Development of environmentally friendly biological algicide and biochemical analysis of inhibitory effect of diatom Skeletonema costatum
Ji Yang,Qingzhng Zhu,Jinlong Chai,Fng Xu,Yuni Ding,Qiang Zhu,Zhaoxin Lu,Kuan Shiong Khoo,Xiaoying Bian,Shujun Wang,∗,Pau Lok Show,∗∗
a Jiangsu Key Laboratory of Marine Bioresources and Environment/Jiangsu Key Laboratory of Marine Biotechnology,Jiangsu Ocean University,Lianyungang 222005,China
b Jiangsu Marine Resources Development Research Institute,Lianyungang 222005,China
c Co-Innovation Center of Jiangsu Marine Bio-industry Technology,Jiangsu Ocean University,Lianyungang 222005,China
d Helmholtz Institute of Biotechnology,State Key Laboratory of Microbial Technology,Shandong University,Qingdao 266237,China
e College of Food Science and Technology,Nanjing Agricultural University,Nanjing 210095,China
f Department of Chemical and Environmental Engineering,Faculty of Science and Engineering,University of Nottingham Malaysia,Jalan Broga,43500 Semenyih,Malaysia
Keywords:Bacillomycin D Skeletonema costatum Transcriptome Growth inhibition Photosynthesis
ABSTRACT Skeletonema costatum is a diatom widely distributed in red tide microalgae blooms and as one of the main algae causing harmful algal blooms,because of their rapid reproduction and production of toxic and harmful substances,often play a negative role in aquatic ecosystems,and human health and wellbeing.Bacillomycin D is a nonribosomal cyclic antifungal lipopeptide in the iturins family.In this study,Bacillomycin D was tested for its ability to inhibit the growth of S.costatum.The EC50 24h of Bacillomycin D on S.costatum was 24.70 μg/mL.The chlorophyll fluorescence parameters Fv/Fm,Fv/Fo,and yield of the diatoms decreased significantly with increasing concentrations of Bacillomycin D.Study of the mechanism showed that Bacillomycin D induced cell death by changing cell membrane permeability,promoting the release of cellular contents.In this study,transcriptomic analysis showed Bacillomycin D significantly inhibited the photosynthesis and metabolism of S.costatum.These findings investigated the inhibitory effect of Bacillomycin D on the growth of S.costatum and provided a theoretical foundation for the development of new environmentally friendly biological algicide.
Harmful algal blooms (HABs) or red tides frequently occur all over the world and have become a difficult problem in the field of water ecology.Due to natural environmental factors,human activities,and climate change,HABs cause massive fish kills,destroy or poison shellfish,which result in wildlife death and disrupting ecosystem [1-3].In terms of number,frequency and intensity,the occurrence of harmful algal blooms have dramatically increased in coastal regions throughout the world,more toxic algal species,algal toxins,and geographic areas were affected [4].Over a 35-year period of tracking HABs,diatoms have been the most dominant phytoplankton group (>90%) [5],and diatoms cause HABs to be a significant problem for the environment and the fishery industry[6].
S.costatumis one of the most abundant diatoms occurring internationally in coastal marine phytoplankton.Therefore,it is of great significance to study the inhibition ofS.costatumgrowth.Physical,chemical,and biological methods have been used to control HABs [7].Chemical methods affect a broad-spectrum of species,killing beneficial phytoplankton and fish while killing the target algae.Therefore,the direct use of chemical controls can damage and disrupt the original water ecosystem,resulting in adverse effects of regional ecosystems [8].Physical methods are cumbersome,costly and not suitable for large-scale applications [9].Biological control is preferred because of its decreased ecological and environmental impact,which has become a more sought-after method and a dynamic research field [10].
Bacillomycin D is an antibacterial lipopeptide secreted from the bacteriaBacillus subtilisand is formed by linking a heptapeptide with aβ-amino aaliphatic hydrocarbon,forming a cyclic lipopeptide [11].Studies have showed that Bacillomycin D has an inhibitory effect on many fungi,such asAspergillus flavus[11,12],Fusarium graminearum,Alternaria alternata,Rhizoctonia solani,Cryphonectria parasitica,Phytophthora capsici,andCandida albicans[13].The literature indicate Bacillomycin D can cause elevated levels of reactive oxygen species and DNA damage in cancer cells and apoptosis by cell contraction,nuclear agglutination,and rupture[14],and it also resists oxidization and has free radical scavenging activity [15].To sum up,Bacillomycin D,as a secondary metabolite with a variety of biological activities,has have application in many field and it has the potential to be developed into a lead compound for food preservation as well as uses in medicine.However,the prevention and treatment of HABs by Bacillomycin D has not yet been reported.
In the present study,Bacillomycin D isolated fromB.subtilisstrain fmbJ was used to inhibitS.costatum.In order to better revealS.costatumin response to Bacillomycin D and improve the efficacy of Bacillomycin D as a biological control,finally to determine the feasibility of commercial application of Bacillomycin D.The data of chlorophyll fluorescence parametersFv/Fm,Fv/Fo,SEM,TEM and transcriptomics were used to analyze the dynamic changes in global gene expression inS.costatumafter treatment with Bacillomycin D.
S.costatum(GY-H11) used in this study was supplied by Shanghai Guangyu Biotechnology Co.,Ltd.(Shanghai,China).S.costatumculture was grown in an f/2-Si medium at pH 8 at 25 °C under cycles of 12 h of darkness and 12 h of cool white fluorescent light (60 μmol photons m−2s−1),transferred to fresh medium once a week to ensure that experiments were always conducted with cultures during the exponential growth phase.
Bacillomycin D was obtained fromB.subtilisfmbJ (CGMCC No.0943) [16].Its biological activity was determined by the addition of Bacillomycin D (final concentrations 6.25,12.5,25,or 50 μg/mL)to cultures ofS.costatumin 100 mL f/2-Si medium in an exponential growth phase.The flasks were incubated for up to 7 days in an environmentally controlled cabinet at 25 °C with continuous illumination of the mean photon flux density of 60 μmol photons m−2s−1agitated two times daily.The algae inhibition rate was calculated using the following Eq.1.
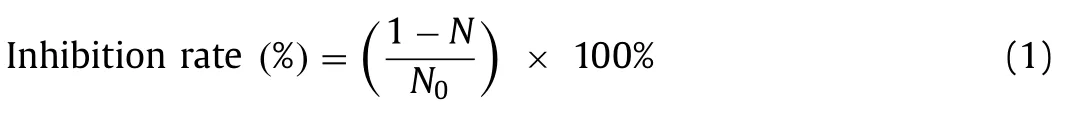
where,Nis the treatment andN0is the control of the algae density ofS.costatumwith and without treatment,respectively [17].
The median effective concentration (EC50) was calculated by the probit unit method using SPSS 23 software [18].
The changes in the cell shape and ultrastructure ofS.costatumwere investigated by scanning electron and transmission electron microscopy.Cells treated with 24.70 μg/mL of Bacillomycin D for 24 h were centrifuged at 10,000×g for 10 min at 4 °C before washing twice with Na-phosphate buffer (50 mmol/L,pH 7.2).For scanning electron microscopy,the washed samples were pre-fixed with 2.5% glutaraldehyde.The cells were subsequently dehydrated using sequential ethanol concentrations ranging from 30% to 100%.The ethanol was replaced with tertiary butyl alcohol.The samples were coated in gold by cathodic spraying.The morphology of the bacterial cells was observed using scanning electron microscopy (SEM) (S-3000N,Hitachi,Japan).For transmission electron microscopy (TEM) (H-7800,Hitachi,Japan),the cells were fixed with 1% osmic acid solution for 1 h,followed by infiltration with 2.5% glutaraldehyde overnight at 4 °C,and then sectioned with an ultramicrotome (Lica,Germany) before observation using a Hitachi HT-7800 TEM [19].
The chlorophyll a content was determined as previously described [19].Algal cells incubated in the absence or presence of Bacillomycin D were harvested at the indicated times.Algal culture samples (10 mL) were centrifuged at 10,000×gfor 10 min.The supernatant was discarded.The remaining algal cells were mixed with 5 mL of absolute ethanol,shaken,and place in a refrigerator at 4 °C in the dark for 24 h.After the overnight incubation,the mixture was centrifuged at 10,000×gfor 10 min.The supernatant was collected for spectroscopic analysis at 632 nm,649 nm,665 nm,and 696 nm.The chlorophyll a content (μg/L) was calculated using Eq.2.

Chlorophyll fluorescence parameters were measured using an AP-C 100 handheld phytoplankton fluorometer (PSI,Czech Republic).The initial chlorophyll fluorescence (Fo) and maximal chlorophyll fluorescence (Fm) were measured after dark adaptation for 3 h.The variable fluorescence (Fv) was calculated as (Fm/Fo).The effective PSⅡactual quantum yield and the maximal quantum effi-ciency of PSⅡ(Fv/Fm) were also calculated [16].
For transcriptome analysis,the effects of Bacillomycin D onS.costatumwere studied.In the exponential growth phases (4-7 days),24.70 μg/mL of Bacillomycin D was added toS.costatumculture in f/2 medium.The flasks were incubated 24 h at 25 °C with continuous illumination of 60 μmol photons m−2s−1and were agitated two time daily.The control sample was incubated without Bacillomycin D.The samples used for transcriptome sequencing are divided into two groups and each group were three parallel experiments.
Sequencing libraries were generated using NEBNext® UltraTMRNA Library Prep Kit for Illumina® (NEB,USA).Every sample was prepared from 1.5 μg total RNA.The mRNA was purified from the total RNA using poly-T magnetic beads and then fragmented.First strand cDNA was synthesized using random hexamer primers and M-MuLV Reverse Transcriptase.Second-strand cDNA was synthesized using DNA Polymerase I and RNase H.According to the standard Illumina protocol,the cDNA libraries of 250–300 bp in length were constructed.
The cDNA libraries were sequenced on an Illumina HiSeq platform to generate paired-end reads of 150 bp.Clean reads were obtained by removing adapters,reads containing poly-N stretches,and low-quality reads from the raw data.De novoassembly was carried out using Trinity with the min_kmer_cov set to 2,with all other parameters set to default.The longest transcript of each subcomponent was defined as a ‘unigene’for functional annotation.The function of each unigene was annotated using the NCBI nonredundant protein (Nr),the NCBI non-redundant nucleotide (Nt),the Swiss-Prot protein,the Protein family (Pfam),the Clusters of Orthologous Groups of Proteins (KOG/COG),the Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthologue (KO),and Gene Ontology (GO) databases.The raw RNA sequence data deposited in the National Centre for Biotechnology Information (NCBI,accession SRP309218).
The effect of different concentrations Bacillomycin D onS.costatumgrowth is shown in Fig.1.At 24 h,concentration-dependent inhibition ofS.costatumwas observed.The 24 h median effective concentration value (EC5024h) was 24.70 μg/mL.While current research on Bacillomycin D has mainly focused on its fungistatic and antitumor activities [12,20,21],there have been few studies on algae inhibition.

Fig.1.Growth inhibition curve of the algae S.costatum treated with different concentrations of Bacillomycin D.

Fig.2.The effect of Bacillomycin D on cell morphology of S.costatum.(A) Normal cells of S.costatum in use of TEM;(B) Cells of S.costatum were under the condition of Bacillomycin D in use of TEM;(C) Normal cells of S.costatum in use of SEM;(D)Cells of S.costatum were under the condition of Bacillomycin D in use of SEM.
To determine the mechanism by which Bacillomycin D inhibitsS.costatumgrowth,SEM and TEM were used to observe changes at the ultrastructural level.Fig.2 shows the structural changes ofS.costatumcaused by Bacillomycin D.Normal cells ofS.costatumare cylindrical,with a very regular cell wall and uniform space between the cell membrane and cell wall (Fig.2A).In addition,regular nuclear area,vesicles,and other cell organelles were observed(Fig.2C).However,exposure to Bacillomycin D caused cell damage(Figs.2B and D).In comparison to the untreated control,the surface of the cell was collapsed,and the siliceous shell was opened.Furthermore,the content of the cell had flowed out,and finally,the cell was ruptured.Bacillomycin D-treated cells had nuclei with a smaller nucleolus,and the cells had lost their basic structure,leaving only a siliceous shell.These images showed that Bacillomycin D induced cell death by disrupting cell membrane permeability and promoting cellular contents.
Chlorophyll is essential to photosynthesis due to its ability to absorb light energy.The chlorophyll a content ofS.costatumwas measured in the absence and presence of different concentrations of Bacillomycin D incubated after 24 h (Fig.3A).The chlorophyll a content of the control was 1.79 μg/L.In the presence of the lowest concentration of Bacillomycin D,the chlorophyll a content dropped sharply to 1.05 μg/L (P <0.05).The chlorophyll a content decreased with increasing concentrations of Bacillomycin D.
Chlorophyll a is a significant pigment of diatom chlorophyll[22],and previous studies have shown that phytoplankton has the highest chlorophyll a content after 24 h [23].The water pollutant 2,4-dichlorophenol (at 3 and 6 mg/L) reduces the chlorophyll a content (by 24.1% and 20.4%) and the chlorophyll c content (by 15.7% and 14.4%),respectively [24].In another study,the chlorophyll a content ofS.costatumdecreased by 34% after 48 h exposure to phenol and returned to control level after 96 h of treatment[25].This may be due to reactive oxygen species (ROS) scavenging by antioxidant enzymes such as catalase (CAT) and superoxide dismutase (SOD) [26].
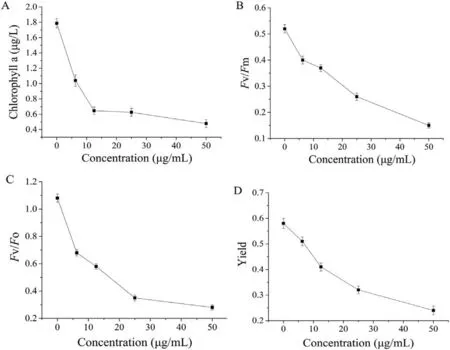
Fig.3.Effects of different concentrations of Bacillomycin D on the photosynthetic characteristics of S.costatum.(A) Chlorophyll a content in S.costatum.(B) Maximum light quantum yield (Fv/Fm).(C) Potential activity (Fv/Fo).(D) Actual quantum yield.
The measurement of chlorophyll fluorescence is a non-invasive method to evaluate photosynthesis under environmental stress,especially the changes of PSⅡfunction and electron transfer rate[19].TheFv/Fmratio indicates the maximum light energy conversion efficiency of the PSⅡreaction center [27].In this study,theFv/Fmvalue inS.costatumdecreased with increasing Bacillomycin D concentrations (Fig.3B).Under treatment with 6.25 μg/mL to 50 μg/mL Bacillomycin D,theFv/Fmvalue decreased from 77.46% to 27.94% of the control value,respectively.
TheFv/Foratio indicates the potential activity of the photoreaction center PSII,and its value decreases when stressed by environmental conditions.As shown in Fig.3C,theFv/Fovalue decreased with increasing Bacillomycin D concentrations.Under treatment with 6.25 μg/mL to 50 μg/mL Bacillomycin D,theFv/Fovalue decreased from 62.96% to 25.92% of the control value,respectively.
Yield represents the paramount light energy capture efficiency of the PSII reaction center under light adaptation conditions.It is one of the most influential parameters for evaluating the PSⅡphotosynthetic efficiency of plants [28].In the present study,yield decreased as the concentration of Bacillomycin D increased (Fig.3D).Undertreatment with 6.25 μg/mL to 50 μg/mL Bacillomycin D,the yield value decreased from 88.14% to 42.37% of the control value,respectively.The significant decrease in yield indicates the adverse effect of Bacillomycin D on the photosynthetic efficiency of algae,which may be due to the impaired cell affecting carbon fixation and assimilation [28].
RNA-seq was conducted forS.costatumgrowing in Bacillomycin D-treated and untreated conditions to characterize the differentially expressed genes (DEGs) related to the growth inhibition effect of Bacillomycin D.A total of 51,062,058 raw reads were generated,and 48,674,630 clean reads (7.30 G clean bases) remained after filtering of the raw data and removal of low-quality sequences and adaptor sequences (Table S1 in Supporting information).
The clean reads were assembled using Trinity into 20,008 unigenes with a mean length of 1866 bp.Among the unigenes obtained,17,804 (88.98%) matched with homologs with known functions in at least one database,and 2,998 unigenes were successfully annotated across all databases shown in Table S2 (Supporting information).Functional classification of 6,096 unigenes was annotated using the KOG database and classified into 25 categories based on sequence homology (Fig.S1 in Supporting information).Among these categories,864 unigenes corresponded to translation,ribosomal structure and biogenesis,327 corresponded to carbohydrate transport and metabolism,and 69 corresponded to cell wall/membrane/envelope biogenesis (Fig.S1A).According to GO annotation,13,564 unigenes were classified into three ontologies and 55 sub-categories.Among the biological processes,metabolic process and cellular process were the two dominant groups,followed by cellular process,metabolic process,single-organism process,and catalytic activity (Fig.S1B).Besides,based on analysis of the KEGG pathways,most of the unigenes were associated with translation,carbohydrate metabolism,and amino acid metabolism,followed by energy metabolism,folding,sorting and degradation,metabolism of cofactors and vitamins,and nucleotide metabolism(Fig.S1C).

Fig.4.Volcano map of differentially expressed genes (DEGs) in S.costatum between Bacillomycin D-treated and untreated groups.The abscissa indicates the fold change of gene expression in different samples (log2FoldChange),and the ordinate indicates the significance level of expression difference (-log10padj).Each dot represents a gene (red dots: up-regulated DEGs;green dots: down-regulated DEGs;and blue dots: non-DEGs).
The transcriptomic analysis is an effective method to discover biological functions of differentially expressed genes (DEGs) and corresponding metabolic pathways.Transcriptomic analysis ofS.costatumgrown in the presence of Bacillomycin D allowed further exploration of the possible physiological and molecular responses ofS.costatumto this antimicrobial molecule.GO function enrichment and KEGG pathway enrichment analyses were performed on the genes differentially expressed (padj<0.05|log2FoldChange|>1) between the Bacillomycin D-treated and untreated samples.In total,7,331 DEGs were identified,including 5,279 upregulated and 2,052 downregulated genes under Bacillomycin D treatment (Fig.4 and Table S3 in Supporting information).As shown in Fig.5,the GO enrichment analysis indicated that upregulated DEGs were enriched in small molecule metabolic process,singleorganism metabolic process,biosynthetic process,organonitrogen compound metabolic process,organic substance biosynthetic process,organic cyclic compound metabolic process,catalytic activity,organic cyclic compound binding,and heterocyclic compound binding (Fig.5A).The downregulated DEGs were enriched in cellular protein metabolic process,protein metabolic process,intracellular organelle,cytoplasmic part,and structural constituent of ribosome (padj<0.05) (Fig.5B).

Fig.5.GO analysis of differentially expressed genes (DEGs) between Bacillomycin D-treated and untreated conditions.DEGs were classified into three functional categories:biological process,cellular component,and molecular function.

Fig.6.A possible mechanism related to Bacillomycin D resistance in S.costatum.(A) The pathway of carbon fixation;(B) The pathway of photosystem.(The red text denotes up-regulated genes,the green text denotes down-regulated genes,the yellow text denotes up- and down-regulated genes).The enzymes or proteins corresponding to all genes were provides in Table S4.
Photosynthesis is the most important metabolic process in algae.Algae utilize light energy to convert inorganic matter (CO2),and water into organic compounds (sugars) that store energy and release oxygen.As shown in Table S4 (Supporting information),many regulatory genes encoding key enzymes of photosynthesis changed significantly with Bacillomycin D treatment.This overview shows that a large number of genes encoding key enzymes in carbon fixation were inhibited by Bacillomycin D,indicating that Bacillomycin D had a significant effect on the photosynthetic system ofS.costatum.This transcriptomic data are in accordance with the fluorescence parameters measured above.
Bacillomycin D are commonly reported to have a powerful antifungal activity [29].In the present study,Bacillomycin D showed a potent inhibitory effect on the growth ofS.costatum.The photosynthesis was important in metabolism ofS.costatum.Ribulose bisphosphate carboxylase (RuBisco),composed of the large (rbcL)and small (rbcS),subunits,is a crucial enzyme for fixing CO2and limiting the rate of enzyme activity (Fig.6A) [30,31].At the beginning of the carbon fixation pathway in photosynthesis,RuBisco catalyzes the oxidation of ribulose bisphosphate in the Calvin cycle and photorespiration pathways [32-34].After adding Bacillomycin D,some transcripts encoding RuBisco were upregulated,indicating thatS.costatumenhanced the expression of critical genes in photosynthesis in order to ensure average growth in response to environmental stress.Also,in the photosynthetic pathway,ferredoxin-NADP+reductase (PetH),a flavoprotein with a flavin adenine dinucleotide as an auxiliary base that catalyzes the reduction of ferredoxin to reduce NADP+to form NADPH,was upregulated.Transcripts of Cytochrome c6 (PetJ),F-type H+/Na+-transporting ATPase subunit beta,and other enzymes related to ATP formation were upregulated in the Bacillomycin D-treated groups (Fig.6B) [33,35].These results indicate thatS.costatumproduces more ATP to provide energy for photosynthesis and other activities in response to the stress of Bacillomycin D [36,37].As shown in Fig.6B,P700 and P680 are central pigment proteins in Photosystem I and II,and several transcripts related to P700 and P680 were upregulated,this result indicates that under the influence of Bacillomycin D,the light-harvesting function and photosynthesis ofS.costatumcells are activated at the transcriptional level,which may be a response mechanism for cells to capture more energy from the outside in responses to environmental stress.Physiological response results also showed that after adding Bacillomycin D,theFv/Fmand photosynthetic efficiency ofS.costatumdecreased.Therefore,cells need to highly express photosynthetic enzymes in enhancing the conversion of light energy to chemical energy so as to provide more energy for cells in a restricted state.However,components of the light-harvesting complex (LHC),which plays an essential role in light absorption and energy transfer to the photosystem center,showed both up-regulated and down-regulated expression.These results were due to the fact thatS.costatumincreased expression of related genes in response to Bacillomycin D stress.However,as time goes by,multiple metabolic pathways and organelles are destroyed and cannot be repaired,leading to gradual decreased expression of the up-regulated genes initially.
Bacillomycin D inhibits the carbon fixation pathway of photosynthesis by inhibiting the nitrogen metabolism pathway (Fig.6A).Alanine aminotransferase (ALT) is an important enzyme in nitrogen metabolism.In the presence of pyridoxal phosphate,ALT can catalyze the reaction of alanine andα-ketoglutarate to generate pyruvate and glutamic acid.Alanine is essential to the C4 metabolic pathway,and pyruvate plays a significant role in carbon fixation [38,39].The rate-limiting steps of CO2fixation in photosynthetic organisms are mainly concentrated in the C3 metabolic pathway.Many of the genes encoding these enzymes were downregulated by Bacillomycin D.ALDO is the first enzyme that catalyzes the conversion of C-3 compounds to C-6 compounds after CO2fixation.In the C3 metabolic pathway,the absorption of CO2by ribulose-1,5-bisphosphate to produce fructose-6-phosphate is continuously catalyzed by six enzymes,including glyceraldehyde 3-phosphate dehydrogenase (GAPA) and phosphoglycerate kinase (PGK).Oxaloacetic acid (OAA),an important compound in the C4 pathway,is converted to malic acid by malate dehydrogenase(MDH) in mesophyll cells and decarboxylated in chloroplasts of bundle-sheath cells (BSCs) to release CO2and produce pyruvate[40].Pyruvate orthophosphate dikinase (PPDK) converts pyruvate into phosphoenolpyruvate (PEP),which then enters the next C4 cycle [41].Ransketolase plays a vital role in carbon fixation,mainly catalyzing the reaction of glyceraldehyde triphosphate and fructose hexaphosphate.Its activity directly affects the photosynthetic rate of plants,and the weak change of its activity can significantly inhibit the metabolites of aromatic amino acids and phenylalanine.Eventually,this slows the growth rate of plants [42].
In summary,based on these results,Bacillomycin D acts on the surface ofS.costatumcells and changes the permeability of the cell membrane.Bacillomycin D enters the algae cells and inhibits the photosynthetic system,decreasing chlorophyll a level,maximum photochemical quantum yield (Fv/Fm),the potential activity(Fv/Fo) of the photoreaction center PSII,and the actual photon yield in theS.costatumcells.The damage to membrane lipids and the degradation of the photosynthetic pigments further affects normal photosynthesis and energy metabolism in the cells.The algal cells could not prepare enough material and energy to repair the oxidative damage.The cell membranes ruptured,the contents gradually flowed out,and cell vacuolation was severe,which resulted in cell death eventually.
Although Bacillomycin D show more significant inhibitionS.costatum.However,the low yield of Bacillomycin D limited application as efficient and safe biological algae inhibitor.Therefore,with the following development,increase the production of Bacillomycin D by biological and chemical methods should be considered in future studies.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was financially supported by grants from the National Natural Science Foundation of China (No.32100037,51971100),China Postdoctoral Science Foundation (No.2019M661767),Jiangsu Planned Projects for Postdoctoral Research Funds (No.2019K015),Natural Science Research General Project of Jiangsu Higher Education Institutions (No.20KJB550008),Jiangsu Ocean University Research Funds (No.KQ17028),Priority Academic Program Development of Jiangsu Higher Education Institutions(PAPD).
Supplementary materials
Supplementary material associated with this article can be found,in the online version,at doi:10.1016/j.cclet.2021.09.053.
 Chinese Chemical Letters2022年3期
Chinese Chemical Letters2022年3期
- Chinese Chemical Letters的其它文章
- Direct catalytic nitrogen oxide removal using thermal,electrical or solar energy
- Construction and applications of DNA-based nanomaterials in cancer therapy
- Recent research progress of bimetallic phosphides-based nanomaterials as cocatalyst for photocatalytic hydrogen evolution
- Nanostructured materials with localized surface plasmon resonance for photocatalysis
- Recent progress of Pd/zeolite as passive NOx adsorber: Adsorption chemistry,structure-performance relationships,challenges and prospects
- Microfluidic methods for cell separation and subsequent analysis
