescence microscope (Fig.3A).It is seen that the single cell can be caught by the capture unit according to the expected hydrodynamic behavior,and it exhibits a capture efficiency of>90% during the operation.The efficiency single-cell capture is closely related to the cell suspension density and the aspiration time.Generally a higher cell suspension density and a longer aspiration time may increase the probability of cell capture.After the single cell transfer operation (as illustrated in Movie S1),the captured single cell can be quickly released from the capture unit.At the same time,we performed AO staining on the captured cells.The fluorescence image in Fig.3B clearly indicated that the captured cells exhibit favorable viability.This demonstrated the fact that theoretical simulation is coincide with the experimental results.The mSCP is feasible to capture and release single cells conveniently with no damage to the cells.

Fig.3.The illustration for capturing individual cells on the mSCP.(A) Microscopic image of the captured cell on the mSCP at the cell number density of 1 × 106 cells/mL.(B) Fluorescence microscopic image of the captured cell after AO staining(4 °C for 20 min in the dark).The capture unit structure has less damage to the captured cells.Scale bar,100 μm.
Electrothermal atomic absorption spectrometry (ETAAS) is an elemental analysis method with high sensitivity and tiny sample demand.It relies on high-temperature ashing of the sample to realize the analysis of biological samples without pretreatment.However,due to its own sampling limitation,ETAAS has not been applied for single-cell analysis.In the present study,ETAAS is adopted for the first time to perform single-cell analysis,by using mSCP as single cell sampler to accurately quantify intracellular CdTe QDs.Full details for the preparation of CdTe QDs,and the scrutiny of experimental and instrumental conditions are given in the Supporting Information.
In brief,the logarithmic growth phase of HepG2 cells is incubated with 20 mg/L MPA-CdTe QDs.Subsequently,mSCP captures individual cell from the incubated cell suspension and releases a single cell directly into the graphite tube of ETAAS.The single cell is detected under the optimal experimental conditions.First,3 μL MgNO3(0.06%,wt%) as the matrix modifier,∼1 μL unincubated single HepG2 cell obtained through mSCP as a matrix matching sample,and 5 μL cadmium standard solution (a series of 0.05,0.1,0.15,0.2,0.25 μg/L) are added into the graphite tube for analytical performance testing,deriving a detection limit of 1.8 ng/L for intracellular Cd quantification (Fig.S3 in Supporting information).In comparison with the biomedical applications and toxicology of cadmium cations,it is more important to study the biological impact of cadmium containing QDs.The uptake of QDs by cells is highly variable due to the physical and chemical characteristics of QDs,culture conditions,and cell heterogeneity.In this experiment,the single cells are captured by mSCP followed by their introduction into ETAAS for quantification.70 individual cells are captured,injected and analyzed by ETAAS,and the results indicated that the content of Cd in a single cell falls into a range of 14 to>1000 fg.
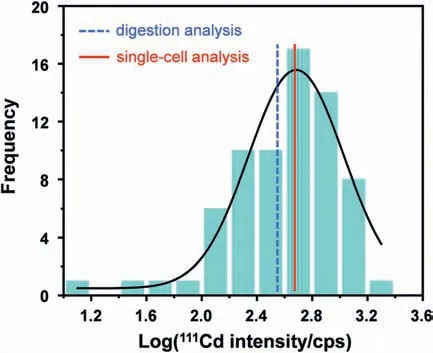
Fig.4.Histogram showing the log-normal distribution of Cd mass for the detection of 70 individual cells.The two averages from single-cell probabilistic analysis (the red solid line) and cell digestion analysis (the blue dot line) are 482 fg and 353 fg,respectively.
In order to verify the accuracy of the method,all these samples are further analysed by subjecting to wet digestion.It indicated that the results derived by wet digestion and ETAAS are in favourable agreement.The mass value derived from the analysis results of 70 single cells by fitting the logarithmic distribution histograms with the Gaussian function is 482 fg Cd per cell (the average mass of the 70 single cells is 533 ± 387 fg),while that from wet digestion analysis of 4.5 × 106cells is derived to be 353 ± 27 fg Cd per cell.Fig.4 shows that a log-normal distribution approximates the Cd mass of the 70 individual cells.It can be seen that through the mSCP strategy,ETAAS can well ensure the detection of CdTe-QDs in a single-cell.In comparison with the average value of wet digestion,ETAAS-based single cell detection can reflect heterogeneity in cell population.Previous studies have shown that cell heterogeneity is an inherent condition of cell populations,possibly as a result of stochasticity in gene expression and protein synthesis [22].The microenvironment,due to the different deposition or aggregation states of QDs particles around a single cell,it may also affect the absorption of nanoparticles by a single cell.Finally,cell cycle phase has a significant effect on the uptake of QDs [23].Therefore,the combination of the affecting factors ultimately leads to a wide distribution of QDs in the individual cells,as shown in Fig.4.The data provide significant evidence that seemingly identical single cells respond very differently to QDs after exposure.The mSCP-ETAAS method has the capability to quantitatively analyse QDs in single cells and will aid better understanding of cell-tocell variation,giving deeper insights into basic biological processes.Simultaneously,this study expands the scope of single-cell analysis methods,helping to meet the needs of basic laboratories for single-cell analysis.
The mSCP platform can also conveniently inject single-cell droplet directly into the continuous carrier flow of TRA-ICP-MS to achieve precise single-cell ICP-MS (sc-ICP-MS) analysis.Fig.S4A(Supporting information) clearly illustrated that only a spike is achieved for the single cell,and also proved that mSCP has excellent single-cell capture/release capabilities.In comparison with the traditional methods,the present approach ensures the detection of a specific single cell,and it is feasible to obtain the relationship between a specific single-cell and the cell population,which is very important for accurate and in-depth analysis of single-cells.This is further illustrated in Fig.S4C (Supporting information) wherein the single-cell data represented by the green dot line has a relative positional relationship in the cell population.Thus,the mSCP platform is an effective tool for capturing specific single cells.
In summary,we describe a modular single-cell pipette (mSCP)microfluidic chip for the capture and release of single cells directly from cell suspension.This novel strategy exhibits key features of simple operation,high efficiency,stable capture,and maintenance of cell viability.This direct and stand-alone approach can be easily operated without the assistance of any additional equipment.The extremely small single-cell suspension droplets are ideal microsystem for performing single-cells chemical and biological analyses.Furthermore,the mSCP can be used as a live single-cell sampling platform to achieve accurate single-cell sampling and detection based on ETAAS and ICP-MS.This strategy can not only expand the quantitative detection methods of single cells,but also is essential for the precise detection of single cells in the future.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors are grateful for the financial support of the National Natural Science Foundation of China (Nos.21727811,21922402),the Fundamental Research Funds for the Central Universities (Nos.N2005003,N2105017),the Liaoning Revitalization Talents Program (No.XLYC1802016),and Scientific Research Funding Project of the Education Department of Liaoning (No.LJKZ0007).Special thanks are due to the instrumental analysis from the Analytical and Testing Center of Northeastern University.
Supplementary materials
Supplementary material associated with this article can be found,in the online version,at doi:10.1016/j.cclet.2021.08.024.
 Chinese Chemical Letters2022年3期
Chinese Chemical Letters2022年3期
- Chinese Chemical Letters的其它文章
- Direct catalytic nitrogen oxide removal using thermal,electrical or solar energy
- Construction and applications of DNA-based nanomaterials in cancer therapy
- Recent research progress of bimetallic phosphides-based nanomaterials as cocatalyst for photocatalytic hydrogen evolution
- Nanostructured materials with localized surface plasmon resonance for photocatalysis
- Recent progress of Pd/zeolite as passive NOx adsorber: Adsorption chemistry,structure-performance relationships,challenges and prospects
- Microfluidic methods for cell separation and subsequent analysis
