Progress on In-situ DNA Self-assemblies for Cellular Fluorescence Imaging Analysis
WANG Si-cheng,HAN Ting*,WANG Guang-feng
(1.Key Laboratory of Chem-Biosensing and Key Laboratory of Functional Molecular Solids of Anhui Province,College of Chemistry and Materials Science,Anhui Normal University,Wuhu 241000,China;2.Department of Chemistry,University of North Texas,Denton 76201,America)
Abstract:DNA self-assemblies play an increasingly significant role in biosensing,drug delivery and therapy due to their minimal toxicity,high biocompatibility and built-in features.Compared to most natural polymers or synthetic fibers,these DNA molecules could be relatively strong while also modifiable through sequence variation.By simply attaching to a polymer as a side chain,the secondary structure formed could endow the DNA structure with designable responsiveness to something like metal ions,proteins,pH,DNA,RNA,and some other small signal molecules(such as ATP)response to achieve some functional self-assemblies.In this review,the most recent progress of DNA self-assembly is introduced.The focus is on the initiator to distinguish the trigger mechanism and their fluorescence imaging applications.
Key words:in-situ DNA self-assembly;fluorescence imaging;living cell;trigger mechanism
Molecular self-assembly can be seen everywhere in nature,providing information about the structural and functional aspects of cells[1].In the past few decades,biomaterial engineers have been inspired by the natural assembling principles to design artificial structures such as linear,crosslinked,branched,dendritic polymers,and synthetic inorganic molecules with the ambition to explore suitable techniques within living cells such as therapeutic,target molecules,and fluorescent probes,etc.With its excellent biocompatibility,adaptable programmability,simple synthesis and modification,and strong physical and chemical stability,deoxyribonucleic acid(DNA)has been considered as one of the most powerful biomolecules to control the assembled structure[2].DNA nanomaterials having high programmability can be organized into precise stoichiometry and distributed on dynamic or static scaffold which can range from one-dimensional(1D)to three-dimensional(3D)structures[3].This arrangement provides the ability to improve molecular recognitionin vivo,construct universal templates to understand biomolecular interactions,and enhance cell imaging,especially in fluorescence imaging analysis.The whole concept has opened new opportunities to promote the development of nano-therapy.
In-situ DNA self-assemblies for cellular fluorescence imaging analysis can be classified into five categories according to the trigger types.
Category 1:Metal ion dependent DNAzyme catalysis DNA self-assembly.DNAzymes have outstanding adaptability to a large number of fluorophores,they can be labeled with a variety of organic fluorescent dyes to achieve simultaneous detection and fluorescence imaging.They are able to be labeled with different organic fluorophores to recognize the simultaneous detection and imaging of multiple metal ions[4].This feature has been utilized to design DNAzyme-responsive fluorescence image detection systems.
Category 2:mRNA responsive DNA self-assembly.mRNA-responsive DNA self-assembly can be triggered by products of rolling circle amplification and functional hairpins.As a single-stranded ribonucleic acid,the mRNA is the smallest genetic carrier in cells[5].However,tumor-related mRNA has high recognition specificity and strong binding affinity with the target.It is an important biomarker in clinical applications and can be used to stimulate response DNA components[6].
Category 3:ATP-mediation through coexisting environmental molecule.There are four major classes of biological macromolecules,carbohydrates,lipids,proteins,and nucleic acids.Each is an important component of the cell and performs a wide array of functions.And ATP is a nucleotide which is upregulated in disease cells.ATP-responsive is expected to open new directions for genome editing and advanced protein delivery for targeted disease treatment[7].
Category 4:pH-environmental conditions changes induced self-assembly.The interactions between DNA and environmental molecules,intervened by static electricity,insertion,and other processes,can act as regulators for DNA self-assembly.For example,by controlling the degradation of ZIF-8 in the tumor environment(low pH),the 3D DNA walker can be achieved with pH-responsive assembly and disassembly[8].
Category 5:Others.The cell membrane is the interface between the extracellular and intracellular environment,which regulates intracellular biological activity through signal transduction between signal molecules and membrane-bound receptors.Due to the unique Watson-Crick base pairing and sequence programmability,oligonucleotides have been widely explored as building blocks for the construction of numerous DNA nanostructures[9].Selection is made through a process called systematic evolution of exponentially enriched ligands(SELEX).Aptamers are single-stranded DNA or RNA that could bind cellspecific biomarkers on the surface of target cells with extremely high affinity.
Since fluorescence assays have gained much attention due to their simplicity,fast response,and noninvasive characteristics,and have been widely used in biological applications[10].Additionally,the signal amplifying element in the biological analysis system is essential to achieving target-detection effectiveness and accuracy based on their characterizationin vivowith excellent spatial and temporal resolution.In this review,our discussions will focus on the trigger mechanisms of DNA self-assembly and their application in fluorescence imaging in cells[11-12].
1 Trigger
1.1 Metal ion dependent DNAzyme catalysis DNA self-assembly
Metal ions are very important for many biological processes.However,the beneficial properties of many metal ions are often offset by their toxic effects when metal ions are in excess,or other toxic metal ions are present in the environment.In 2013,Lu’s group first try to use DNAzymes to achieve the goal of UO2+fluorescence detectionin vivo[13].Subsequently,his group conducted other research based on the utilization of DNAzymes for intracellular imaging.DNAzymes have outstanding adaptability to various fluorophores and could be labeled with a variety of fluorescent dyes to realize the simultaneous detection and in-situ imaging of various metal ions in living cell[14-15].Based on such property,in order to significantly reduce the interference of existing metal ion in living cells,Tang’s group developed an amplification strategy through DNAzyme catalysis to detect intracellular Zn2+and Cu2+imaging,which much improves the sensitivity of imaging based on the functional DNA-assembly[4,16].They prepared metal ion sensors using graphene oxide(GO)as a carrier,allowing Zn2+and Cu2+to be recognized by DNAzyme.Fluorophore-labeled hairpin probes act as an amplification unit and signal transduction.As shown in Fig.1A,fluorophores are quenched by GO because of fluorescence resonance energy transfer(FRET).After the graphene-based sensor enters the cell through endocytosis in the presence of free metal ions inside cell,the substrate chain can be cleaved under the catalysis of DNAzyme.As the interaction between GO and double-stranded DNA decreases,the quenched fluorescence is restored.Zn2+in the cell can be imaged by amplified fluorescence generated by the self-assembly of functional DNA catalyzed by DNAzyme.Functional DNA self-assembly,induced by DNA fragments,leads to the enrichment of a variety of fluorescent molecules,capturing high-sensitivity simultaneous imaging of Cu2+and Zn2+in vivo[17].Their work clarified the effects of low concentrations of Cu2+and Zn2+on cell function and disease development,providing a new strategy for the high-sensitivity simultaneous detection and in-situ imaging of trace metal ions in cells(Fig.1B).
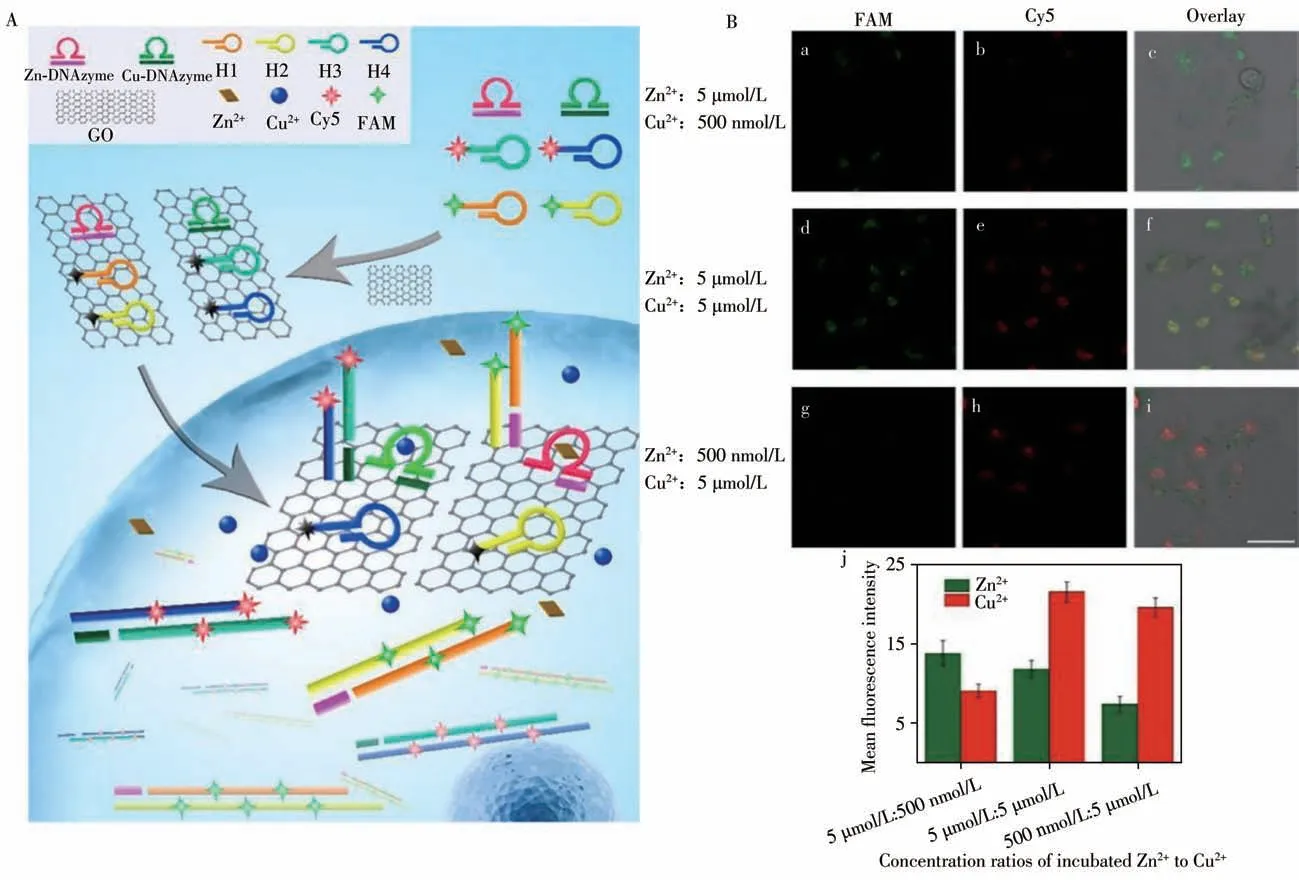
Fig.1 Schematic illustration of the signal amplification sensor of Zn2+and Cu2+in living cells(A);fluorescence images(a-i)and quantitative analysis(j)of Zn2+and Cu2+in MCF-7 cells(B)[4]
1.2 mRNA-responsive DNA self-assembly
There are extremely high demands for nanocarriers with accurate,sensitive,and nondestructive probing of endogenous mRNAin vivo.In order to advance sensing and drug delivery in living cells,the Kanaras’group designed nanoparticulate systems with high specificity and selectivity which can perform multiple synergistic functionsin vi vo[18-19].They combined the sensing and DNA-Au nanoparticles(AuNPs)selfassembly to develop a AuNPs dimer that can show multiple synergies in a cellular environment.
Yang’s group proposed tumor-associated TK1 mRNA-responsive DNA nanospheres(DNA-NS),which contains 1sDNA with a TK1 recognition domain and can be easily prepared by one-pot annealing to indicate fluorescent hairpin(HP)and doxorubicin(DOX)indicators[5].Under the stimulation of endogenous TK1 mRNA,1sDNA of DNA-NS specifically recognizes and hybridizes with TK1 mRNA,leading to the gradual decomposition of DNA-NS and the release of HPs.Subsequently,the released HP is opened by binding withβ-actin mRNA that is widely present invivo[20-21].In this method,a fluorescent signal is generated.Due to the destruction of the binding site of DOX-HPs,DOX is used for the release.Based on this design,coordinated drug delivery is achieved through a precise quantification of mRNA.The DNA-NS construction strategy automatically regulates DOX’s release through mRNA level.The plan provides a reference for the application of self-assembled DNA materials in responsive drug delivery and biological mRNA analysis.
In addition,Zhu’s research group recently developed another way for the amplification and analysis of mRNA in the vivo through a self-assembled DNA tree triggered by a target[10].The probe assembled into a DNA tree is transported into the cell through exosomes,which is beneficial to reduce cell damage and identify nondestructive analysis(Fig.2).The probe is an L-shaped single-stranded DNA(LDNA)that can resist degradation by exonuclease and endonuclease,laying the foundation for accurate analysis.With the induction of the target mRNA,the probes in the cell assemble into plantlets and grow into a tree after several rounds of selfcirculation,achieving exponential fluorescent signal amplification.Compared to 1D DNA trunk self-assembly signal amplification,3D DNA tree exhibits outstanding sensitivity both in-situ or ex-situ.In this method,greater accuracy,sensitivity,and non-destructive analysis are integrated into the system.This DNA tree expands upon the analysis platform and can analyze more biomarkers in an intracellular,non-destructive,and hypersensitive manner.At the genetic level,it has great potential in research applications and clinical diagnosis[22-24].
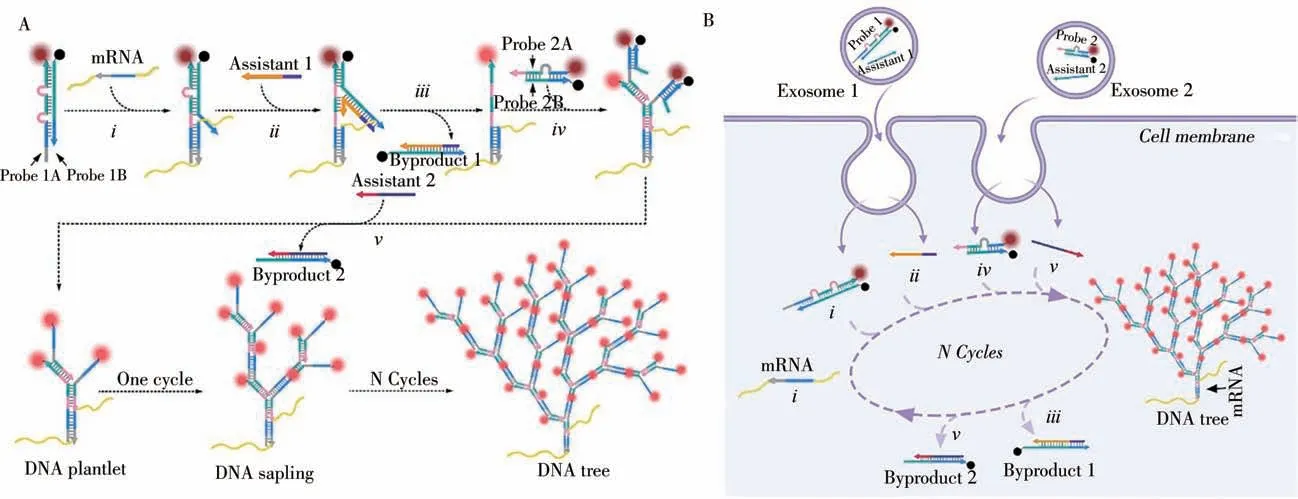
Fig.2 Design and principle of DNA tree for mRNA amplified imaging[10]A:schematic illustration of the DNA tree synthesis progress;B:the exosome-mediated intracellular self-assembled DNA tree
Recently,with dynamic DNA self-assembly(DDSA),enzyme-free amplification techniques for the in-situ detecting mRNAin vivohave been developed by Yang’s group(Fig.3).They proposed a split-new amplifier for FRET imaging of mRNAin vivosuch brought on the designing of an intramolecular catalytic hairpin assembly(intraCHA).Compared to the free-CHA,the probes H1 and H2 in CHA are immobilized on the DNA tetrahedron at the same time.The local concentration of H1 and H2 in CHA is higher than that of free CHA,increasing the initial reaction rate.From the effects of space confinement,the kinetic reaction generated by the target catalytic signal is much improved.The 3D nanostructure helps the H1 and H2 in the CHA amplifier easily enter the cell without any nanocarriers or transfection.The probe and its products are not subject to biological interference.Instead,they can provide a higher signal stability for the reliable imaging of mRNAin vivo[12,25-27].
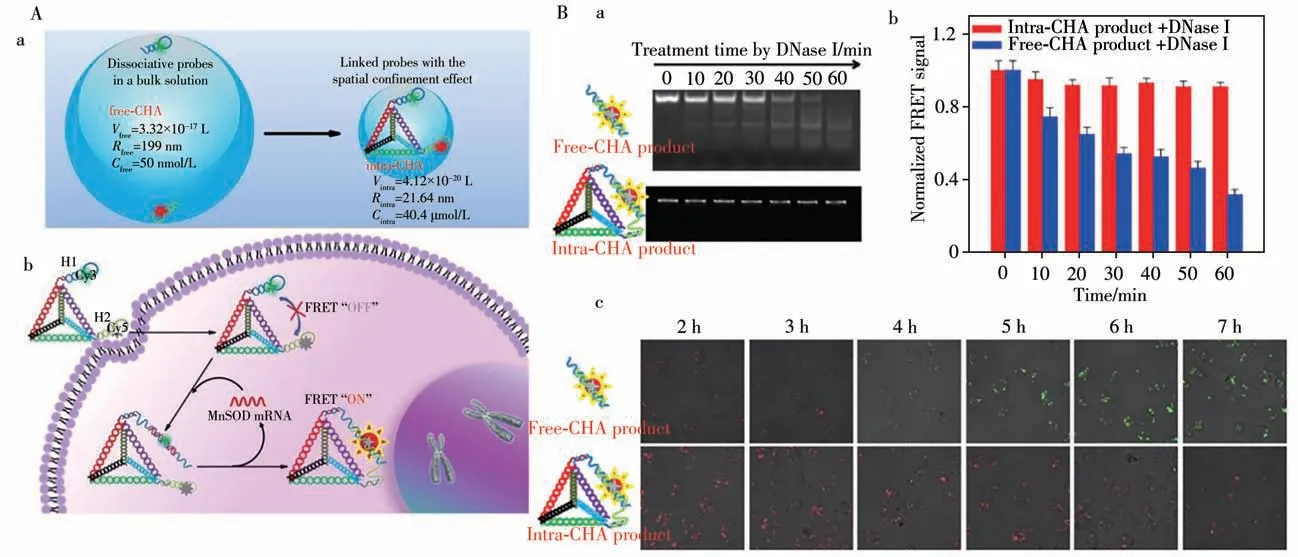
Fig.3 Schematic illustration of the collision model for improving the reaction kinetics(a)and mechanism for the FRET imaging of mRNA in living cell using the intra-CHA system(b)(A);gel electrophoresis diagram(a),FRET signal of CHA products(b),and merge fluorescence images(c)by incubation time(B)[12]
1.3 ATP-mediation through coexisting environmental molecule
ATP is upregulated in diseased cells.ATP-responsiveness is expected to open new possibilities for genome editing and advanced protein delivery for treatment.In 2018,Jiang’s group fabricated a protein scaffold DNA nanohydrogel,the protein was constructed by using self-assembly DNA directly alongside three types of streptavidin(SA)-based DNA tetrads to achieve cancer cell imaging and promote targeted therapy.With the SDH obtained,the Cy5 fluorophore can be effectively quenched by the BHQ3 because they are very close to each other[28].After incubating MUC1 and doxorubicin(Dox)aptamer functionalized SDH(Dox-SDH-Apt),the nanohydrogel specifically attaches to the targeted cancer cells through receptor-mediated endocytosis.Within,SDH is broken down due to its high concentration and its stronger binding to its aptamer.Then the Cy5 fluorophore is restored,and the pre-loaded Dox is quickly released through structural transformation to induce apoptosis of cancer cells.These multifunctional SA scaffold DNA nanohydrogels(SDH)are able to selectively target cancer cells and release preloaded therapeutic agents through structural transformation in the ATP-rich intracellular environment,thereby activating fluorescence and treating cancer cells[29].
Mao’s group reported an ATP-reactive zeolite imidazole framework-90(ZIF-90)as a universal system for CRISPR/Cas9 genome editing and cytosolic protein delivery.It is ultimately encapsulating proteins through proteins formed ZIF-90/protein nanoparticles which synthesize from the self-assembly of Zn2+with imidazole-2-formaldehyde in the presence of ATP.The competitive interactions between ATP and ZIF-90's Zn2+causes ZIF-90/protein NPs to be degraded and release protein[8,30].It has been shown that ZIF-90/protein NPs are able to deliver a variety of proteins to the cytoplasm via intracellular delivery regardless of the size and molecular weight of the protein.The delivery of cytotoxic RNase A inhibits the growth of tumor cells.The effective delivery of the genome editing protein Cas9 knocks out the green fluorescent protein(GFP)expression of HeLa cells with an efficiency of up to 35%.Mao’s team reported the self-assembly of imidazole-2-carboxaldehyde(2-ICA)and Zn2+with proteins forms ZIF-90/protein NPs(Fig.4). The procedure encapsulates proteins with an efficiency of more than 90%.In addition,notice that ZIF-90/protein nanoparticles decompose in the presence of ATP to release protein because of the competitive coordination between Zn2+and ATP is present in the extracellular environment at low concentrations(<0.4 mmol/L),but concentrated in the cytoplasm(1-10 mmol/L).ZIF-90 nanoparticles can be broken down by ATP as a result.At the same time,they will also release proteins into the cell.Intracellular delivery studies have shown that ZIF-90/protein nanoparticles can escape from the endosomes for cytosolic delivery of the genome editing Cas9 nuclease and cytotoxic RNase A[31-32].
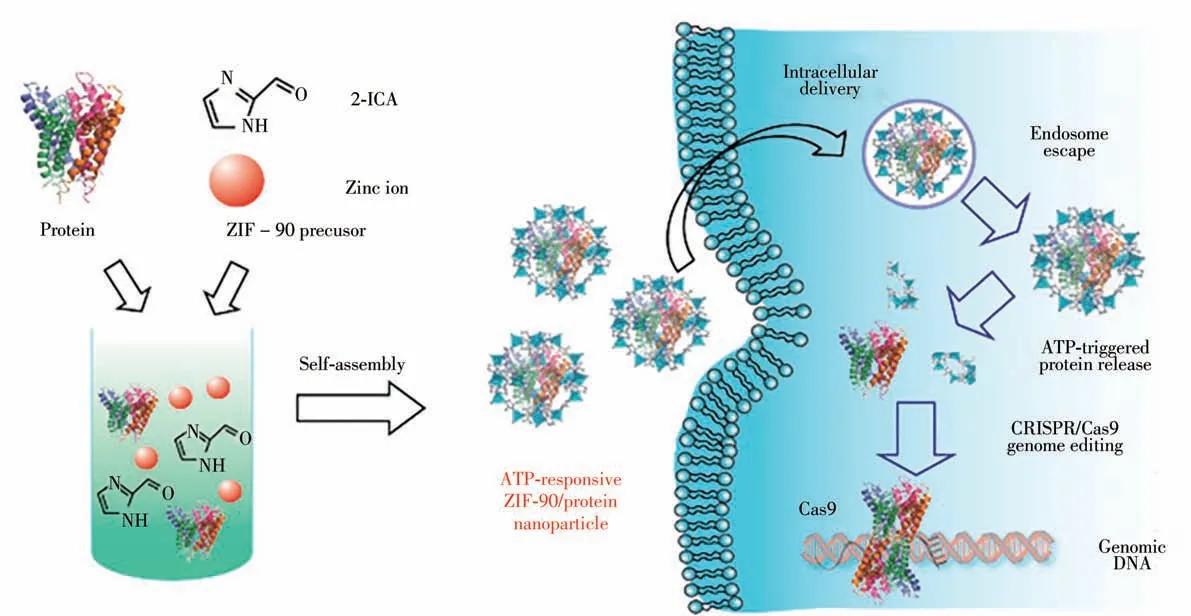
Fig.4 Schematic illustration of the self-assembly of ZIF-90/protein nanoparticle and ATP-triggered protein release from ZIF-90 nanoparticle inside cells[7]
1.4 p H-environmental conditions changes induced self-assembly
Based on the differences of environmental conditions inside and outside the cell,to adjust the DNA selfassembly is necessary.DNA self-assembly that is controlled by pH adjustment has been successfully developed and continuously improved upon.It has been used to construct many DNA-based structures,including 1D fluorescent probes,2D DNA tiles,and 3D DNA hydrogels.In Wang’s group work,they assembled a 3D DNA motor,which includes a Zn2+-dependent DNAzyme and a substrate on ZIF-8[7].Due to protonation,it has special pH-sensitive degradation characteristics and can be used as a carrier for delivery of nucleic acid probes in cells,releasing probes under weakly acidic conditions in the endosome.After entering the living cells,ZIF-8 is effectively degraded in the tumor microenvironment(low pH),locally releasing Zn2+and DNA motors.Soon after,a self-sufficient DNA motor autonomously performs the biological analysis task of imaging miRNA-10b.This self-sufficient 3D walker allows real-time imaging of MDA-MB-231 cells through intracellular manipulation.
The pH-activated protonated and non-protonated cytosine bridge-based(CCH+)triplex structure controls the binding and dissociation of the hydrogel in a weakly acidic or alkaline environment.In order to further explore the possibility of pH-activated pure DNA hydrogels,Yang’s research group proposed a new modular strategy by introducing protonated cytosine-guanine-cytosine(C-GC+)and thymine-adenosine purine-thymine(T-AT)(Fig.5).Based on the triple structure of C-GC+formed at pH 5.0 and decomposed at pH 7.0,the thymine-adenine-thymine(T-AT)-based triplet structure forms at pH 7.0 and decomposes at pH 10.0.Therefore,this pure DNA hydrogel base on the triple DNA structure provides a pH-controlled reversible selfassembly,and it can switch between gel and liquid states.More importantly,the introduction of fluorophore(FAM)and quencher(BHQ1)into the hydrogel provides an innovative way to monitor the selfassembly and disassembly process via fluorescence technology[33].

Fig.5 The principal structures of a series of self-assembled DNA modules(A-F)and the schematic diagram of the formation of three different pH-activated hydrogels(I,II and III)[33]
1.5 Others
The cell membrane is the interface between the intracellular and extracellular environment.Intracellular biological activity is regulated by signal transduction between membrane-bound receptors and signal molecules(including hormones,neurotransmitters or therapeutic agents from complex extracellular environments).Based on aptamer-linked DNA nanodevices(aptNDs),Tan’s group reported upon the anchoring of pre-formed fluorescent aptNDs and the in-situ self-assembly of fluorescent aptNDs on the surface of target living cells[34].They used a strategy based on diacyl lipid-DNA conjugates to decorate specific DNA enzymes on the cell surface to monitor target metal ions in the cell microenvironment.Specifically,by spontaneously inserting the lipophilic tail into the plasma membrane,the diacyl lipid-DNAzyme probe hybridized with the substrate is anchored on the cell surface.The diacyl lipid-DNAzyme probe has three parts.The 3'-terminal region is the DNAzyme sequence.The 5'-terminal region contains the diacyl lipid tail,and the PEG linker is located between the two regions.For signal transduction,DNAzyme and substrate scaffold are labeled with fluorophore and quencher,respectively.In the absence of target metal ions,the fluorescence is quenched due to the close distance between the fluorophore and the quencher.After binding to the target metal ion,DNAzyme can cut the substrate into two fragments.Due to the reduced hybridization stability,the cleaved substrate subsequently dissociates from the DNAzyme strand,separating the quencher from the fluorophore,which in turn leads to the restoration of fluorescence on the cell membrane.Based on the inherent advantages of deoxyribonuclease,including fast kinetics and high sensitivity,this fluorescent membrane-anchored sensor should be able to monitor target metal ions in the cell microenvironment with spatiotemporal resolution[35-40].
2 Summary and perspectives
In this review,we introduced the trigger mechanisms of DNA self-assembly for fluorescence imaging with synthetic nanostructures and DNA molecules in living cells.The functionalization of DNA molecules leads cells to acquire new molecular recognition functions that do not necessarily exist in nature.In addition,the cells do not need to be genetically manipulated,which may cause safety problems for potential future applications.They can recognize not only other cells and organisms,but also substrates without theoretical limitations.Moreover,to enhance ability to recognize the surrounding environment,cells can also achieve unrecognized functions by forming shielding materials on the cell surface through DNA orientation.These functions of promoting and inhibiting cell recognition make the DNA self-assembly engineering method a unique one among many counterparts.Thein vivoengineering of synthetic DNA and its self-assembly shows great promise in applications such as tissue engineering,cancer immunotherapy,biosensing,cell regulation,and cell delivery.However,it is important to realize that there are several issues that need to be further studied and resolved.The first problem is the dissociation of DNA from the cell surface or the internalization of DNA in the cell[41].New strategies are needed to increase the retention time of DNA on the cell surface.Like the first problem,the second problem is DNA degradation.Degradation will definitely shorten the half-life of DNA molecules and nanostructures on the cell surface[42].Click or tap here to enter text.If the modification does not damage the key functions of DNA molecules and nanostructures,chemical modification may help solve this problem.Third,although DNA is generally considered to be biocompatible,large amounts of DNA on the cell surface may stimulate immune cells in a manner similar to the extracellular traps of neutrophils involving tolllike receptors[43].The integration of DNA with other biopolymers(such as alginate)or synthetic polymers(such as polyethylene glycol)may alleviate this potential problem to improve biocompatibility[44].Fourth,DNA aptamers have attracted great attention in the field of cell surface engineering.However,although there are a large number of aptamers available for soluble protein targets,there are not many aptamers that recognize cell membrane proteins.It is necessary to discover more aptamers with high affinity and specificity for cell membrane proteins.Probably because of these problems,most cell surface engineering research is only carried outin vitr o,not in thein vi voenvironment.More research is needed to examine how efficiently these elegant systems work in the body.Of course,the self-assembly of DNAin vi vois still in its infancy.With more efforts and the development of new DNA nanostructures,this field of research will undoubtedly flourish soon.
- 分析测试学报的其它文章
- 阿尔兹海默症生物标志物和早期诊断新技术
- 40周年刊庆 引 言
- 微管纸喷雾质谱法快速筛查血液中5种强极性毒物
- 基于核酸分子光开关的闭管可视化环介导等温扩增检测方法
- Detection of Broad Spectrum Bacteria Using a FITC-Lysozyme and Positively Charged AuNPs Constructed FRET Platform
- Detection of Biomarker Protein PDGF-BB in Esophageal Squamous Cell Carcinoma Using a DNA Biosensor Based on Enzyme Cycle Amplification

