Deciphering the transcriptomic signature of synaptic activity
Guido Hermey
Neurons undergo activity-dependent changes in their molecular composition and structure in order to regulate cellular processes such as dendritic growth, synapse elimination, spine maturation and synaptic strength. Such synaptic plasticity plays an important role during a critical period in brain development and contributes to sensory adaptation and to learning and memory in the mature nervous system. Its dysregulation underlies a number of pathological processes in psychiatric and neurodegenerative disorders, such as addiction, depression,anxiety, schizophrenia, epilepsy and traumatic brain injury. Short-term activitydependent synaptic changes rely mostly on post-translational modifications of pre-existing proteins, whereas the longterm maintenance of synaptic adaptations depends on gene induction. Signals from the synapse to the nucleus activate gene expression. Such signals are thought to be encoded in calcium waves or conveyed by macromolecular signaling complexes translocated retrogradely by motor proteins.The induced gene transcription and protein synthesis alters the composition of synaptic protein networks and provides a mechanism for translating synaptic activity into persistent synaptic changes. In accordance,large sets of genes whose expression levels are regulated by synaptic activity have been described in the past decades. As expected,several of these genes encode proteins that modulate synaptic function and play a role in neuronal plasticity-related processes.However, it has to be considered that altered gene expression levels are only part of the complex activity-regulated transcriptional signature. Alternative splicing, the differential inclusion and exclusion of exonic sequence, in combination with other related processes such as the use of alternative transcriptional initiation sites and alternative polyadenylation sites as well as mRNA editing ensure the transcriptomic and proteomic diversity required for the regulation and diversification of synaptic functions.
Alternative splicing allows a single gene to encode multiple protein isoforms with altered or even antagonistic properties.Transcriptomic and proteomic surveys indicate that more than 90% of mammalian multi-exon genes undergo alternative splicing and several genes express multiple major isoforms. The brain is likely to exhibit the most complex collection of splice variants.Some single neuronal genes, such as neurexins or calcium channels, can give rise to potentially thousands of different mRNA isoforms. Accordingly, alternative splicing of mRNA is expected to have a critical impact on brain function. Defects in alternative splicing have been suggested to affect synaptic, circuit, and brain functions and be causally linked to a substantial fraction of neuropsychiatric and neurodegenerative disorders. The genome-wide analysis of differential neuronal splicing events is still incomplete, but there is an increasing number of studies examining alternative splicing programs of specific neuronal cellular subtypes, brain regions, disease states, or after neuronal activity induction.
Different experimental models have been established to determine neuronal activityregulated transcriptional programs in rodents. These are based on the assumption that electrical or chemical stimulations produce long-lasting changes in neurons,such as synaptic plasticity. Initialin vivoapproaches to identify transcriptional changes in excitatory neurons suffered from the fact that these changes were likely to be obscured or diluted by surrounding cell types not involved in the challenged activity-dependent circuitry.Thus, experimental strategies aimed at an intense global stimulation of an entire brain area. These employed electrophysiological methods such as long-term potentiation or electroconvulsive or chemically induced seizures resulting in a robust, strong, and synchronized induction of synaptic activity in neurons of the hippocampus. Actually,the first successful differential screens to identify activity-regulated genes employed seizure protocols as a model (reviewed in(Benito and Barco, 2015). Alternativein vivoapproaches use the ocular dominance shift in the visual cortex in response to monocular deprivation. Moreover,in vitrostrategies engage dissociated primary neuronal cultures in which neuronal activity is induced by membrane depolarization through high extracellular potassium concentration or by the application of glutamate.
Overall, studies using these models demonstrated altered neuronal activityregulated expression levels of hundreds of genes. Some of these earlier reports observed alternative splicing of activityregulated genes by chance or by specific candidate gene approaches, but most surveys focused only on differential gene expression levels and did not address the expression of specific isoforms. The advancing development of splicing sensitive genome-wide techniques and the generation of splicing sensitive algorithms allow filling this gap in knowledge. Accordingly, the already established models to identify activity-regulated genes are now used to determine alternative splicing in transcriptomic signatures.
We performed a genome-wide survey of neuronal activity-regulated exon usage in the hippocampus of mice at different time points after chemically induced seizures (Denkena et al., 2020). Using exon-specific microarrays,we identified alternative exon usage induced by neuronal-activity in previously described activity-regulated genes, but also of genes so far undescribed in this context. To validate the differential expression of exons and splicing junctions, we generated and analyzed an independent RNA-sequencing data set of hippocampal transcripts isolated at different time points after seizure onset.Moreover, we used RNA-sequencing data derived from primary cultured hippocampal neurons under control-conditions or after KCl induced depolarization and demonstrated identical activity-regulated splicing for a number of genes. Our study aimed at the identification and validation of single splicing events regulated by neuronal activity. Moreover, we provide datasets that will be a rich resource to identify transcriptional alterations after neuronal activity in additional comparative analysis and to dissect enhancers and promotors that drive activity-regulated expression. The detected splicing events comprised diverse categories of alternative splicing, including the differential use of 5′- or 3′-exons,insertion and exclusion of internal exons, and the transition of internal to terminal exons.The differential use of 5′-exons indicates engagement of alternative promotors and has been described previously for BDNF. Now we observed alternative activity-induced 5′-exon usage for Cda, Erffi1, and Inhba. We detected truncated transcripts for other genes, e.g. for Krt75. Also, we found rather complex activity-regulated splicing patterns for Tpm1, Dclk1, and Homer1. Alternative splicing of Homer1 has been frequently described and widely studied after synapticactivity induction. Activity-induced alternative splicing of Homer1 determines a number of molecular and cellular changes thought to underlie synaptic plasticity.
Homer1 is a member of the evolutionary conserved Homer scaffolding protein family(Shiraishi-Yamaguchi and Furuichi, 2007).These cytosolic adaptor proteins share an N-terminal Enabled/Vasp homology (EVH1)-like domain which interacts with prolinerich amino acid sequences in target proteins.The following proline (P-) motif (Ser-Pro-Leu-Thr-Pro) is specific for Homer1. Alternative splicing results in different Homer1 variants (Figure 1AandB). The longer variants comprise a C-terminal coiled-coil domain, which conveys homo- and heterodimerization with other Homer proteins.Multimers of Homer1b/c act as a molecular hub that links through the EVH1-like domain specific postsynaptic scaffolding and signaling proteins, including group I metabotropic glutamate receptors (mGluR1 and mGluR5),inositol 1,4,5-triphosphate receptors, Shank scaffolding proteins, ryanodine receptors,transient receptor potential canonical channels, voltage-gated calcium channels,and Dynamin (Figure 1C). Notably, Homer1b/c cooperate with the postsynaptic scaffolding protein Shank to anchor mGluRs at the post synaptic density, but also connect this complex to Dynamin and confer a molecular link to the postsynaptic endocytic zone(Scheefhals et al., 2019). Since Homer1a and Ania3 lack the dimerization domain but yet bind ligands through the EVH1-like domain, it has been proposed that Homer1a and Ania3 interfere with the interactions of the longer constitutively expressed Homer1 variants containing a coiled-coil domain (Sala et al.,2003).
The short variant Homer1a was the first family member described and initially identified in screens for genes induced by neuronal activity (Brakeman et al., 1997; Kato et al., 1997). Subsequently, a large number of studies demonstrated alternative splicing of Homer1 in experimental models for synaptic plasticity and after more physiological plasticity-inducing stimuli such as contextual fear conditioning. These studies collectively established that neuronal activity causes an exon switch in the Homer1 transcript resulting in an activity-induced alteration within the coding region. The premature termination of transcription results in the use of exon 6 or exon 7 as terminal exons and creates the C-terminally truncated Homer1a and Ania3 proteins, respectively (Figure 1A,B). Surveys using RNase protection, Northern blot assays, and microarray analysis to assess transcript levels following experimentally induced seizures demonstrated a bimodal expression of Homer1 splice variants(Bottai et al., 2002; Hermey et al., 2013).The variants Homer1b and Homer1c are constitutively expressed in neurons, whereas expression levels of Homer1a and Ania3 are low under control conditions and markedly induced by neuronal activity but differ in their time course of induction. The encoded shorter Homer1a isoform acts dominantnegative, induces disassembly of multimeric complexes formed by the longer variants and induce structural and functional reorganization of dendritic spines critical for synaptic plasticity (Sala et al., 2003;Figure 1C). Ania3 is likely to play a similar role.
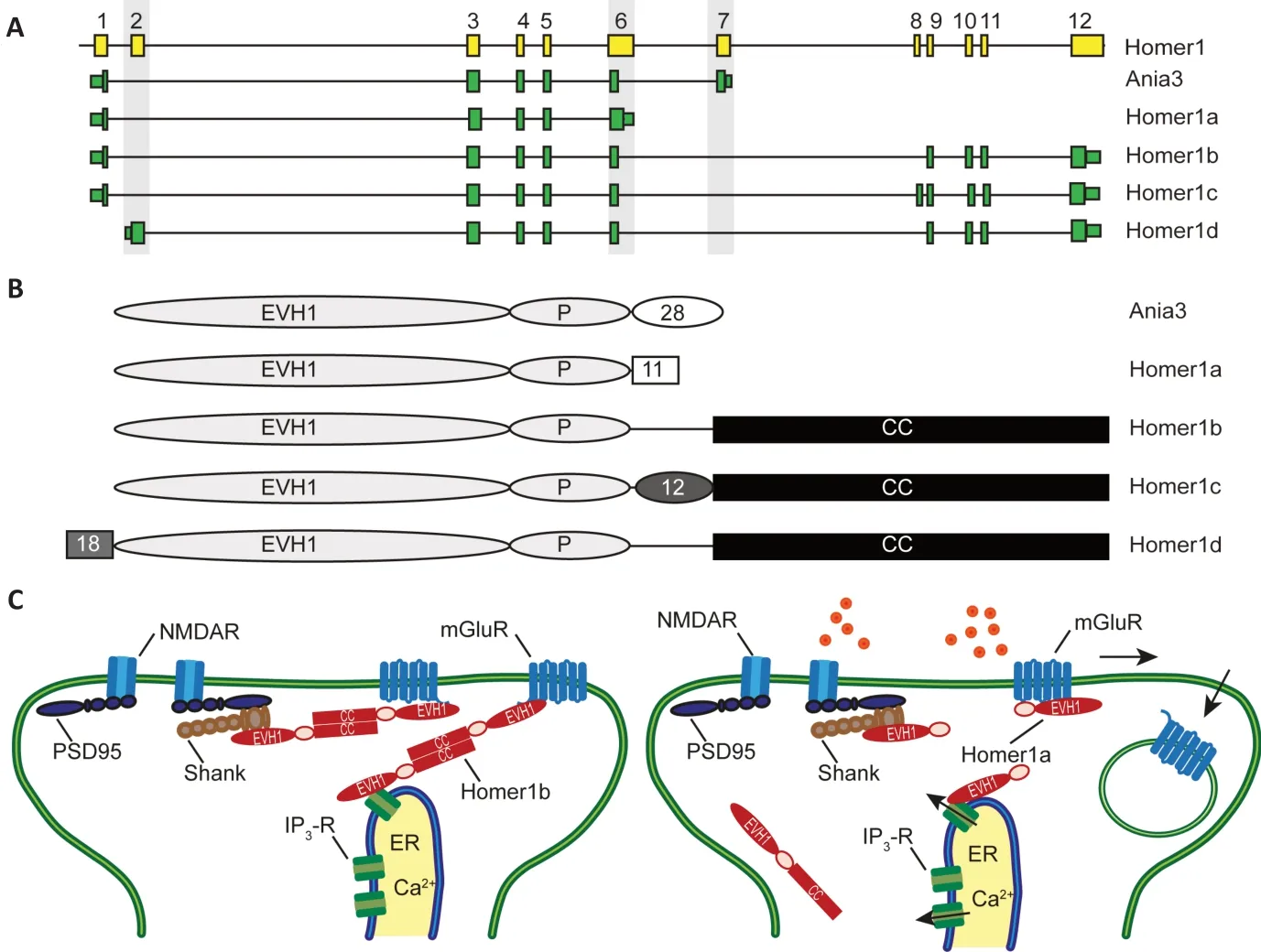
Figure 1|Activity-dependent alternative splicing of Homer1.
Homer1a has been most intensely studied,employed as a marker for highly active neurons and related to the local regulation of synaptic strength. Moreover, converging observations strongly suggest that Homer1a protects against traumatic brain injury through regulating group I metabotropic glutamate receptors and against NMDAinduced neuronal injury (Luo et al., 2014;Wang et al., 2015). Preventing activityinduced upregulation of Homer1a in micein vivoexacerbates inflammatory pain. Due to its crucial involvement in pain plasticity,Homer1 has been suggested a promising therapeutic target for the treatment of chronic inflammatory pain (Tappe et al.,2006). In addition, Homer1 has been implicated in homeostatic scaling. Homer1a initiates a homeostatic scaling-down response conveyed by alterations in the signaling of protein kinase A and mGluR1/5 and weakens excitatory synapses through removal and dephosphorylation of synaptic AMPA-type glutamate receptors (Diering et al., 2017).
In accordance with previous studies, we observed in our recent survey neuronalactivity regulated splicing of Homer1(Denkena et al., 2020). Chemically-induced seizures led to the immediate expression of exons specific for Homer1a and Ania3 in murine hippocampi. However, we also found an increased expression of exon 2 of Homer1, which is specific for Homer1d(Figure 1A). Exon 2 is used in this variant as the initial 5′-exon suggesting alternative promoter usage. Importantly, we observed an induction of Homer1d after seizures and after potassium induced depolarization in primary cultured neurons, demonstrating that the induction is not specifically seizurerelated (Denkena et al., 2020). Still, additional studies are needed to determine whether it is induced after any synaptic activity or this effect is stimulus-specific and if it is induced under more physiological conditions such as contextual fear conditioning. The usage of exon 2 as an initial exon results in an alternative N-terminus, which is prolonged(18 amino acids) compared to all other variants. It is yet unclear if this prolongation conveys alternative functions. So far, the specific impact of the Homer1d protein has been poorly studied, but one report relates Homer1d to the trafficking of metabolic glutamate receptors (Saito et al., 2002). The induction of Homer1d is rapid and the kinetic appears different from that of the short isoforms. Hence, Homer1d could transiently modify postsynaptic metabolic glutamate receptor localization or anchorage by competing with the constitutively expressed variants.
Taken together, Homer1 serves as a prototype for activity-induced alternative splicing and different categories of alternative splicing determine distinct Homer1 isoforms.The functional relevance of Homer1a expression has been demonstrated, whereas the function of Ania3 and Homer1d is still poorly studied. Future investigations should determine if Homer1d and the other long isoforms are isofunctional or if Homer1d interferes with their function.
Despite the importance of Homer1 splicing for synaptic plasticity, alternative splicing of a large number of other genes is regulated by synaptic plasticity. Generation and analysis of additional datasets will be needed to encompass the differential activity-regulated expression of specific isoforms, their induction kinetics and stimulus specificity.Most genome-wide analysis of differential splicing relied so far on junction reads from short-read RNA-sequencing, where isoform detection can be compromised by reads that cannot be mapped unambiguously. Although short-read platforms are currently dominant,because they provide higher throughput,alternative long-read technologies that enable single-molecule sequencing of complete individual RNA molecules are expected to be considered for future surveys focusing on isoform discovery. Such longread RNA-sequencing methods revealing fulllength transcript reads can deliver improved isoform-level data and will substantially advance knowledge of differentially spliced genes. The generation of such comprehensive data sets from different experimental models and species will serve future comparative studies to provide an integrated view of the transcriptomic signatures. It will improve our understanding of regulatory and epigenetic mechanisms underlying alternative splicing, its evolutionary conservation and contribution to synaptic plasticity.
The author apologizes to all colleagues whose important contributions to this field could not be cited due to space constrains.
Guido Hermey*
Institute for Molecular and Cellular Cognition,Center for Molecular Neurobiology Hamburg,University Medical Center Hamburg-Eppendorf,Hamburg, Germany
*Correspondence to:Guido Hermey, PhD,
guido.hermey@zmnh.uni-hamburg.de.https://orcid.org/0000-0003-4762-5262(Guido Hermey)
Date of submission:December 11, 2020
Date of decision:January 29, 2021
Date of acceptance:March 17, 2021
Date of web publication:June 7, 2021
https://doi.org/10.4103/1673-5374.315229
How to cite this article:Hermey G (2022)Deciphering the transcriptomic signature of synaptic activity. Neural Regen Res 17(1):82-84.
Copyright license agreement:The Copyright License Agreement has been signed by the author before publication.
Plagiarism check:Checked twice by iThenticate.
Peer review:Externally peer reviewed.
Open access statement:This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
Open peer reviewer:Edgar R Kramer, University of Plymouth, UK.
- 中国神经再生研究(英文版)的其它文章
- Genes for RNA-binding proteins involved in neuralspecific functions and diseases are downregulated in Rubinstein-Taybi iNeurons
- Research advances on how metformin improves memory impairment in “chemobrain”
- Dendritic spine density changes and homeostatic synaptic scaling: a meta-analysis of animal studies
- Optogenetic activation of intracellular signaling based on light-inducible protein-protein homo-interactions
- Presenilin mutations and their impact on neuronal differentiation in Alzheimer’s disease
- Growth differentiation factor 5: a neurotrophic factor with neuroprotective potential in Parkinson’s disease

